Team:Bielefeld-Germany/Project/Phenazine
From 2013.igem.org
Phenazine
Overview
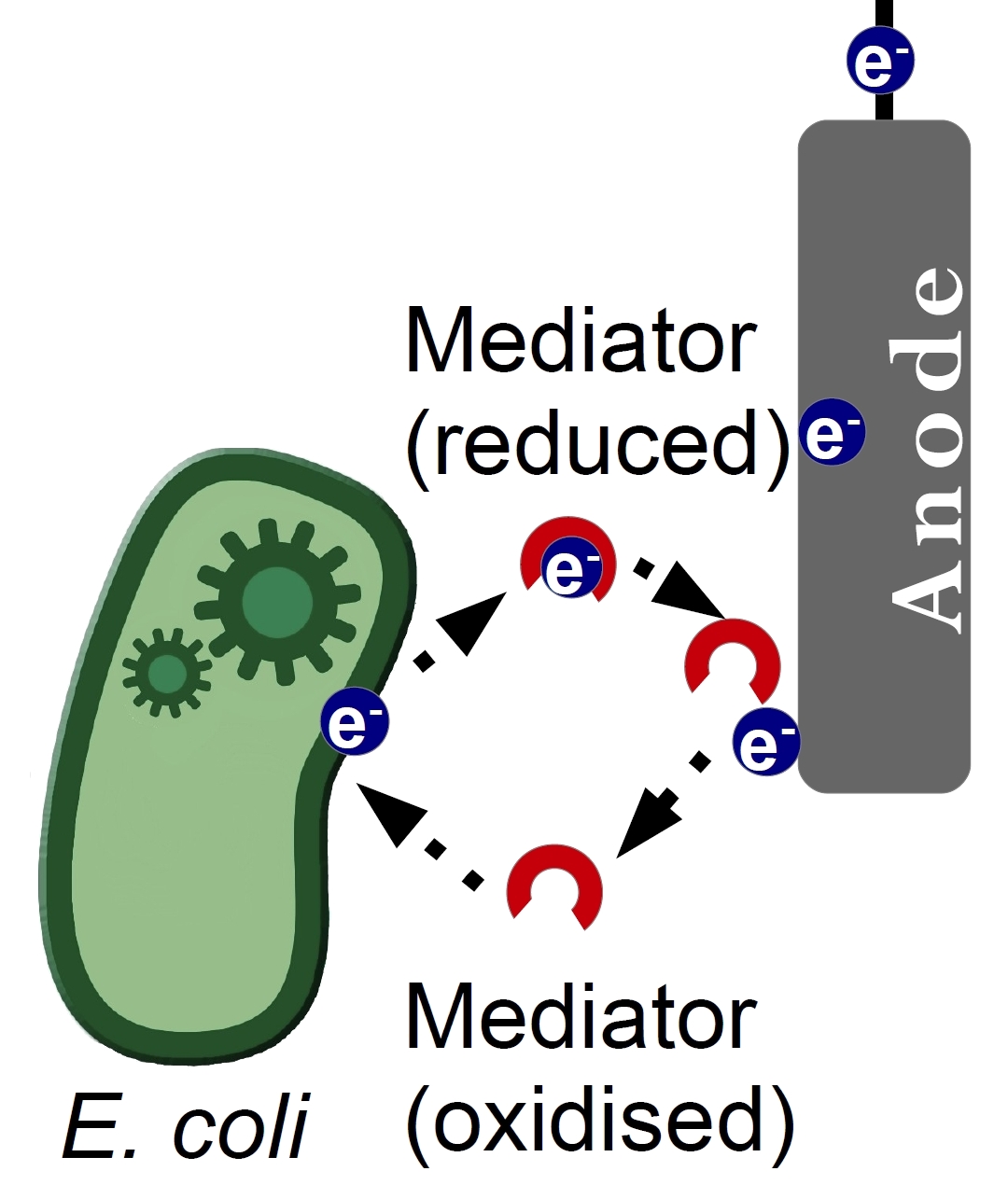
Of great interest is the production of endogenous mediators. The overexpression of glyceroldehydrogenase in E. coli is a promising approach. Because many derivates of glyceroldehydrogenase are small, water-soluble redoxmolecules, they have the properties of a mediator. Futhermore, it will be tested, if there is a possibility of expressing the mediator phenazin. Phenazin is an endogenous mediator of Pseudomonas species.
Theory
Phenazines comprise a large group of heterocyclic nitrogen-containing substances, having a tricyclic ring in its core. They are substituted at different points around their rings, which alters water solubility and other properties. After discovery of naturally occurring phenazines, more than 6000 were chemically synthesized. (Mavrodi, [http://www.ncbi.nlm.nih.gov/pubmed/16719720| 2006]). Neutral red, a well-known pH-indicator, also belongs to this class.
Phenazine derivatives were one of the first secondary bacterial metabolites that were consequently studied since their discovery in 1859. The first substance of this family, a blue pigment of ‘Pseudomonas aeroguinosa‘, was called «pyocyanin». It had antimicrobial properties that raised further interest in these substances.
Phenazines are produced by many bacterial specie, but they are mostly studied in representatives of fluorescent pseudomonads. A single bacterial strain usually produces two or more species-specific phenazines, except Pseudomonas fluorescens, that is known to produce only PCA (phenazine-1-carboxylic acid). (Mavrodi et al., 2001)
Biologically phenazines are known for their broad-spectrum antibiotical, antiparazitar and even anti malaria properties (Handelsmann et al., 1996). They are highly associated with pathogenicity of bacterial species. A broad-range, non-specific mechanism of phenazine action firstly thought to be aimed against an essential metabolic pathway (Leo C. Vining, 1990), but potentiometric measurements on PYO discovered a redox system, that was formed between its reduced and oxidised derivatives.
Consequently it was shown, that almost all properties of phenazines could be attributed to their ability to undergo redox transformations (Muller, 1995). This acknowledgement brought an idea, that these molecules can be used as an electron shuttle in a MFC. Indeed, it was shown that external addition of phenazines, both PCA and PCN alike, causes a significant raise of current (The Hai Pham, 2008). Surprising, a mixed culture of phenazine-secreting and phenazine non-secreting strains showed similar results, proving that phenazine can be used for redox cycling not only by its producer (Hernandez et al. 2004).
A group of soilborne, root-desease controlling microorganisms, such as Pseudomonas fluorescens and Pseudomonas chlororaphis, are known to produce phenazine compounds (PCA and PCN, phenazine-1-carboxamide). They belong to safety class S1, so we saw our chance to try a heterologous expression, taking the coding sequences from one of the known phenazine producers. There were already reports of successful phenazine expression in E.Coli (MacDonald et al. 2001)
The phenazine synthesis is a branch-off of a shikimic acid pathway that delivers aromatic acids. It was discovered and researched in detail for Pseudomonas aeroguinosa and Pseudomonas fluorescens 2-79. A seven-gene cluster was [http://www.ncbi.nlm.nih.gov/nuccore/L48616.1| phzABCDEFG] was identified, that was to almost 95% identical in both organisms.
Also research on phenazine diversity and genetic evolution, caused by additional genes, following or preceding the seven-gene cluster (Mavrodi, 2010 http://www.ncbi.nlm.nih.gov/pubmed/20008172) brought us to a thought, that many species of fluorescent pseudomonas have the corresponding sequences and are able to produce phenazines.
Phenazines are highly associated with pathogenicity of bacterial species. This acknowledgement was a serious concern about biological safety of constructing a strain, overexpressing phenazines. We were uncertain, if this is an appropriate option and what consequences can we await.
Genetic Approach
Results
References
- Mavrodi D.V, Tomashow L.S. (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation.
[http://www.ncbi.nlm.nih.gov/pubmed/16719720| Annual review of Phytopathology 44:417– 445]
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS (2001) Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. [http://www.ncbi.nlm.nih.gov/pubmed/11591691| J Bacteriol 183:6454–6465]
- Handelsman, J. and E. V. Stabb (1996) Biocontrol of soilborne plant
pathogens. [http://www.plantcell.org/content/8/10/1855.long| Plant Cell 8(10): 1855-69.]
- Vining L.C. (1990). Functions of secondary metabolites. [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.44.100190.002143| Annu Rev Microbiol: 44:395-427]
- Muller M. Scavenging of neutrophil-derived superoxide anion by 1-hydroxyphenazine, a phenazine derivative associated with chronic Pseudomonas aeruginosa infection: relevance to cystic fibrosis. (1995) [http://www.ncbi.nlm.nih.gov/pubmed/8541351| Biochim Biophys Acta: 1272(3):185-9.]
- Pham T.H. (1998) Use of Pseudomonas species producing phenazine-based metabolites in the anodes of microbial fuel cells to improve electricity generation. [http://www.ncbi.nlm.nih.gov/pubmed/18688612| Appl Microbiol Biotechnology: 80(6):985-93.]
- Hernandes M.E (2004) Phenazines and other redox-active antibiotics promote microbial mineral reduction [http://www.ncbi.nlm.nih.gov/pubmed/14766572| Appl Environ Microbiol.:70(2):921-8.]
- McDonald M, Mavrodi DV, Thomashow LS, Floss HG. (2001). Phenazine biosynthesis in Pseudomonas fluorescens: branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. [http://www.ncbi.nlm.nih.gov/pubmed/11562236| J. Am. Chem. Soc. 123:9459–60]
- Mavrodi D.V. (2010) Diversity and evolution of the phenazine biosynthesis pathway. [http://www.ncbi.nlm.nih.gov/pubmed/20008172| Appl Environ Microbiol.:866-79.]
</div>
 "
"