Team:Uppsala/results
From 2013.igem.org
Results and achievement"
Achievement

Successfully managed to clone and sequence verify totally new biobricks.

Improved on already existing biobricks.

Successful characterization of several biobricks.

Helped another iGEM team.

Made a successful human practice.

Created a new standard backbone for lactobacillus.

Useful mathematical modelling of metabolic pathways

Completed our chromoprotein collection from previous years
Results
P-coumaric acid
We managed to clone out and biobrick tyrosine ammonia lyase and verify the biobrick by sequencing. Also we did succeful characterization on this part, showing that it works as expected. We managed to express our enzyme and detect it in a western blot, and also detect our metabolite in both spectrophotometry and hplc. The biobrick was characterized in e-coli d5halpha and e-coli nissle. For detailed information about the characterisation methods, see the protocol section.

1. Positive control
2. TAL with Cp8 promotor
3. TAL with J23110 promotor
4. Negative control
2. TAL with Cp8 promotor
3. TAL with J23110 promotor
4. Negative control
Figure 1:SDS-page and western blot. Expression of Tyrosine ammonia lyase with constitutive promotors. The negative control is empty, showing that there is no natural protein in e-coli with the same attributes.
 | Figure 3. Graph showing the HPLC result of a sample prepared from e coli expressing tyrosine ammonia lyase. Reverse phase HPLC with a C18 matrix was used. The peak for p-coumaric acid can be seen ~9 min, as shown by the standard sample below. |
 | Figure 4. Graph showing the HPLC result of a sample standard with p-coumaric acid |
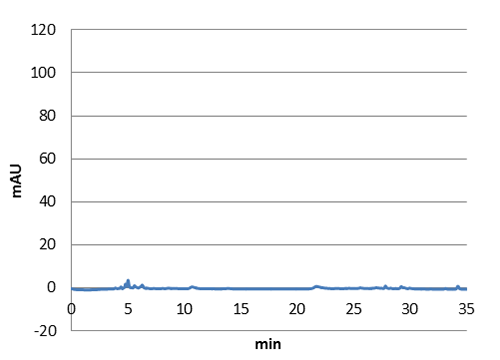 | Figure 5. E-coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peak at 9 minutes. |

 "
"









