Team:Bielefeld-Germany/Project/Cytochromes
From 2013.igem.org
Cytochromes
Overview
To enable transfer of electrons from the general metabolism to the outside of the cell the mtrCAB operon from [http://www.ncbi.nlm.nih.gov/genome/1082?project_id=57949 Shewanella oneidensis MR-1] was heterologously expressed in E. coli. This operon encodes for a minimal set of genes required to build an electron shuttle pathway via different c-type cytochromes. For a correct heme insertion into the decaheme cytochromes MtrA and MtrC the cytochrome c maturation machinery is required. The corresponding genes are naturally expressed in E. coli under anaerobic conditions, for aerobic expression they have to be expressed via plasmid. The mtrCAB cluster contains two illegal restriction sites, which where removed by generating a silent mutation via overlap-extension-PCR. The resulting three fragments were combined and ligated with [http://parts.igem.org/Part:pSB1C3 pSB1C3] by Gibson assembly. Subsequently this gene cluster was combined with three different promotors and ribosome binding sites of varying strength. The functional expressin of the cytochromes could not be experimentaly verified. Furthermore it was attempted to clone the ccmAH cluster as well, which was unsuccesfull. This is, however, a minor issue, since the microbial fuel cell will work under anareobic conditions.
Theory
Cell membranes work as a natural insulator and prevent the flow from electrons out of the cell. To enable transfer of electrons from the general metabolism to the outside of the cell we had to alter the membrane of our organism E. coli without disturbing cell growth, stability and metabolism. Some species from the genera Shewanella and Geobacter have developed different mechanisms to allow extracellular electron transfer. In Shewanella oneidensis MR-1 this is achieved via different c-type cytochromes, which shuttle the electrons along a defined molecular route from the cytoplasma and the inner membrane to the outside of the cell during anaerobic respiration. This pathway is very well understood and characterized.
Members of the Shewanella species are the gram-negative γ-proteobacteria and are known for their respiratory versatility. They are reported to be using over 20 terminal electron acceptors for respiration (Nealson, Scott, 2003). Shewanella oneidensis MR-1 expresses many c-type cytochromes, membrane-bound redox-active proteins or soluble periplasmatic proteins, which play an important in the electron transport in the bacterial respiration and photosynthesis.
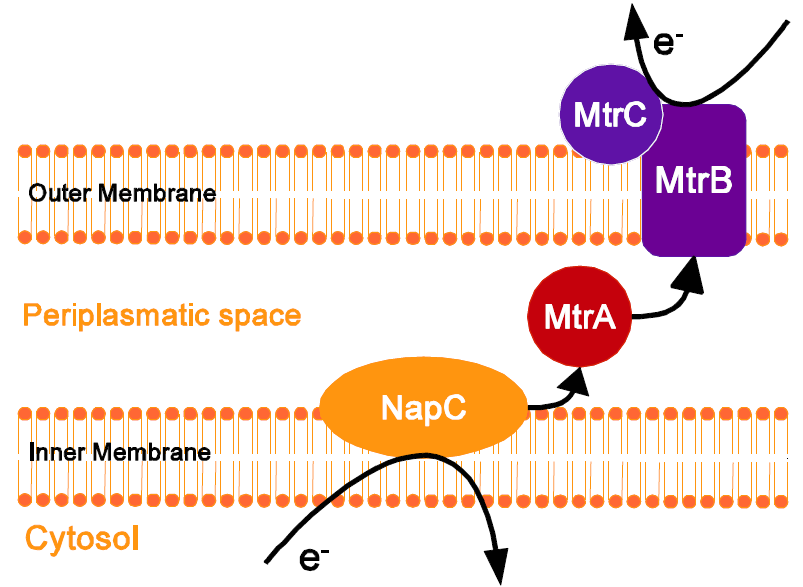
One of the electron-transfer models points out at the c-type cytochromes encoded by the omcA-mtrCAB gene cluster (Myers, Myers, 1997). CymA is the inner membrane cytochrome c and officiates as the electron-acceptor. Analagoical role plays in E. coli NapC and further, to date unidentified, proteins. The electrons are then transferred through from CymA to the MtrA in the periplasm. From MtrA, electrons are transferred to the proteins in the outer membrane, within the MtrCAB complex, and further to extracellular electron acceptors (Goldbeck et al., 2012). MtrA is a periplasmic decaheme cytochrome c of the mass 32-kDa, MtrB is a 72-kD β-barrel outer
membrane protein and MtrC is a 69-kDa membrane-bound decaheme cytochrome c. MtrC requires MtrB for the correct assembly within the outer membrane(Myers, Myers, 2002). In addition, MtrB requires MtrA for its stability.
Furthermore, essential for the biosynthesis of the proteins MtrA and MtrC are the cytochrome c maturation (ccm) genes, which encode eight membrane proteins CcmABCDEFGH.
There have been already numerous approaches, where the complex MtrCAB was introduced into E.coli strains and the genes could be successfully expressed in E. coli under anaerobic cultivation(Goldbeck et al., 2012)
Previous work suggests that a working electron transfer chain can be achieved by a minimal set of three genes, the periplasmatic decaheme [http://www.ncbi.nlm.nih.gov/protein/NP_717386.1 MtrA], the outer membrane β-barrel protein [http://www.ncbi.nlm.nih.gov/protein/NP_717385.1 MtrB] and the outer membrane cytochromes [http://www.ncbi.nlm.nih.gov/protein/NP_717387.1 MtrC]. MtrA interacts with at least one native redox protein, f.e. NapC and can therefore start the transfer of electrons. (Jensen et al. 2010) Additionally another set of genes, the cytochrome c maturation genes (ccmABCDEFGH), are required for correct protein localization and heme insertion into MtrA and MtrC. Under anaerobic conditions these genes are naturally expressed in E. coli, whereas under aerobic conditions we had to co-express them. For aerobic growth the cells were transformed with both plasmids, containing the cytochrome-cluster, as well as the ccmAH-cluster . By this approach extracellular electron transfer should be possible in E.coli and allow the use of this genetically engineered strain in a microbial fuel cell. (Thony-Meyer et al. 1995)
Genetic Approach
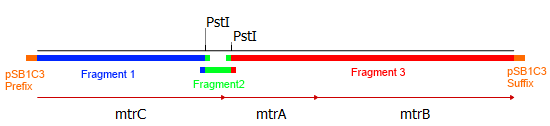
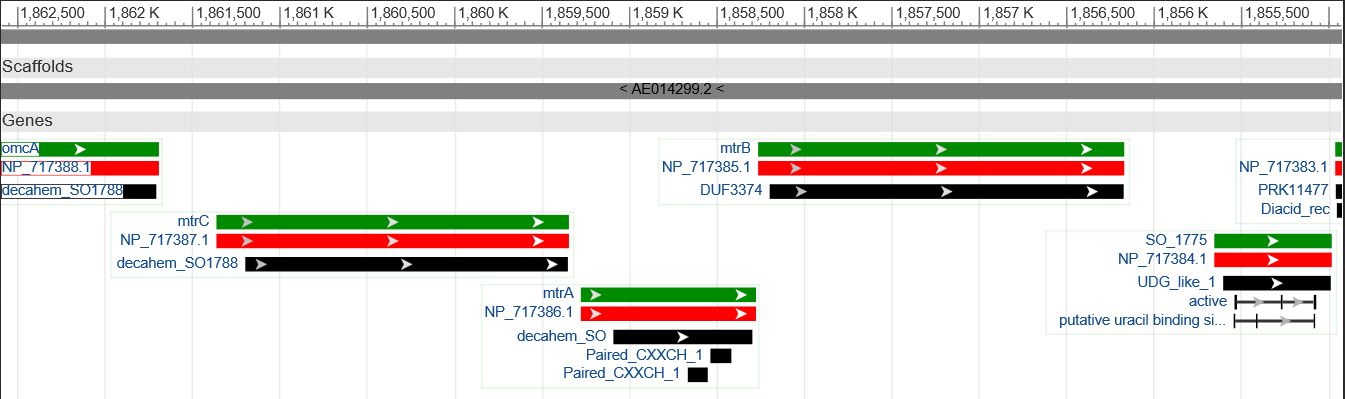
As shown in Figure2 the three corresponding genes are organized in an operon in the donor organism Shewanella oneidensis MR-1 and could therefore amplified altogether. Since two illegal PstI restriction sites are present in this cluster it had to be subdivided into three fragments. The illegal restriction sites were subsequently deleted by incorporating a silent mutation via overlap extension PCR. Eventually the fragments should be joined back together and ligated with the shipping backbone pSB1C3 via Gibson assembly. This approach required specifically designed primers that can bind the targeted genetic sequence and hold an overlap to facilitate ligation with the other fragments and the backbone.Hence, eight primers were designed, two for each fragment of the mtrCAB operon and two for the pSB1C3 vector to generated proper backbone-overlaps as well. The resulting fragments are visualized in Figure 3. The colored overlaps indicate homologous sequences,that facilitate the religation via Gibson assembly. The parts labeled 'pSB1C3 prefix' and 'pSB1C3 suffix' are overlaps homologous to the vectors prefix and suffix, respectivly. The genes and resulting fragments are listed in the table below. The fragment sizes incorporate the aforementioned primeroverlaps of 20 bp each.
| Gene | Size | Fragment | Size |
|---|---|---|---|
| [http://www.ncbi.nlm.nih.gov/gene/1169552 mtrC] | 2016 bp | Frag 1 | 1833 bp |
| [http://www.ncbi.nlm.nih.gov/gene/1169551 mtrA] | 1002 bp | Frag 2 | 321 bp |
| [http://www.ncbi.nlm.nih.gov/gene/1169550 mtrB] | 2094 bp | Frag 3 | 3057 bp |
Protein Overview
Shewanella oneidensis MR-1 expresses many c-type cytochromes, membrane-bound redox-active proteins or soluble periplasmatic proteins, which play an important in the electron transport in the bacterial respiration and photosynthesis. One of the electron-transfer models is based on c-type cytochromes encoded by the mtrCAB gene cluster (Myers, Myers, 1997). Although additional proteins like OmcA and NapC are involved in the electrone transport chain, a minimal set of the three proteins MtrC, MtrA and MtrB is sufficient to heterologously express this pathway in E. coli (Jensen et al. 2010). Figure 4 illustrates the process of the electron transport. NapC ist naturally expressed from E.coli and interacts very well with the proteins from S. oneidensis. The inner membrane cytochrome c NapC accepts electrons generated by the reduction ofmenaquinone to menaquinol. The electrons are then passed from CymA to the periplasmic decaheme cytochrome c [http://www.ncbi.nlm.nih.gov/protein/NP_717386.1 MtrA] in the periplasm, which transports them to [http://www.ncbi.nlm.nih.gov/protein/NP_717385.1 mtrB], a 72-kD β-barrel outer membrane protein, which is physically connected with [http://www.ncbi.nlm.nih.gov/protein/NP_717387.1 MtrC], a 69-kDa membrane-bound decaheme cytochrome c. From MtrC the corresponding electrons can be transfered to extracellular electrone acceptors such as iron oxid, or, as in our case, an anode in a microbial fuel cell. The CcmAH complex composed of the eight membrane proteins CcmABCDEFGH is essential for the biosynthesis of the proteins decaheme cytochromes MtrA and MtrC. The system first loads the heme into the periplasm and catalizes the formation of thioester bonds that link the heme to two cystein residues in the apocytochrome. Afterwards the axial ligands are located towards the heme and the holocytochrome is folded.(Sanders et al. 2010). Naturally these proteins are expressed by E. coli solely under anaerobic conditions.
| [http://www.ncbi.nlm.nih.gov/gene/1169552 mtrC] | [http://www.ncbi.nlm.nih.gov/gene/1169551 mtrA] | [http://www.ncbi.nlm.nih.gov/gene/1169550 mtrB] | [http://www.ncbi.nlm.nih.gov/gene/1169550 CcmAH] |
|---|---|---|---|
| outer membrane decaheme type c cytochrome | periplamatic decaheme type c cytochrome | 28 strand β-barrel membrane protein | cytochrome c maturation machinery |
| 671 aa | 333 aa | 697 aa | |
| 69 kDa | 32 kDa | 72 kDa |
 "
"