Characterization of recombinant effector proteins
For the development of a transgenic water filter it is important to create a collection of well described and functional effector proteins which are either able to bind (BioAccumulation) or to degrade (BioDegradation) xenobiotics present in the aquatic environment. This task was completed by the production of relevant effector proteins in E. coli and to complete their subsequent purification and characterisation. Deliberately we have chosen some well established BioBricks from the last years such as a laccae (<partinfo>BBa_K1159002</partinfo>) or the catechol dioxigenase (<partinfo>BBa_K648011</partinfo>) to improve these BioBricks. Beside these improvements we also added new BioBricks to the registry which we characterized in vitro such as the erythromycin esterase (EreB) (<partinfo>BBa_K1159000</partinfo>) or the NanoLuc luciferase (<partinfo>BBa_K1159001</partinfo>) which will be a useful tool for subsequent generations of iGEM students. For technical questions on our experiments, please see protein biochemical methods for further information.
Table 1: Investigated Proteins
| Protein
| BioBrick
| RFC
| Affinity tag
| Size [kDa]
| Disulphid bridges
| comment
|
| Eryhtromycin esterase (EreB)
| <partinfo>BBa_K1159000</partinfo>
| RFC[25]
| c-term. Streptag II
|
|
|
|
| Laccase
| <partinfo>BBa_K1159002</partinfo>
| RFC[25]
| c-term. Streptag II
|
|
|
|
| Nano Luciferase
| <partinfo>BBa_K1159001</partinfo>
| RFC[25]
| c-term. Streptag II
|
|
|
|
| XylE
| <partinfo>BBa_K648011</partinfo>
| RFC[25]
|
|
|
|
|
| PP1
| <partinfo>BBa_K1159004</partinfo>
| RFC[25]
|
|
|
|
|
| YFP_TEV_CFP
| <partinfo>BBa_K1159112</partinfo>
| RFC[10]
|
|
|
|
|
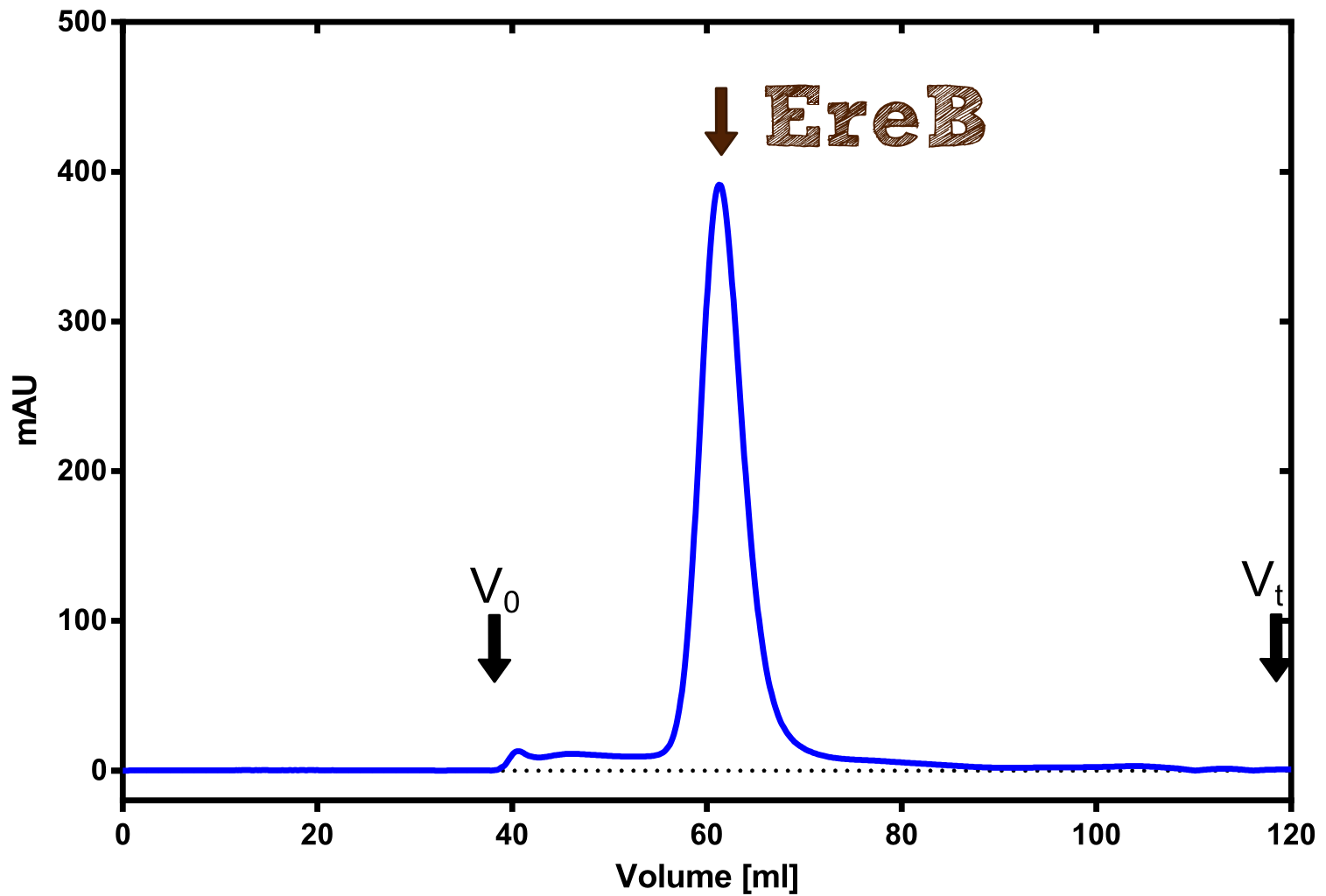
Eryhtromycin Esterase
[...] description
[...] reaction
|
Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)
ATGGCCGGCAGGTTCGAA ... GTTTATGAAACCGGT
ORF from nucleotide position -8 to 1260 (excluding stop-codon) |
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| 1 | MAGRFEEWVKDKHIPFKLNHPDDNYDDFKPLRKIIGDTRVVALGENSHFIKEFFLLRHTLLRFFIEDLGFTTFAFEFGFAEGQIINNWIHGQGTDDEIGR | | 101 | FLKHFYYPEELKTTFLWLREYNKAAKEKITFLGIDIPRNGGSYLPNMEIVHDFFRTADKEALHIIDDAFNIAKKIDYFSTSQAALNLHELTDSEKCRLTS | | 201 | QLARVKVRLEAMAPIHIEKYGIDKYETILHYANGMIYLDYNIQAMSGFISGGGMQGDMGAKDKYMADSVLWHLKNPQSEQKVIVVAHNAHIQKTPILYDG | | 301 | FLSCLPMGQRLKNAIGDDYMSLGITSYSGHTAALYPEVDTKYGFRVDNFQLQEPNEGSVEKAISGCGVTNSFVFFRNIPEDLQSIPNMIRFDSIYMKAEL | | 401 | EKAFDGIFQIEKSSVSEVVYETG* |
|
Sequence features: (with their position in the amino acid sequence, see the list of supported features) | None of the supported features appeared in the sequence |
|
Amino acid composition:| Ala (A) | 27 (6.4%) | | Arg (R) | 16 (3.8%) | | Asn (N) | 20 (4.7%) | | Asp (D) | 29 (6.9%) |
| | Cys (C) | 3 (0.7%) | | Gln (Q) | 14 (3.3%) | | Glu (E) | 30 (7.1%) | | Gly (G) | 31 (7.3%) |
| | His (H) | 15 (3.5%) | | Ile (I) | 36 (8.5%) | | Leu (L) | 34 (8.0%) | | Lys (K) | 29 (6.9%) |
| | Met (M) | 12 (2.8%) | | Phe (F) | 31 (7.3%) | | Pro (P) | 14 (3.3%) | | Ser (S) | 22 (5.2%) |
| | Thr (T) | 19 (4.5%) | | Trp (W) | 4 (0.9%) | | Tyr (Y) | 19 (4.5%) | | Val (V) | 18 (4.3%) |
|
|
Amino acid counting | Total number: | 423 | | Positively charged (Arg+Lys): | 45 (10.6%) | | Negatively charged (Asp+Glu): | 59 (13.9%) | | Aromatic (Phe+His+Try+Tyr): | 69 (16.3%) |
| Biochemical parameters | Atomic composition: | C2204H3348N568O636S15 | | Molecular mass [Da]: | 48459.2 | | Theoretical pI: | 5.55 | | Extinction coefficient at 280 nm [M-1 cm-1]: | 50310 / 50498 (all Cys red/ox) |
|
Codon usage | Organism: | E. coli | B. subtilis | S. cerevisiae | A. thaliana | P. patens | Mammals | | Codon quality (CAI): | good (0.74) | good (0.77) | good (0.75) | good (0.80) | good (0.76) | good (0.66) |
|
The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation.
If you have any questions, comments or suggestions, please leave us a comment. |
Analytical preparation
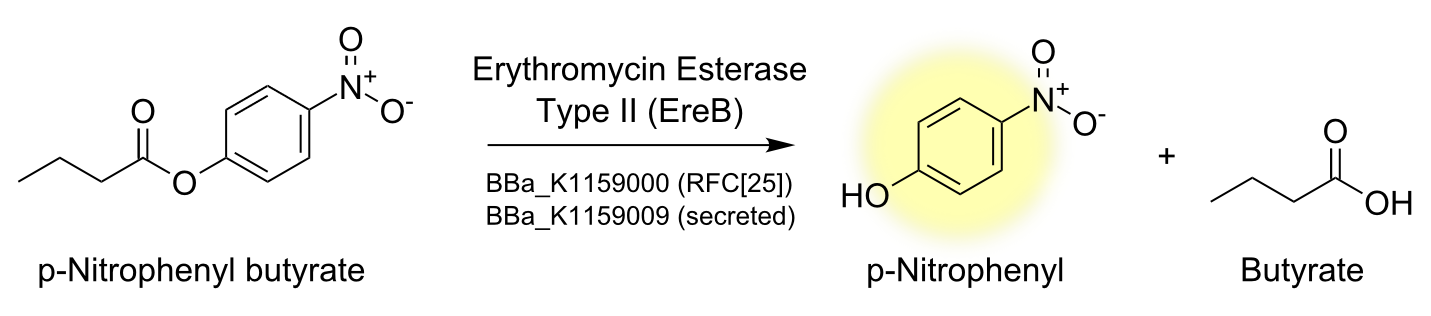
Degradation of a chromogenic esterase substrate: 4-Nitrophenyl butyrate
Degradation of Erythromycin
Reaction conditions
HPLC
Kirby Bauer-Assay
[...] Characterization
Laccase
[...] description
[...] reaction
[...] production
|
Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)
ATGGCCGGCAACCTAGAA ... GATATCATCACCGGT
ORF from nucleotide position -8 to 1530 (excluding stop-codon) |
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| 1 | MAGNLEKFVDELPIPEVAKPVKKNPKQTYYEIAMEEVFLKVHRDLPPTKLWTYNGSLPGPTIHANRNEKVKVKWMNKLPLKHFLPVDHTIHEGHHDEPEV | | 101 | KTVVHLHGGVTPASSDGYPEAWFSRDFEATGPFFEREVYEYPNHQQACTLWYHDHAMALTRLNVYAGLAGFYLISDAFEKSLELPKGEYDIPLMIMDRTF | | 201 | QEDGALFYPSRPNNTPEDSDIPDPSIVPFFCGETILVNGKVWPYLEVEPRKYRFRILNASNTRTYELHLDNDATILQIGSDGGFLPRPVHHQSFSIAPAE | | 301 | RFDVIIDFSAYENKTITLKNKAGCGQEVNPETDANIMQFKVTRPLKGRAPKTLRPIFKPLPPLRPCRADKERTLTLTGTQDKYGRPILLLDNQFWNDPVT | | 401 | ENPRLGSVEVWSIVNPTRGTHPIHLHLVQFRVIDRRPFDTEVYQSTGDIVYTGPNEAPPLHEQGYKDTIQAHAGEVIRIIARFVPYSGRYVWHCHILEHE | | 501 | DYDMMRPMDIITG* |
|
Sequence features: (with their position in the amino acid sequence, see the list of supported features) | RFC25 scar (shown in bold): | 130 to 131 |
|
Amino acid composition:| Ala (A) | 27 (5.3%) | | Arg (R) | 29 (5.7%) | | Asn (N) | 23 (4.5%) | | Asp (D) | 31 (6.0%) |
| | Cys (C) | 5 (1.0%) | | Gln (Q) | 14 (2.7%) | | Glu (E) | 37 (7.2%) | | Gly (G) | 30 (5.8%) |
| | His (H) | 23 (4.5%) | | Ile (I) | 31 (6.0%) | | Leu (L) | 41 (8.0%) | | Lys (K) | 27 (5.3%) |
| | Met (M) | 10 (1.9%) | | Phe (F) | 24 (4.7%) | | Pro (P) | 47 (9.2%) | | Ser (S) | 18 (3.5%) |
| | Thr (T) | 33 (6.4%) | | Trp (W) | 8 (1.6%) | | Tyr (Y) | 22 (4.3%) | | Val (V) | 33 (6.4%) |
|
|
Amino acid counting | Total number: | 513 | | Positively charged (Arg+Lys): | 56 (10.9%) | | Negatively charged (Asp+Glu): | 68 (13.3%) | | Aromatic (Phe+His+Try+Tyr): | 77 (15.0%) |
| Biochemical parameters | Atomic composition: | C2671H4072N718O760S15 | | Molecular mass [Da]: | 58883.0 | | Theoretical pI: | 6.03 | | Extinction coefficient at 280 nm [M-1 cm-1]: | 76780 / 77093 (all Cys red/ox) |
|
Codon usage | Organism: | E. coli | B. subtilis | S. cerevisiae | A. thaliana | P. patens | Mammals | | Codon quality (CAI): | good (0.72) | good (0.75) | good (0.68) | good (0.74) | good (0.77) | good (0.67) |
|
The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation.
If you have any questions, comments or suggestions, please leave us a comment. |
Laccase a is a secreted enzyme
Analytical präparation
[...] Characterization
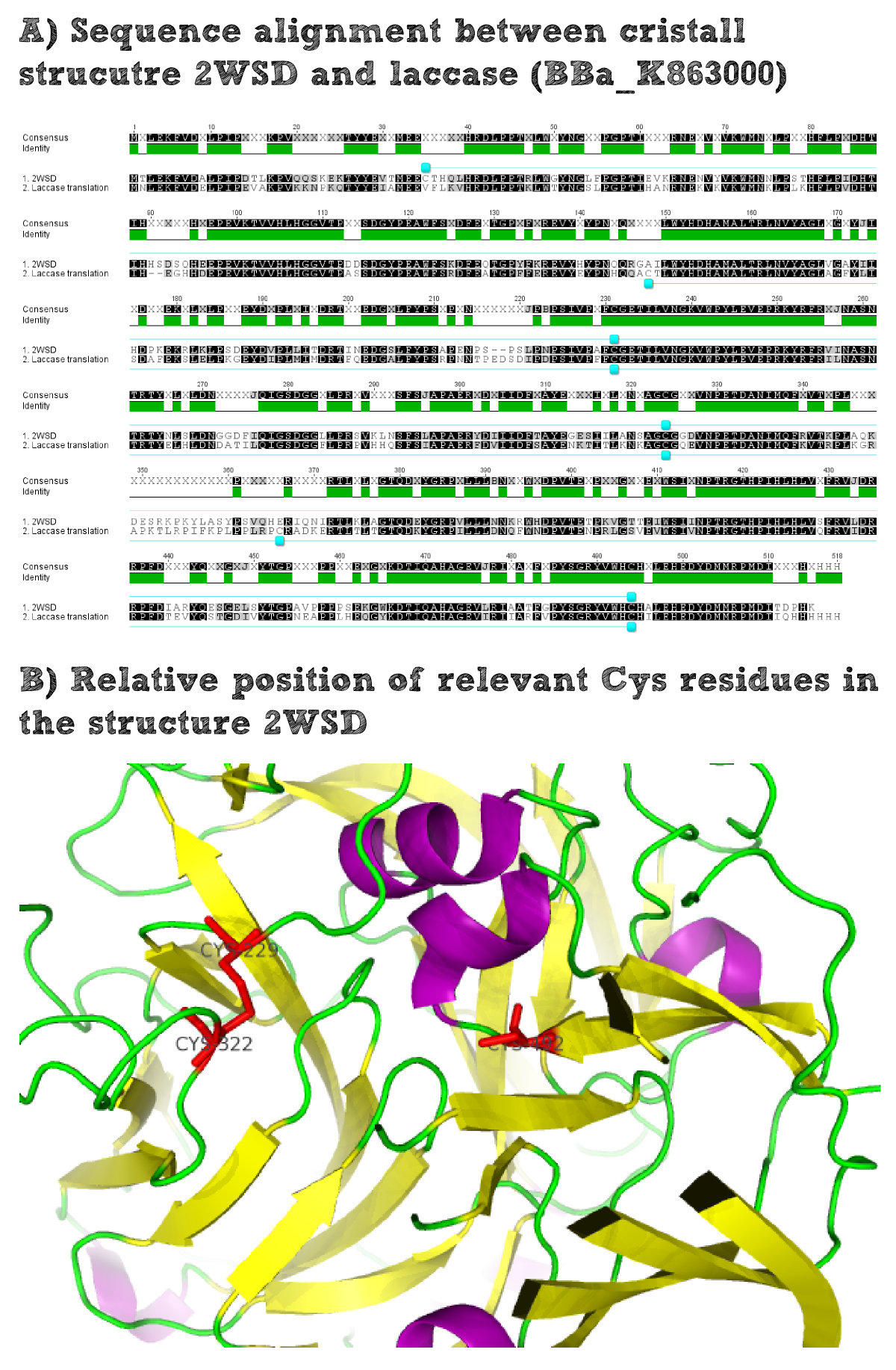
Structural consideration of Laccase from B. pumilus
Activity determination using ABTS
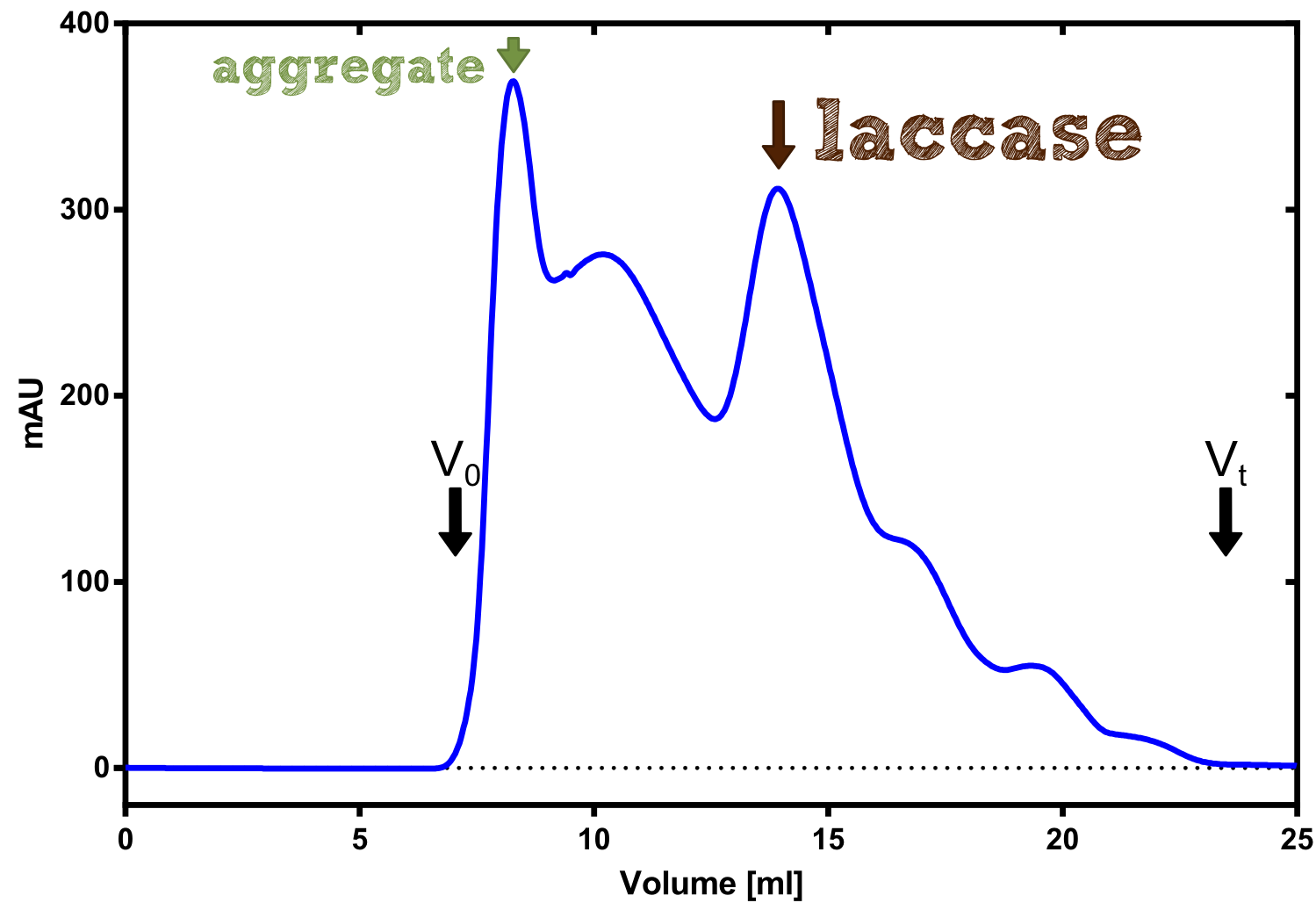
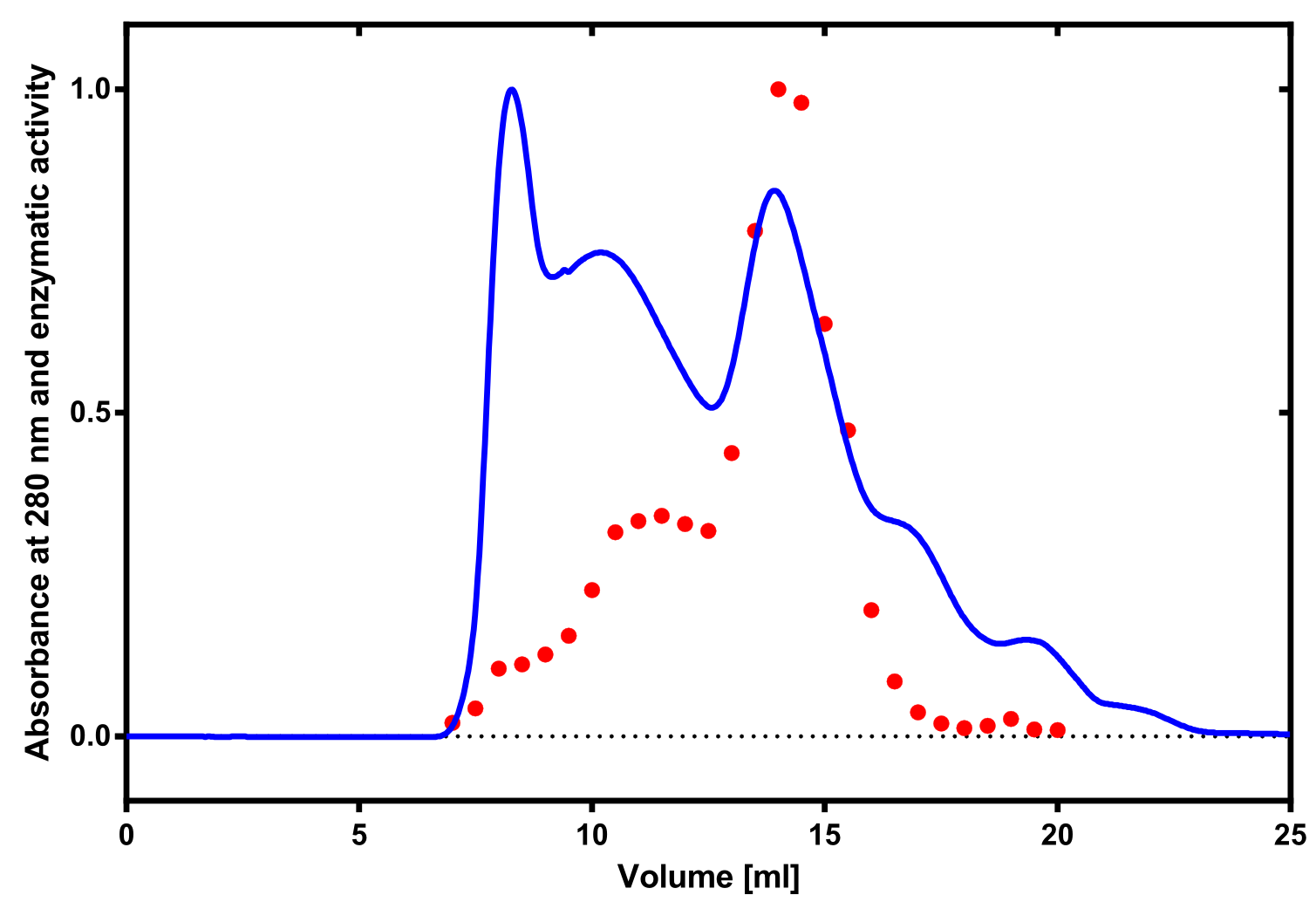
The enzymatic activity of the purified laccae was determined by the ABTS-assay. In a first pre experiment the appropriate dilution factor was determined to 100-fold. The elution fractions obtained from size exclusion chromatography were diluted 1:100 in PBS and in an ELISA plate 100 µl of the enzyme and 100 µl of ABTS substrate were mixed and a kinetic measurement at 405 nm was performed. The absorption at 280 nm in the SEC chromatogramm (blue) identifies three main protein peaks, with a first peak corresponding to aggregated protein, a shoulder which also corresponds to higher molecular protein and a single peak which was proposed to be the monomeric laccase. The relative activity obtained for the different elution fractions was plotted in the same diagramm and shows a clear peak which matches the laccase peak in the SEC. Beside this major peak a second smaller peak of active fraction was visible which appeared in earlier elution fractions and might correspond to dimerized laccase. As the laccase is a secreted enzyme which also bears disluphide bonds it was produced in the cytoplasm and subsequently it was oxidized to form the proper disulphide bond. As this process might be only partial there is a possiblity for the formation of disulphid dimers. Never the less the fractions 14 to 17 were pooled for further experiments as they showed the highest enzymatic activity. The protein concentration of the pooled fraction was determined to 0.48 mg/ml after SEC.
Oxidation of relevant xenobiotics
Diclofenac
Estradiol
Nano Luciferase
The Nano Luciferase (NanoLuc) which was introduced in 2013 by Promega is a new member of the luciferase reporter gene/protein familiy and shows some advantages compared to the other family members. The NanoLuc is very small (19 kDa) compared to the firefly luciferase (61 kDa) and the Renilla luciferase (36 kDa). On the other hand it is also said that the specific activity of the NanoLuc is about 150-fold stronger compared to conventional luciferases and the background caused by autoluminescense of the substrate shel be smaller.
Production in E. coli and purification

Figure 3:Analytical size exclusion chromatography on a Superdex 200 10/30 column showing a single elution peak for the NanoLuc.
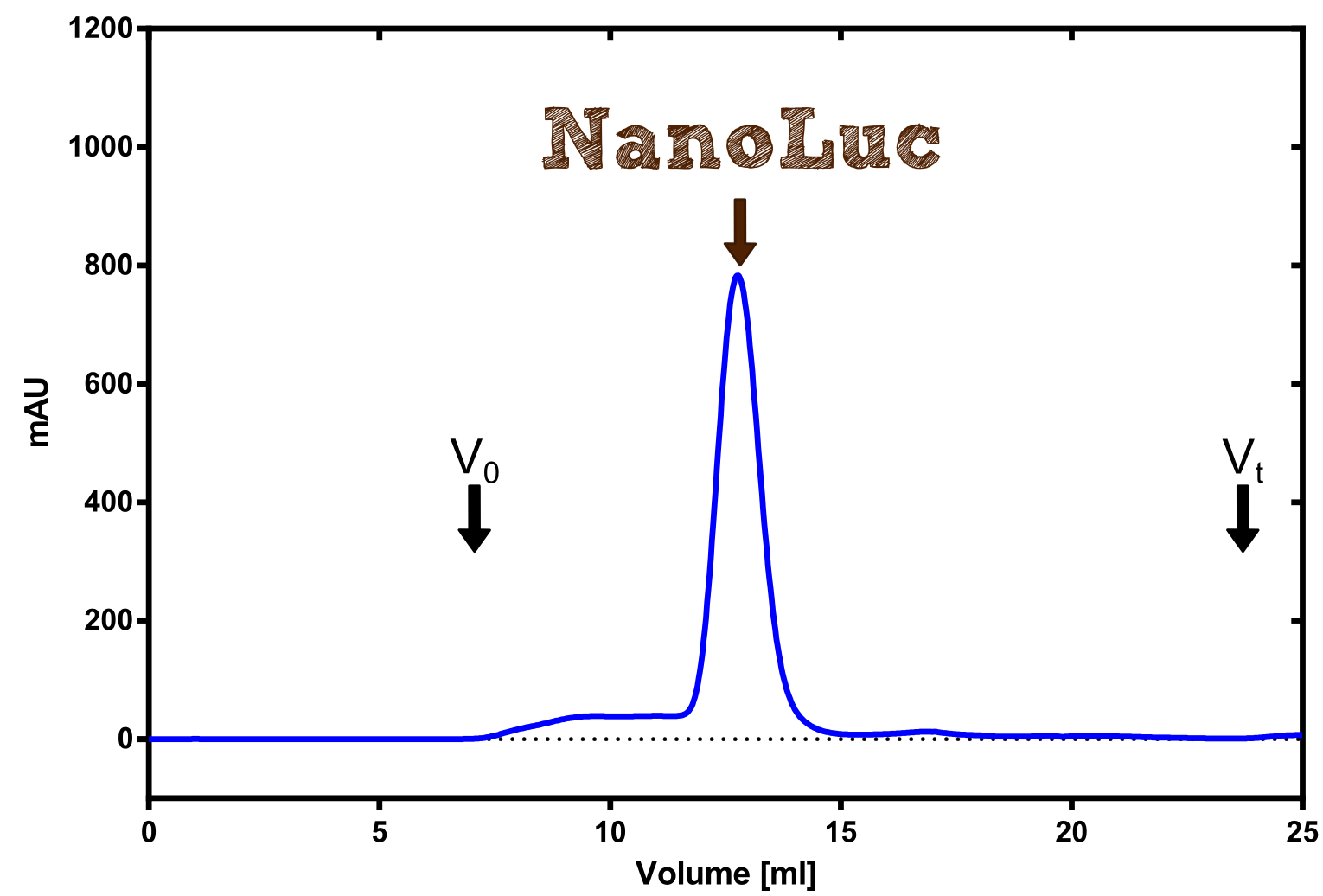
Figure 3:Preperative size exclusion chromatography on a Superdex 75 10/30 column showing a single elution peak for the NanoLuc.
Therefore the NanoLuc was synthesized as a BioBrick in RFC[25] and was produced in E. coli using the pBad expression system with a C-terminal Strep-tag. After the production (2 l of LB-media for analytical and 12 l for preparative preparations) the cells were disrupted using sonification and the lysate was dialysed against 5 l of 1x SA-buffer. Afterwards the lysate was applied to a Streptavidin-Affinity (SA) column and was subsequently washed using SA-Buffer until a baseline was reached and the protein was then eluted using 5 mM of biotin (Attention: These are special columns which are not availible commercially. If you are using commercial colum material you have to use d-Desthiobiotin because usual biotin will elute your protein but you will not be able to regenerate the column after your chromatography). After the SA-chromatography the protein was concentrated using centrifugal concentration units (MWCO: 10 kDa). The concentrated protein was then applied on a Superdex S200/75 size exclusion chromatography.
Structure of the Nano Luciferase
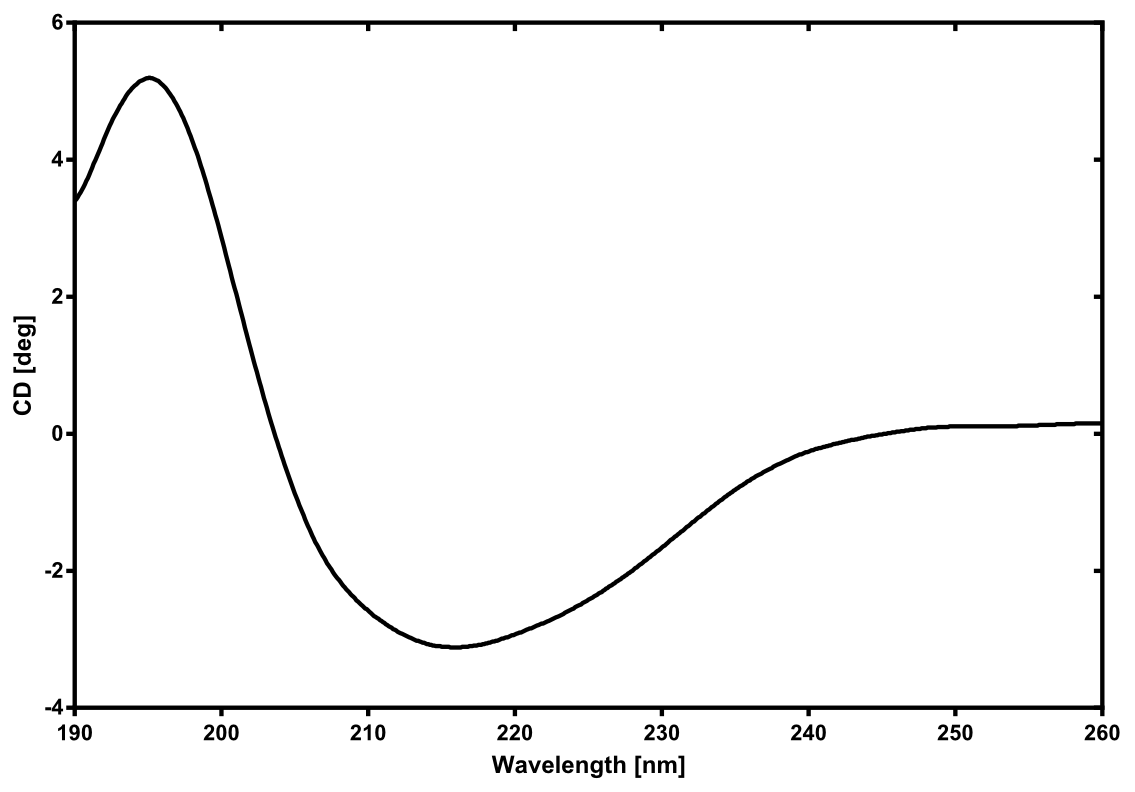
There is no structure availible for the NanoLuc in the [http://www.rcsb.org/pdb/home/home.do Protein Data Bank]. In our protein modelling we used homolgy search and identified the structure [http://www.rcsb.org/pdb/explore/explore.do?structureId=3PPT 3ppt_A] as the solved structure with the highest homology to the NanoLuc which has only 21% identify with a similarity of only 0.359. The result of the homology search is shown as annimated gif in Figure xx (please see our How To for an introduction). The protein was dialysed against 1x CD-buffer and subsequently a circular dichroism spectroscopy was tanken (learn about CD spectroscopy). The CD spectrum was used to predict the secondary structure content of the NanoLuc which could be determined to 35.1% helix, 27.6% b-strand, 18.5% turn and 18.8% random. As there is only a poor homology present, a detailed comparison of the determined and the predicted secondary structure is not possible. But it can be stated that both show a balanced content of different secondary structures and that the produced protein is present in an folded conformation. The mixed secondary structure content is also in consistance with the predicted secondary structure shown in the AutoAnnotator sequence window (click on show).
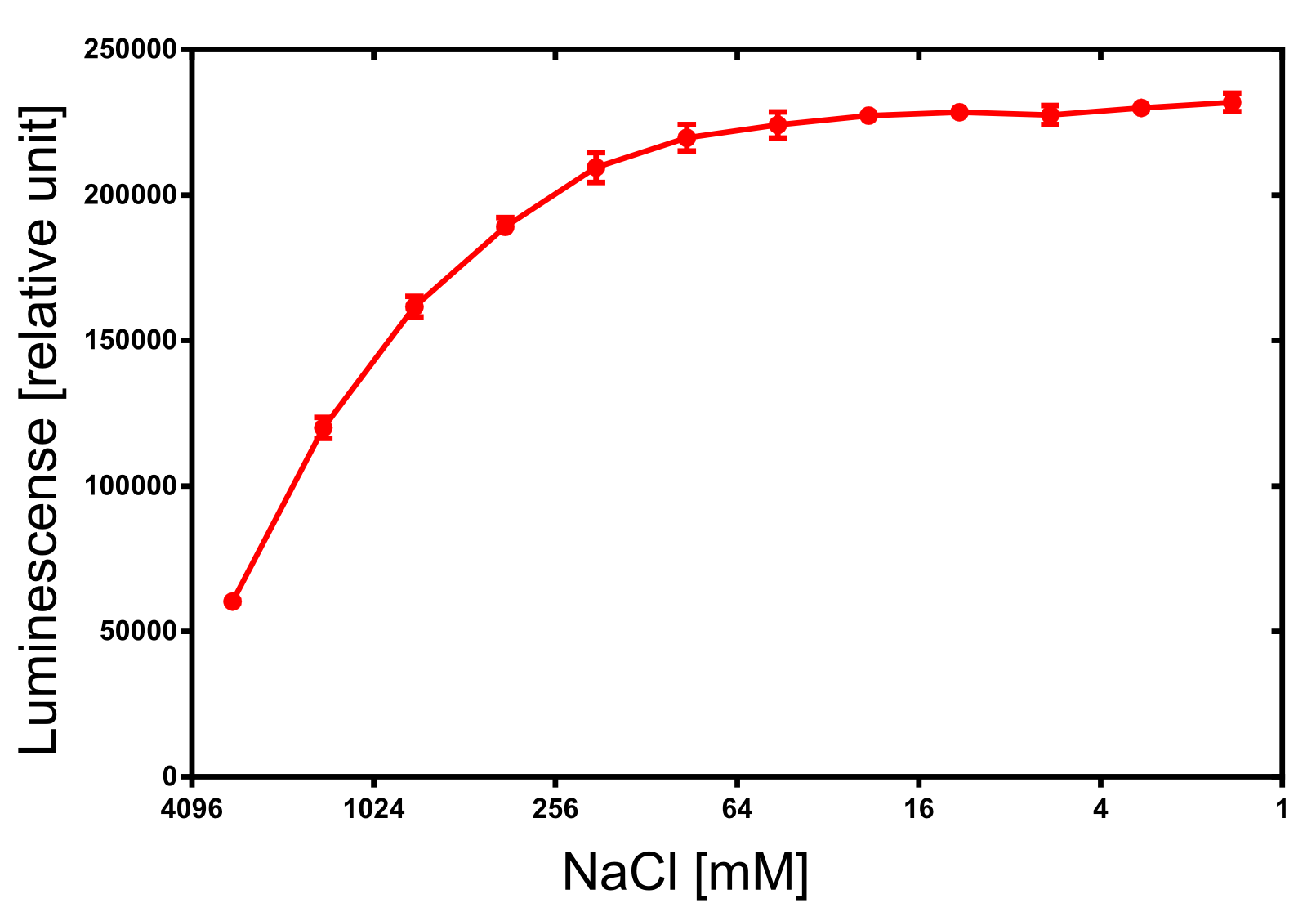
Activity determination of Luminescense
The activity of the produced NanoLuc was investigated by its luminescense. The luminescense
XylE
[...] description
[...] reaction
[...] production
|
Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)
ATGGCCGGCAACAAAGGT ... GTGCTGACCACCGGT
ORF from nucleotide position -8 to 924 (excluding stop-codon) |
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| 1 | MAGNKGVMRPGHVQLRVLDMSKALEHYVELLGLIEMDRDDQGRVYLKAWTEVDKFSLVLREADEPGMDFMGFKVVDEDALRQLERDLMAYGCAVEQLPAG | | 101 | ELNSCGRRVRFQAPSGHHFELYADKEYTGKWGLNDVNPEAWPRDLKGMAAVRFDHALMYGDELPATYDLFTKVLGFYLAEQVLDENGTRVAQFLSLSTKA | | 201 | HDVAFIHHPEKGRLHHVSFHLETWEDLLRAADLISMTDTSIDIGPTRHGLTHGKTIYFFDPSGNRNEVFCGGDYNYPDHKPVTWTTDQLGKAIFYHDRIL | | 301 | NERFMTVLTTG* |
|
Sequence features: (with their position in the amino acid sequence, see the list of supported features) | None of the supported features appeared in the sequence |
|
Amino acid composition:| Ala (A) | 22 (7.1%) | | Arg (R) | 19 (6.1%) | | Asn (N) | 9 (2.9%) | | Asp (D) | 27 (8.7%) |
| | Cys (C) | 3 (1.0%) | | Gln (Q) | 8 (2.6%) | | Glu (E) | 21 (6.8%) | | Gly (G) | 26 (8.4%) |
| | His (H) | 15 (4.8%) | | Ile (I) | 8 (2.6%) | | Leu (L) | 34 (10.9%) | | Lys (K) | 14 (4.5%) |
| | Met (M) | 11 (3.5%) | | Phe (F) | 16 (5.1%) | | Pro (P) | 12 (3.9%) | | Ser (S) | 10 (3.2%) |
| | Thr (T) | 18 (5.8%) | | Trp (W) | 5 (1.6%) | | Tyr (Y) | 12 (3.9%) | | Val (V) | 21 (6.8%) |
|
|
Amino acid counting | Total number: | 311 | | Positively charged (Arg+Lys): | 33 (10.6%) | | Negatively charged (Asp+Glu): | 48 (15.4%) | | Aromatic (Phe+His+Try+Tyr): | 48 (15.4%) |
| Biochemical parameters | Atomic composition: | C1585H2418N434O465S14 | | Molecular mass [Da]: | 35442.1 | | Theoretical pI: | 5.41 | | Extinction coefficient at 280 nm [M-1 cm-1]: | 45380 / 45568 (all Cys red/ox) |
|
Codon usage | Organism: | E. coli | B. subtilis | S. cerevisiae | A. thaliana | P. patens | Mammals | | Codon quality (CAI): | good (0.73) | good (0.70) | acceptable (0.55) | good (0.63) | excellent (0.82) | good (0.78) |
|
The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation.
If you have any questions, comments or suggestions, please leave us a comment. |
Analytical präparation
[...] Characterization
DDT-Dehydrochlorinase
[...] description
[...] reaction
[...] production
|
Nucleotide sequence in RFC 10: (underlined part encodes the protein)
ATGGACTTT ... TTCCTGAGCTAGTAG
ORF from nucleotide position 1 to 627 (excluding stop-codon) |
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| 1 | MDFYYLPGSAPCRAVQMTAAAVGVELNLKLTDLMKGEHMKPEFLKLNPQHCIPTLVDNGFALWESRAIQIYLAEKYGKDDKLYPKDPQKRAVVNQRLYFD | | 101 | MGTLYQRFADYHYPQIFAKQPANPENEKKMKDAVGFLNTFLEGQEYAAGNDLTIADLSLAATIATYEVAGFDFAPYPNVAAWFARCKANAPGYALNQAGA | | 201 | DEFKAKFLS* |
|
Sequence features: (with their position in the amino acid sequence, see the list of supported features) | None of the supported features appeared in the sequence |
|
Amino acid composition:| Ala (A) | 30 (14.4%) | | Arg (R) | 6 (2.9%) | | Asn (N) | 11 (5.3%) | | Asp (D) | 13 (6.2%) |
| | Cys (C) | 3 (1.4%) | | Gln (Q) | 10 (4.8%) | | Glu (E) | 11 (5.3%) | | Gly (G) | 12 (5.7%) |
| | His (H) | 3 (1.4%) | | Ile (I) | 6 (2.9%) | | Leu (L) | 20 (9.6%) | | Lys (K) | 16 (7.7%) |
| | Met (M) | 6 (2.9%) | | Phe (F) | 13 (6.2%) | | Pro (P) | 13 (6.2%) | | Ser (S) | 4 (1.9%) |
| | Thr (T) | 8 (3.8%) | | Trp (W) | 2 (1.0%) | | Tyr (Y) | 13 (6.2%) | | Val (V) | 9 (4.3%) |
|
|
Amino acid counting | Total number: | 209 | | Positively charged (Arg+Lys): | 22 (10.5%) | | Negatively charged (Asp+Glu): | 24 (11.5%) | | Aromatic (Phe+His+Try+Tyr): | 31 (14.8%) |
| Biochemical parameters | Atomic composition: | C1070H1618N272O304S9 | | Molecular mass [Da]: | 23444.8 | | Theoretical pI: | 5.94 | | Extinction coefficient at 280 nm [M-1 cm-1]: | 30370 / 30558 (all Cys red/ox) |
|
Codon usage | Organism: | E. coli | B. subtilis | S. cerevisiae | A. thaliana | P. patens | Mammals | | Codon quality (CAI): | excellent (0.87) | excellent (0.85) | good (0.62) | good (0.71) | excellent (0.80) | good (0.69) |
|
The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation.
If you have any questions, comments or suggestions, please leave us a comment. |
Analytical präparation
[...] Characterization
Protein phosphatase 1 (from Homo sapiens) <partinfo>BBa_K1159004</partinfo>
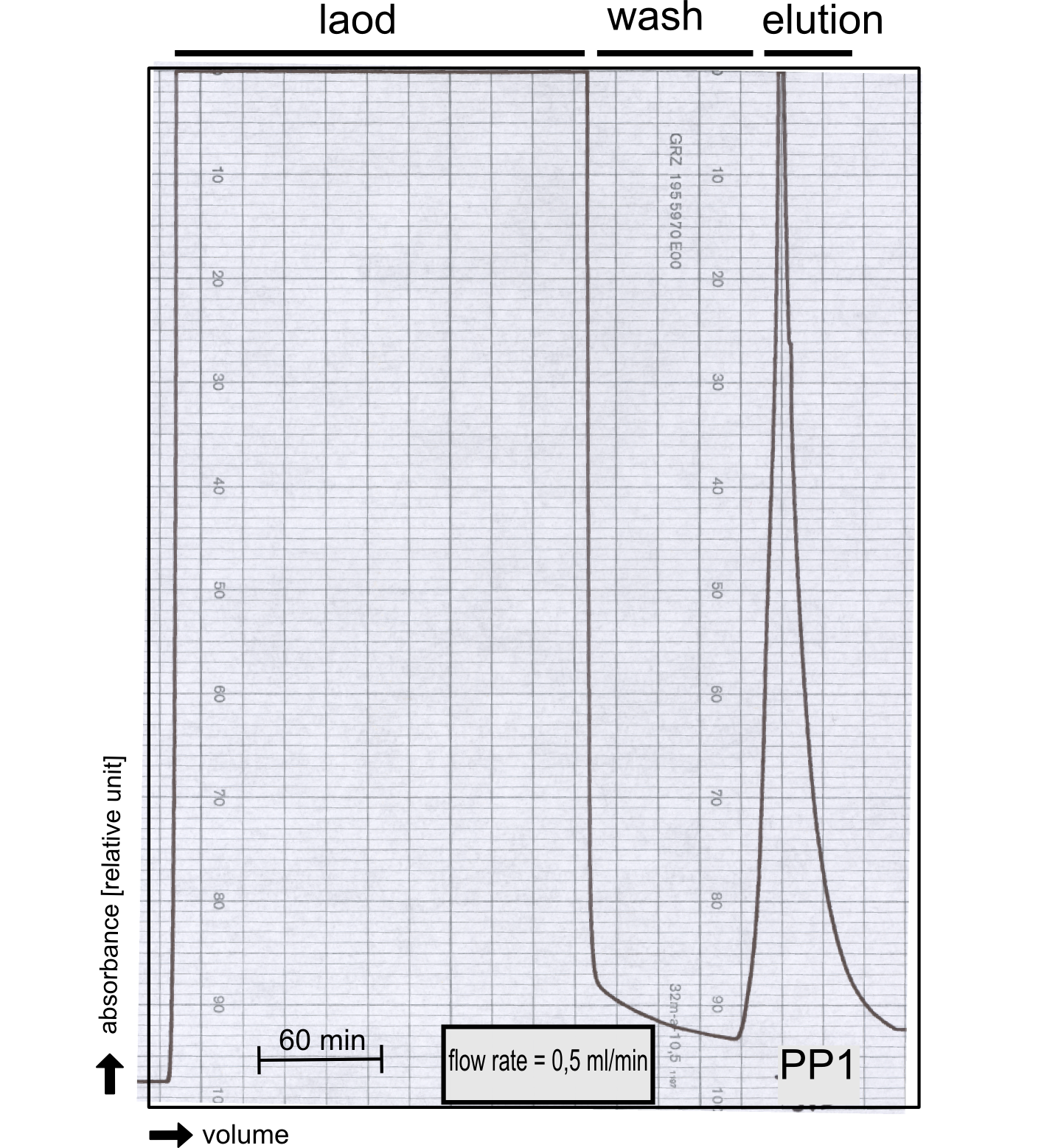
The production of recombinant protein phosphatase 1 was part of our collaboration with Dundee iGEM team 2013. They developed this BioBrick which naturally binds [http://de.wikipedia.org/wiki/Microcystine microcystine], an important environmental toxin. We converted this BioBrick to RFC[25] and cloned it afterward into the expression vector pBad_C-terminal_Strep. Beside the recombinant characterisation we also created a transgenic moss transformed with a receptor containing PP1 in its extracellular domain. The recombinant protein production was carried out in E. coli BL-21 which was grown in 2 L of LB-media. The protein production was induced at OD = 0.8 by addition of arabinose to a concentration of 5 mM. The cells were harvested after 4 h and subsequently resuspended in 20 mL SA-buffer with 5 mM 2-mercapto ethanol. Cell disruption was performed using ultrasonic sound. The dialysed cell extracte was applied to a streptavidin affinity column which was washed until a base line was reached and was subsequently eluted using SA-buffer containing 5 mM of biotin and 5 mM of 2-mercapto ethanol. The elution peak recorded by the continuous measurement of the absorbance at 280 nm indicated a good yield of recombinant protein (Fig. A). As a second purification step after the streptavidin affinity chromatography we concentrated the protein in centrifugal filter units in order to apply it to a size exclusion chromatography. During the concentration process there were clear signal for precipitated protein which appeared as white flakes in the concentration filter unit. This effect was by far the most drastic precipitation of recombinant that was detected during this iGEM project. Anyhow the concentrated protein was centrifuged for 5 min at 13 200 RPM to remove particles of precipitated protein and was then applied to an ÄKTA purified with a Superdex 200 10/30 column (Fig. B). There were four different peaks present in the chromatogramm which most probably correspond to (1) a peak created by aggregated protein which runs in the voit volume of the size exclusion colum, (2) a diffuse peak which might correspond to multimeric PP1, (3) a sharp peak at the elution volume where the recombinant PP1 was expected and (4) a peak which is most probably caused by low molecular buffer substances. The signal intensity is
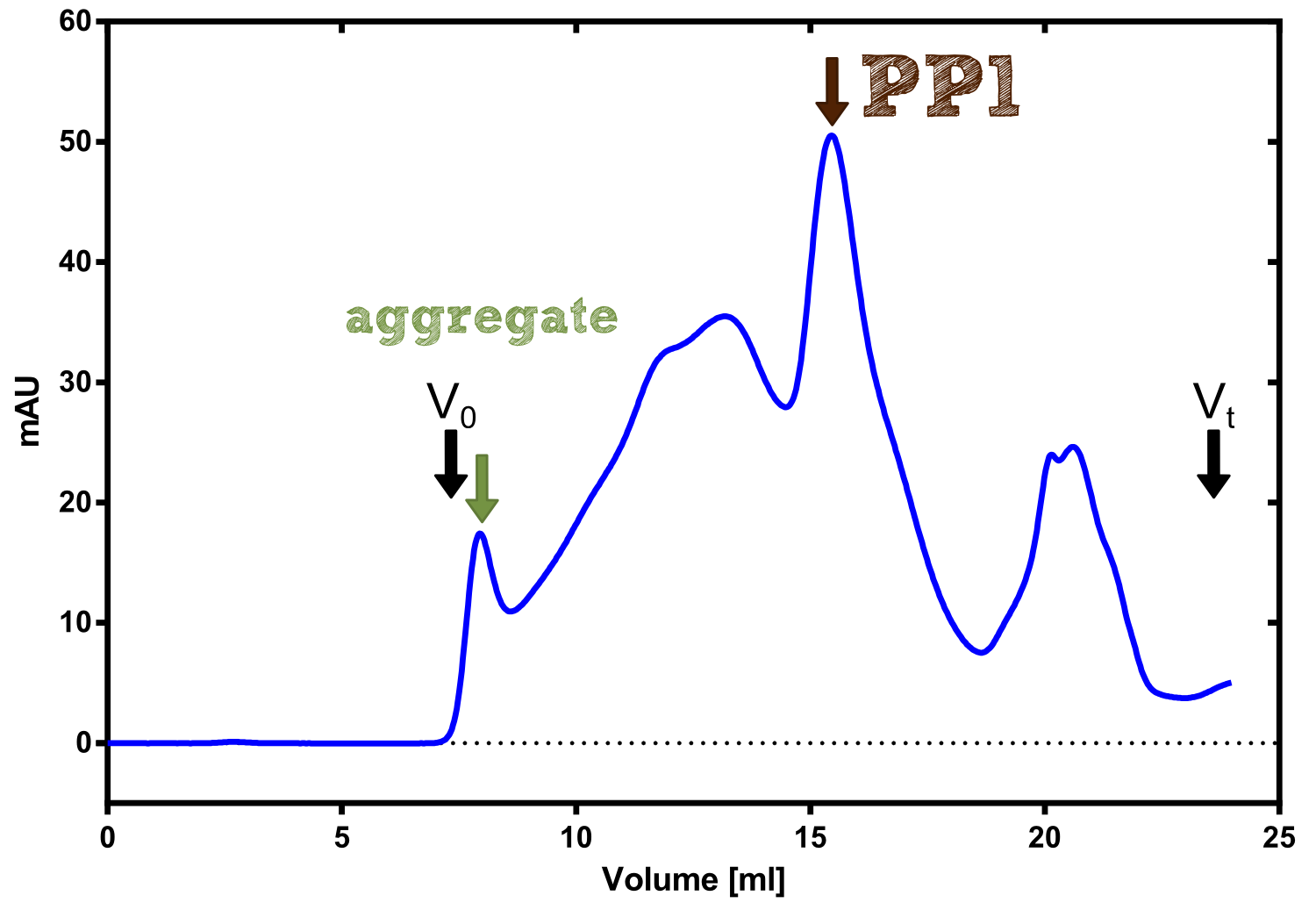
Figure B: Size exclusion chromatography of protein phosphatase 1 on Superdex 200 10/30 column
SpyCatcher & SpyTag
[...] description
[...] reaction
[...] production
|
Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)
ATGGCCGGCGTTGATACC ... GCTCATATTACCGGT
ORF from nucleotide position -8 to 345 (excluding stop-codon) |
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| 1 | MAGVDTLSGLSSEQGQSGDMTIEEDSATHIKFSKRDEDGKELAGATMELRDSSGKTISTWISDGQVKDFYLYPGKYTFVETAAPDGYEVATAITFTVNEQ | | 101 | GQVTVNGKATKGDAHITG* |
|
Sequence features: (with their position in the amino acid sequence, see the list of supported features) | None of the supported features appeared in the sequence |
|
Amino acid composition:| Ala (A) | 10 (8.5%) | | Arg (R) | 2 (1.7%) | | Asn (N) | 2 (1.7%) | | Asp (D) | 10 (8.5%) |
| | Cys (C) | 0 (0.0%) | | Gln (Q) | 5 (4.2%) | | Glu (E) | 9 (7.6%) | | Gly (G) | 14 (11.9%) |
| | His (H) | 2 (1.7%) | | Ile (I) | 6 (5.1%) | | Leu (L) | 5 (4.2%) | | Lys (K) | 8 (6.8%) |
| | Met (M) | 3 (2.5%) | | Phe (F) | 4 (3.4%) | | Pro (P) | 2 (1.7%) | | Ser (S) | 10 (8.5%) |
| | Thr (T) | 14 (11.9%) | | Trp (W) | 1 (0.8%) | | Tyr (Y) | 4 (3.4%) | | Val (V) | 7 (5.9%) |
|
|
Amino acid counting | Total number: | 118 | | Positively charged (Arg+Lys): | 10 (8.5%) | | Negatively charged (Asp+Glu): | 19 (16.1%) | | Aromatic (Phe+His+Try+Tyr): | 11 (9.3%) |
| Biochemical parameters | Atomic composition: | C543H848N144O192S3 | | Molecular mass [Da]: | 12561.7 | | Theoretical pI: | 4.49 | | Extinction coefficient at 280 nm [M-1 cm-1]: | 11460 / 11460 (all Cys red/ox) |
|
Codon usage | Organism: | E. coli | B. subtilis | S. cerevisiae | A. thaliana | P. patens | Mammals | | Codon quality (CAI): | excellent (0.83) | good (0.76) | good (0.78) | good (0.80) | good (0.78) | good (0.67) |
|
The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation.
If you have any questions, comments or suggestions, please leave us a comment. |
Analytical präparation
[...] Characterization
References:
http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984
- http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984 Edens, L., Bom, I., Ledeboer, A. M., Maat, J., Toonen, M. Y., Visser, C., and Verrips, C. T. (1984). Synthesis and processing of the plant protein thaumatin in yeast. Cell, 37(2):629–33.
- http://udel.edu/~gshriver/pdf/Pimenteletal1997.pdf Pmentel et al., 1997 Pimentel, D., Wilson, C., McCullum, C., Huang, R., Dwen, P., Flack, J. Tran, Q., Saltman, T., Cliff, T. (1997). Economic and environmental benefits of biodiversity. BioScience, Vol. 47, No. 11., pp. 747-757.
 "
"



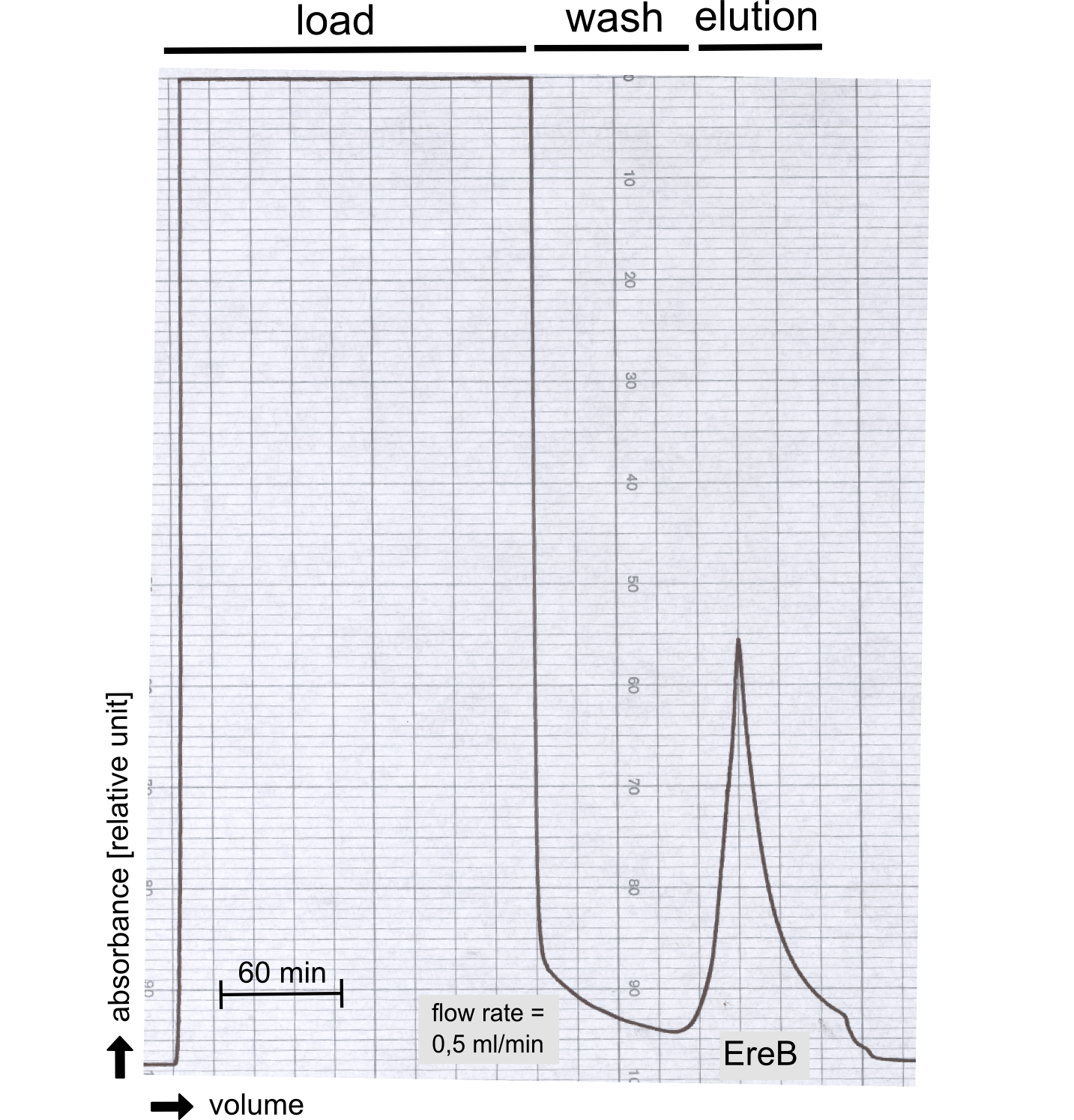
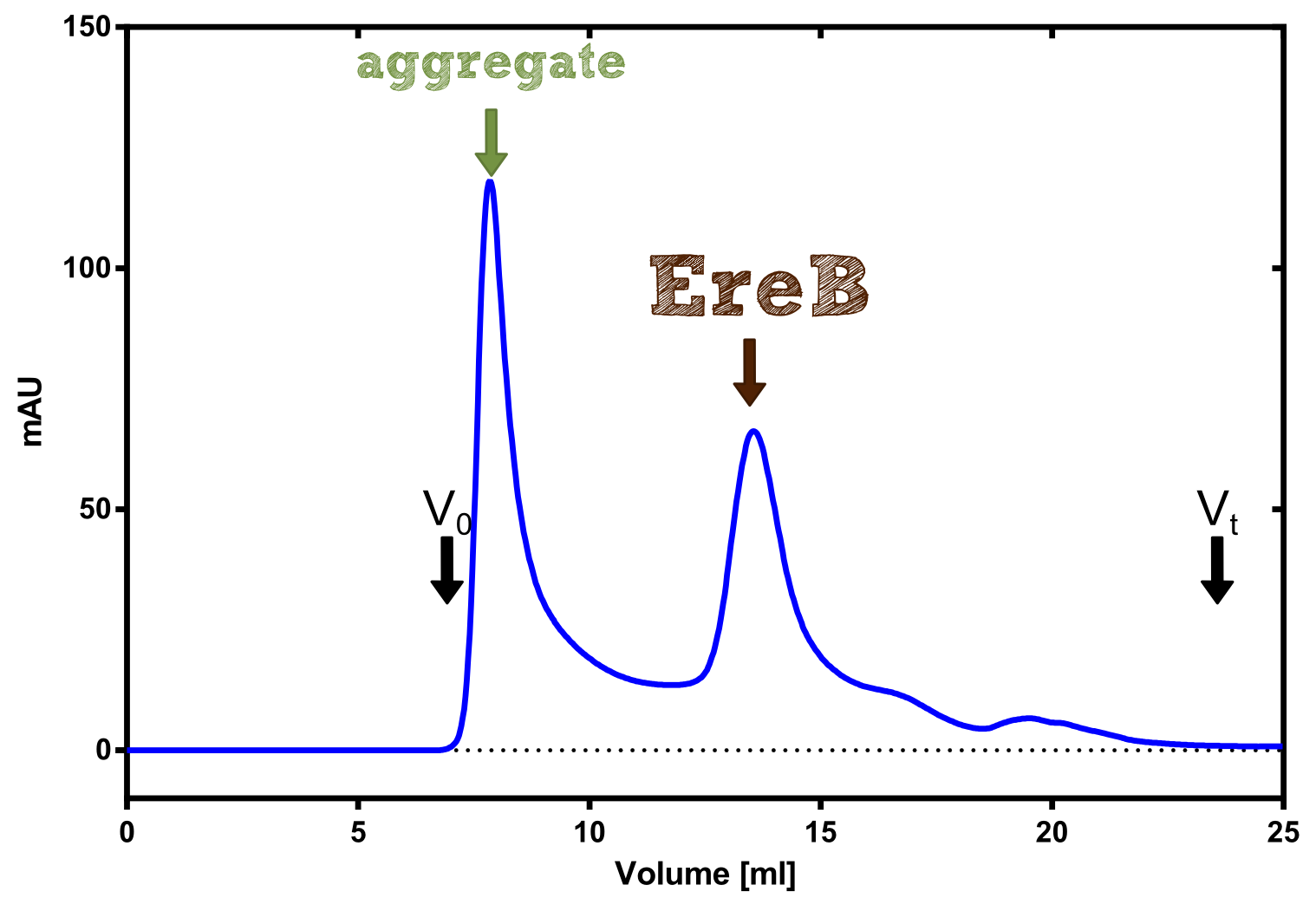






AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351