Team:Bielefeld-Germany/Labjournal/July
From 2013.igem.org
July
Milesstones
9.Week
Organization
- We’ve presented us at the congress ‘Next generation of biotechnological processes 2020+’ in Berlin at 27. June 2013. There we have met an expert (Dr. Falk Harnisch) on the topic of MFC and we have decided to participate the German Synthetic Biology Day.
MFC
Mediators
Cytochromes
- Design and order of new Primers
- The mtrCAB fragment has two illegal PstI restriction sites at x xbp and yy bp, so we had to design new primers to remove them. We replaced one base in each restriction site, without affecting the coding triplett, by respective primer overlaps . We ended up with three different fragments, which will be ligated back together via Gibson Assembly.
- mtrCAB_Frag1_rev:
- mtrCAB_Frag2_fwd:
- mtrCAB_Frag2_rev:
- mtrCAB_Frag3_fwd:
Biosafety
Porines
10.Week
Organization
- We can present new sponsors of iGEM-Team Bielefeld: Stockmeier, Baxter, Promega, BIO.NRW and Applichem will support our team.
MFC
Mediators
Cytochromes
Biosafety
Porines
11.Week
Organization
- We’ve presented us at the congress “BioNRW pHD Student Convention” in Düsseldorf at 13. July 2013 with a short presentation about iGEM and our project.
- We proudly present our first press release. It was amazing to see how often our press release was picked up.
MFC
Mediators
Cytochromes
Biosafety
Porines
12.Week
Organization
- Dr. Falk Harnisch will have a presentation at CeBiTec colloquium as an expert for our team.
- Preparing experiments for the ‘Day of Synthetic Biology’. Our ideas are to have different experiments like DNA isolation from fruit and vegetables, pipetting of bright colors, chromatography with markers, a potato battery or microscopy.
- The TV station WDR would like to contribute with us on our topic
MFC
Mediators
Cytochromes
- Amplification of Fragment 1
- Size: 1800 bp
- Program: PhusionPCR
- Gradient: 54°C - 71°C over 8 steps
- Primer: mtrC_fwd & mtr_Frag1_rev
- Template: S. oneidensis PCR Template from genomic DNA
- Notes: Annealing temperature has no significant impact on the PCR
- Amplification of Fragment 2
- Size: 330 bp
- Program: PhusionPCR
- Primer: mtr_Frag2_fwd & mtr_Frag2_rev
- Template: S. oneidensis PCR Template from genomic DNA
- Notes:
- Amplification of Fragment 3
- Size: 3000 bp
- Program: PhusionPCR
- Gradient: 54°C - 71°C over 8 steps
- Primer: mtr_Frag3_fwd & mtrB_rev
- Template: S. oneidensis PCR Template from genomic DNA
- Notes: Annealing temperature has no significant impact on the PCR
- Amplification of ccmAH cluster
- Size:6311 bp
- Program: PhusionPCR
- Gradient: 51.5°C - 68°C over 8 steps
- Primer: ccmAH_fwd & ccmAH_rev
- Template: E. coli PCR Template from genomic DNA
- Notes: A high annealing temperature of 68°C or more increases yield as well as reduces unspecific bands and is therefore recommended.
- Gelextraction and cleanup of Fragment 1, 2, 3 and ccmAH
- Gelextraction and clean up with the AnalytikJena GelExtraction-Kit
- Elution buffer was preheated to 50°C
- We eluted each column twice, with 20 ul each and combined it afterwards
- Measurement of nucleid acid concentration via NanoDrop
- Fragment1: 4-2607-304: 10.5 ng/ul</p>
- Fragment2: 4-2707-003: 44.6 ng/ul</p>
- Fragment2: 4-2707-004: 13.5 ng/ul</p>
- Fragment3: 4-2607-301: 11.1 ng/ul</p>
- Fragment3: 4-2607-302: 8.9 ng/ul</p>
- Fragment3: 4-2607-303: 9.2 ng/ul</p>
- ccmAH: 4-2807-302: 11.0 ng/ul</p>
- ccmAH: 4-2807-303: 11.5 ng/ul</p>
Cytochromes
Biosafety
Porines
5.Week
Organization
- Spontaneous visit of Radio Bielefeld in our laboratory. In addition to a radio interview a
short video clip was filmed.
MFC
Mediators
Cytochromes
Biosafety
Amplification of different fragments:
Recipe: PhusionBuffer: 5,5µL dNTPs: 1µL Primer1: 1µL Primer2: 1µL Template: 1,5 µL DMSO: 0,5 µL Phusion: 0,5 µL Dest. H2O: 39 µL
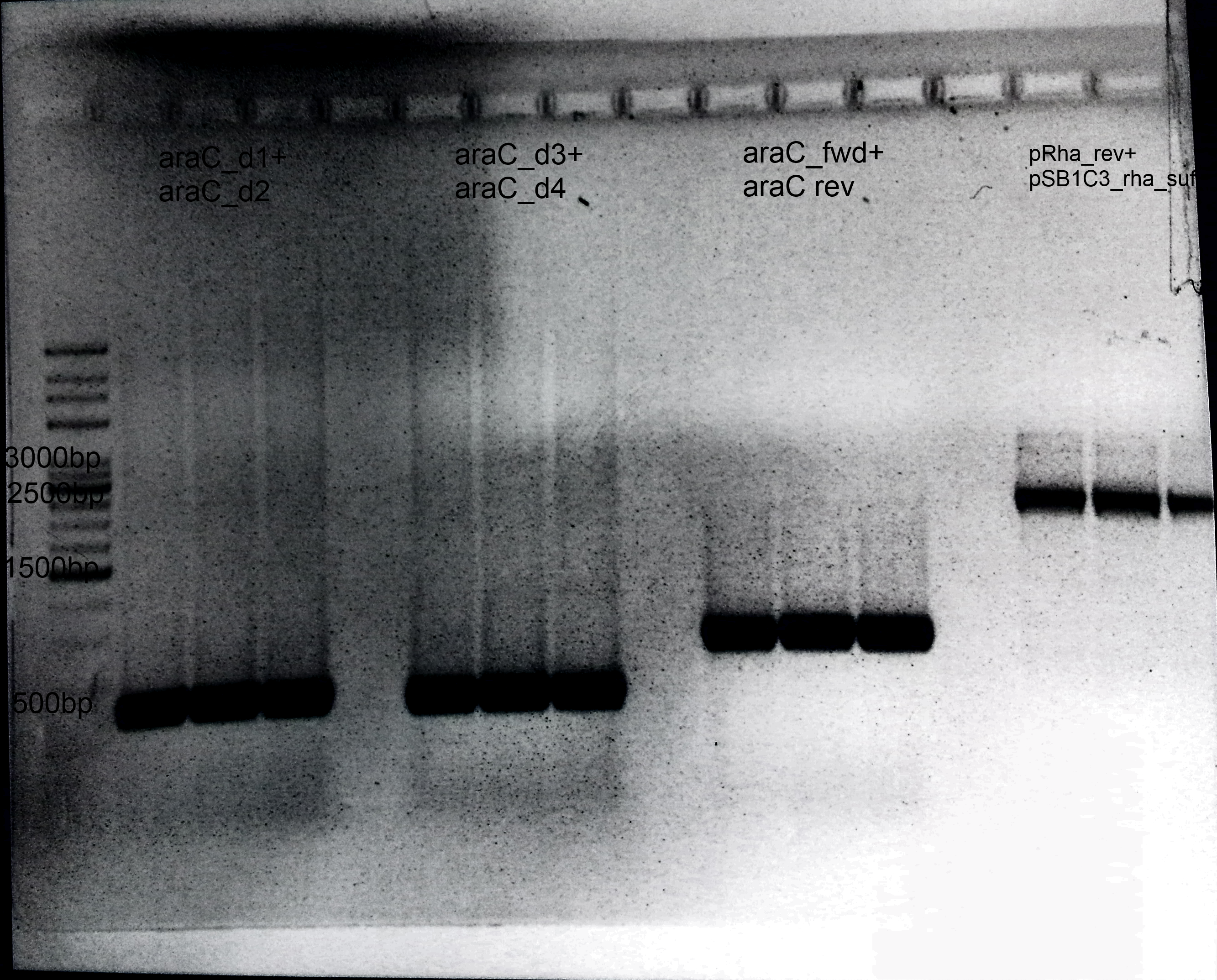
1. AraC: Primer: araC_d1+ araC_d2 Annealing: 62°C Extension: 1min Template: E.coli Genome Size: 600 bp
2. Deletion of araC: Primer: araC_d3+ araC_d4 Annealing= 62°C Extension: 1min Template: E.coli Genome Size: 600 bp
3. Amplification of araC into our Biosafety system: Primer: araC_fwd+ araC rev Annealing= 62°C Extension: 1min Template: BBa_I13458 Size: 900 bp
4.Präparation of pSB1C3: Primer: pRha_rev+ pSB1C3_rha_suf Annealing: 62°C Extension: 1.30 min Template: BBa_I13541 Size: 3 kb
5.Präparation of pSB1C3: Primer: pSB1C3_alr_fwd+ pSB1C3_alr_rev Annealing: 62°C Extension: 1 min Template: E.coli Genome Size: 1kb
6.Präparation of pSB1C3: Primer: pSB1C3_plac_alr+ pSB1C3_alr_rev Annealing: 62°C Extension: 1min Template: E.coli Genome Size: 1kb
7.Präparation of pSB1C3: Primer: pSB1C3_alr_pre+ pSB1C3_alr_suf Annealing: 62°C Extension: 1.30 min Template: pSB1C3 Size: 2kb
8.Präparation of pSB1C3: Primer: pSB1C3_plac_pre+ pSB1C3_alr_suf Annealing: 62°C Extension: 1.30min Template: pSB1C3 Size: 2kb
Analysis by agarosegel
Result:failed
New PCR: Program: 06 phy_cyt mit 62°C Annealing
10µL 5xHF-Buffer 4µL dNTPs 0,5µL Primer1 0,5µL Primer2 1µL Template 1,5µL DMSO 1µL Enzyme 31,5µL dest. H2O
Analysis by agarosegel:
PCR-Purifikation:
BS1a (9-38-451) 113,6ng/µL
BS1b (9-38-452) 75,1 ng/µL
BS2a (9-38-453) 85,5 ng/µL
BS3a (9-38-454) 107,6 ng/µL
BS4a (9-38-455) 14,4 ng/µL
BS5a (9-38-456) 44,5 ng/µL
BS5b (9-38-457) 49,0 ng/µL
BS6a (9-38-458) 63,6 ng/µL
BS7a (9-38-459) 6,9 ng/µL
BS8a (9-38-460) 8,6 ng/µL
 "
"