Team:Evry/Protocols/08
From 2013.igem.org
| (12 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<h1> Genomic DNA extraction </h1> | <h1> Genomic DNA extraction </h1> | ||
| - | <h2> | + | <h2> Goal </h2> |
<p>The aim of the extraction step is to recover <i>Escherichia coli</i> genomic DNA.<br> | <p>The aim of the extraction step is to recover <i>Escherichia coli</i> genomic DNA.<br> | ||
| - | The different step are use to throw other bacterial components off (proteins, cell wall, plasmids,etc).</p> | + | The different step are use to throw other bacterial components off (proteins, cell wall, plasmids,etc). |
| + | Once we have <i>E.coli</i> genomic DNA</p> and <a href=""></a>specific primers, we will obtain sequences we need, doing a <a href="https://2013.igem.org/Team:Evry/Protocols/07" target='_blank'>PCR</a>. | ||
<h2> Preparation </h2> | <h2> Preparation </h2> | ||
| Line 19: | Line 20: | ||
<b><p>1. Cell culture</b><br> | <b><p>1. Cell culture</b><br> | ||
| - | Cultivate cells in LB medium overnight. | + | Cultivate cells in LB medium overnight.<br></p> |
<p><b>2. Cell harvesting</b><br> | <p><b>2. Cell harvesting</b><br> | ||
| - | Set saturated E.coli LM culture into 2 mL tubes. Centrifuge at 8 000 x g for 5 minutes. Discard as much as supernatant as possible. | + | Set saturated E.coli LM culture into 2 mL tubes. Centrifuge at 8 000 x g for 5 minutes. Discard as much as supernatant as possible.<br><p> |
<p><b>3. Cell lysis</b> <br> | <p><b>3. Cell lysis</b> <br> | ||
| Line 32: | Line 33: | ||
Add 20 μL of RNase A solution, mix by vortexing and incubate the tubes for 10 minutes at room temperature.<br> | Add 20 μL of RNase A solution, mix by vortexing and incubate the tubes for 10 minutes at room temperature.<br> | ||
Add 200 μL of Lysis Solution to the sample. Mix thoroughly by vortexing until a homogeneous mixture is obtained. (~15 secondes)<br> | Add 200 μL of Lysis Solution to the sample. Mix thoroughly by vortexing until a homogeneous mixture is obtained. (~15 secondes)<br> | ||
| - | |||
Add 400 μL of 50% ethanol and mix with a vortex or a pipette.<br> | Add 400 μL of 50% ethanol and mix with a vortex or a pipette.<br> | ||
| + | |||
| + | <p><b>4. DNA Binding<br></b> | ||
Transfer the lysate to a GeneJET Genomic DNA Purification Column inserted in a collection tube. Centrifuge the column at 12 000 x g for 1 minute.<br> Discard the collection tube containing flow-through solution and place the column into a new 2 mL tube.<br> | Transfer the lysate to a GeneJET Genomic DNA Purification Column inserted in a collection tube. Centrifuge the column at 12 000 x g for 1 minute.<br> Discard the collection tube containing flow-through solution and place the column into a new 2 mL tube.<br> | ||
| - | <p><b> | + | <p><b>5. Membrane washing<br></b> |
Add 500 μL of Wash Buffer I (previously added with ethanol). Centrifuge at 1 600 x g. Discard the flow-through and place the purification column back into the collection tube. | Add 500 μL of Wash Buffer I (previously added with ethanol). Centrifuge at 1 600 x g. Discard the flow-through and place the purification column back into the collection tube. | ||
Add 500 μL of Wash Buffer II (previously added with ethanol)to the purification column. | Add 500 μL of Wash Buffer II (previously added with ethanol)to the purification column. | ||
| + | <br> | ||
| - | <p><b> | + | <p><b>6. Dry membrane<br></b> |
Centrifuge at 16 000 x g for 3 minutes. | Centrifuge at 16 000 x g for 3 minutes. | ||
| - | + | <br> | |
| - | <p><b> | + | <p><b>7. DNA Elution<br></b> |
Add 200 μL of Elution Buffer to the center of the purification column membrane to elute genomic DNA. Incubate for 2 minutes at room temperature and centrifuge at 8 000 x g for 1 minute. | Add 200 μL of Elution Buffer to the center of the purification column membrane to elute genomic DNA. Incubate for 2 minutes at room temperature and centrifuge at 8 000 x g for 1 minute. | ||
Discard the purification column and store the purified DNA in TrisHCl at -20°C or use it immediatly. | Discard the purification column and store the purified DNA in TrisHCl at -20°C or use it immediatly. | ||
| - | < | + | <h2> Test </h2> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <div class="center"> |
| - | + | <div class="thumb tnone"> | |
| + | <div class="thumbinner" style="width:502px;"> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/f/f0/Nanodrop.png" class="image"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2013/f/f0/Nanodrop.png" width="500" class="thumbimage" /></a> <div class="thumbcaption"> | ||
| + | <div class="magnify"> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/f/f0/Nanodrop.png" class="internal" title="Enlarge"> | ||
| + | <img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a> | ||
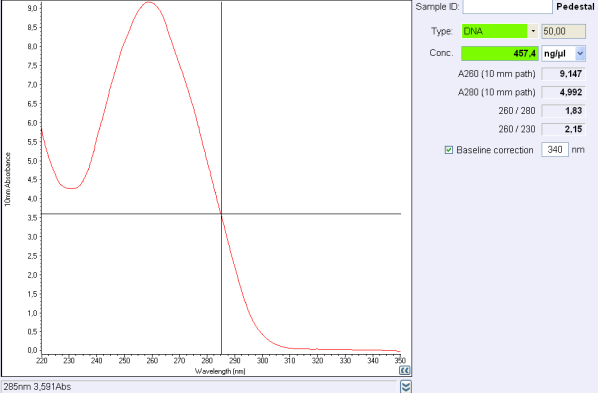
| + | </div> align='center'Figure 1: Nanodrop test after a plasmid purification. | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | If the concentration is between .. and .. ng/μL, concentrate your DNA with SpeedVac concentrator or any other method.<br> | |
| - | + | If the concentration is below .. ng/μL or if 260/280 ratio or 260/230 ratio is not correct, make another purification.<br><br> | |
| + | <i>260/280 ratio and 260/230 indicate the purity of DNA (or RNA). <br> | ||
| + | Nucleic acids absorb at 260 nm while proteins and phenols absorb at 280 nm and carbohydrates at 230 and other contaminents at 230 nm.<br> | ||
| + | For DNA, a 260/280 ratio around 1,8 is concider to be pure and ange 260/230 ratio must be between 2,0 and 2,2.</i><br></p> | ||
Latest revision as of 09:57, 2 September 2013
Genomic DNA extraction
Goal
The aim of the extraction step is to recover Escherichia coli genomic DNA.
The different step are use to throw other bacterial components off (proteins, cell wall, plasmids,etc).
Once we have E.coli genomic DNA
Preparation
Protocol adapted from Thermo Scientific Genomic extraction notebook1. Cell culture
Cultivate cells in LB medium overnight.
2. Cell harvesting
Set saturated E.coli LM culture into 2 mL tubes. Centrifuge at 8 000 x g for 5 minutes. Discard as much as supernatant as possible.
3. Cell lysis
Add 180 μL of Digestion Solution and 20 μL of Proteinase K Solution. Resuspend the cells thoroughly with a vortex or a pipette.
Incubate the tubes at 56°C while vortexing occassionally until the cells are completely lysed(~30 minutes).
Add 20 μL of RNase A solution, mix by vortexing and incubate the tubes for 10 minutes at room temperature.
Add 200 μL of Lysis Solution to the sample. Mix thoroughly by vortexing until a homogeneous mixture is obtained. (~15 secondes)
Add 400 μL of 50% ethanol and mix with a vortex or a pipette.
4. DNA Binding
Transfer the lysate to a GeneJET Genomic DNA Purification Column inserted in a collection tube. Centrifuge the column at 12 000 x g for 1 minute.
Discard the collection tube containing flow-through solution and place the column into a new 2 mL tube.
5. Membrane washing
Add 500 μL of Wash Buffer I (previously added with ethanol). Centrifuge at 1 600 x g. Discard the flow-through and place the purification column back into the collection tube.
Add 500 μL of Wash Buffer II (previously added with ethanol)to the purification column.
6. Dry membrane
Centrifuge at 16 000 x g for 3 minutes.
7. DNA Elution
Add 200 μL of Elution Buffer to the center of the purification column membrane to elute genomic DNA. Incubate for 2 minutes at room temperature and centrifuge at 8 000 x g for 1 minute.
Discard the purification column and store the purified DNA in TrisHCl at -20°C or use it immediatly.
Test
If the concentration is between .. and .. ng/μL, concentrate your DNA with SpeedVac concentrator or any other method.If the concentration is below .. ng/μL or if 260/280 ratio or 260/230 ratio is not correct, make another purification.
260/280 ratio and 260/230 indicate the purity of DNA (or RNA).
Nucleic acids absorb at 260 nm while proteins and phenols absorb at 280 nm and carbohydrates at 230 and other contaminents at 230 nm.
For DNA, a 260/280 ratio around 1,8 is concider to be pure and ange 260/230 ratio must be between 2,0 and 2,2.
 "
"