Team:Paris Saclay/Notebook/July/11
From 2013.igem.org
Contents |
Notebook : July 11
Summary:
For régulation system:
- 1.For those transformation products(of yesterday), RBS+LacS+terminator in PSB1C3 plasmid, the liquid culture has been performed for further experiments. The oligopetides for PCR amplification was made bioinformaticly. The transformation for the terminator BBa_B0010 was still fail.
- 2. The plasmid which contains fnr+RBS+LacZ+terminator and fnr+RBS+AmilCP+terminator was extracted. The restiction digest was performeed for them.
For sensor system:
- 3. The selection of bacterian colonies was achieved by analyzing PCR product. The baterian selected are BphR1 c5 and c6; BphR2 c3 and c4; BphA1 c5 and c6,c7,c8.
Lab work
A.aero/anaerobic regulation system:
- BioBrick RBS+LacZ+terminator in plasmid PSB1C3
Transformation for BBa_I732019 terminator
After ong night culture, we observed 2 tiny colonies on the medium. We continued the experiments by performing another liquid culture at 37°C with ampicillin.
Transformation for BBa_B0010 was always no go
Verification the transformation of BBa_I004450 in PSB3K3 by digestion and eletrophoresis
We performed 2 types of digestion:
Simple digestion:
- DNA: 3µl
- Buffe:r 3 µl
- Enzyme: 1µl
- H2O: 23µl
- Total: 30µl
Double digestion:
- DNA: 5µl
- Buffer: 3 µl
- Enzyme: 2µl
- H2O: 20µl
- Total: 30µl
Buffer used:
- Ecor I+ PST I -> orange
- Xho I -> green
- Sac II -> blue
- XhoI+Sac II -> green
After the digestion, we performed a eletrophoresis for verification:
|
Estimed size and observed size:
| enzyme | estimed size | observed size |
| Ecor I+PST I | 1069bp and 2750 bp | 1000bp and 2700bp |
| Xho I | 2976bp+842bp | 4000bp |
| Sac II | 3819bp | 4000bp |
| Xho I+Sac II | 843bp,616bp,2367bp | 1200bp and 2000bp |
We confimed the existence of BBa_I04550 in plasmid PSB3K3.
B.PCBs sensor system:
- Construction for BioBrick promoter promoter BphR1, BphR2, BphA1
Colony PCR
From the culture of cloning for promoter BphR1, BphR2, BphA1 which we seeded yesterday, we had chosen 8 colonies in total for further test. And like we did for promoter fnr, we used 8 primers for PCR amplification, they were: BphR1 up/down, BphR2 up/down, BphA1 up/down and Vf/VR.
In order to make clear this large number of PCR tubes, we classified them into 4 lots. they were:
Mix A : promoter BphR1
- buffer go ta:(1X) : 5µl
- MgCL2: 2µl
- dNTP: 1µl
- primers(R1_up/VR or VF/R1_down): 0.125µl
- DNA: 2µl
- Enzyme: 0.25µl
- H2O: about 14.5µl
- total: 25µl
Mix B : promoter BphR2
- buffer go ta:(1X) : 5µl
- MgCL2: 2µl
- dNTP: 1µl
- primers(R2_up/VR or VF/R2_down): 0.125µl
- DNA: 2µl
- Enzyme: 0.25µl
- H2O: about 14.5µl
- total: 25µl
Mix C : promoter BphA1
- buffer go ta:(1X) : 5µl
- MgCL2: 2µl
- dNTP: 1µl
- primers(A1_up/VR or VF/A1_down): 0.125µl
- DNA: 2µl
- Enzyme: 0.25µl
- H2O: about 14.5µl
- total: 25µl
Mix D :
- buffer go ta:(1X) : 5µl
- MgCL2: 2µl
- dNTP: 1µl
- primers(VF/VR): 0.125µL
- DNA: 2µl
- Enzyme: 0.25µl
- H2O: about 14.5µl
- total: 25µl
PCR program:
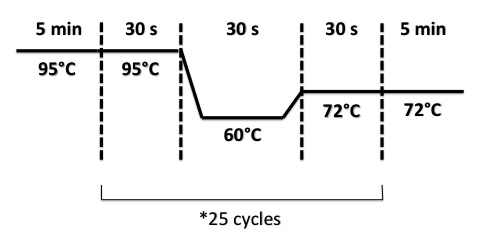
Result : can not find images <p>We considered that promoter BphR1 clone 5 and 6, promoter BphR2 clone 3 and 4, promoter BphA1 clone 5 to 8 accords our estimation. We got them in stock.
Notebook : August 23
Lab work
B - PBC sensor system
Objective : obtaining Bba_K1155001, Bba_K1155002, BphR2
'1 - Colony PCR of Bba_K1155001, Bba_K1155002 and BphR2 in DH5α
Anaïs, Zhou
|
Transformation of Bba_K1155001, Bba_K1155002 and BphR2 protein in DH5α of 07/10/13 work. We will do a Colony PCR. |
COLONIES REPIQUEE DANS 10µL d'eau
Used quantities :
- DNA : 2µL
- Mix A : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- BphR1_Up/VR : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix B : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- VF/BphR1_Down : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix C : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- BphR2_Up/VR : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix D : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- VF/BphR2_Down : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix E : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- BphA1_Up/VR : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix F : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- VF/BphA1_Down : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix G : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 125µL
- MgCl2 : 50µL
- dNTP : 25µL
- VF/VR: 3µL for each oligo
- Enzyme : 6.25µL
- H2O : 362.75µL
PCR program :
| Previous day | Back to calendar | Next day |
 "
"
