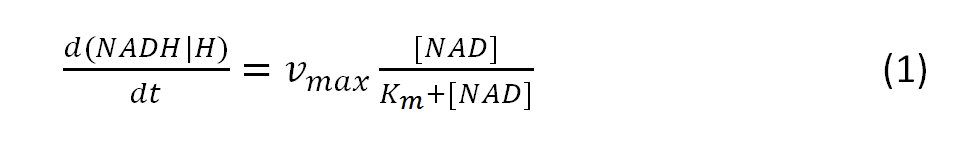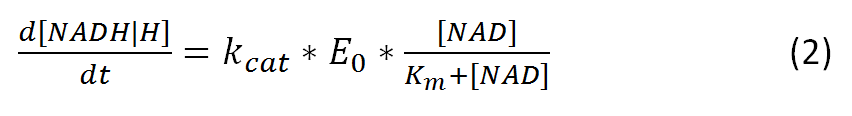Team:Bielefeld-Germany/Modelling/Inter
From 2013.igem.org
m |
|||
| Line 47: | Line 47: | ||
<html> | <html> | ||
| - | <h1>Modelling | + | |
| + | <h1>Modelling</h1> | ||
| + | |||
<div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left:45px; clear:both;"> | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left:45px; clear:both;"> | ||
<div class="bigbutton"> | <div class="bigbutton"> | ||
<a href="https://2013.igem.org/Team:Bielefeld-Germany/Modelling">Overview</a></div> | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Modelling">Overview</a></div> | ||
| - | |||
| - | |||
<div class="bigbutton"> | <div class="bigbutton"> | ||
<a href="https://2013.igem.org/Team:Bielefeld-Germany/Modelling/Inter">Intermediates</a></div> | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Modelling/Inter">Intermediates</a></div> | ||
| + | <div class="bigbutton"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Modelling/Reduction">Mediator Reduction</a></div> | ||
</div> | </div> | ||
| - | |||
| + | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left:45px; clear:both;"> | ||
| + | <div class="bigbutton"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Modelling/Optimal">Optimal conditions</a></div> | ||
| + | </div> | ||
| + | |||
| + | </html> | ||
==Intermediates== | ==Intermediates== | ||
Revision as of 14:00, 27 October 2013
Modelling
Intermediates
The electrons that will be eventually transferred to the anode are generated during the biochemical processes within the bacterial cell. The central pathway in E.coli is the glycolysis and one of its bottleneck reactions is the reaction catalyzed by the Glyceraldehyde 3-phosphate dehydrogenase (GAP-DH). In the course of this reaction the NAD+ is transformed to the reducing intermediate NADH/H+. Thus this intermediate takes part in the electron flow needed for the generation of electricity.
The resulting amount of the intermediate NADH/H can be calculated with the modeled Michaelis-Menten equation:
, where vmax is the maximum rate achieved by the system
and Km describes the substrate concentration at the half-maximal rate, that can be achieved by the system.
Further the vmax can be also represented as:
, where kcat represents the rate-limiting turnover number
and E0 represents the enzyme concentration.
The resulting equation, alternative to (1), would be:
The data of the kinetics have been obtained from the internet database BRENDA. For E.coli the Km value for the NAD as a substrate is 0.045 for the wild-type enzyme. Unfortunately neither the turnover number nor the vmax for this organism could be found. Also the in depth research of the enzyme GAP-DH on the maximum rate of the reaction gave no satisfactory results – the only found value of vmax was the one from the ciliate Tetrahymena thermophile and was obtained from the method, where the GAP-DH was inhibited by stress reagents. Hence, there are two possible solutions how the model could be assessed:
- Applying the turnover number of another bacteria for the wild-type enzyme (e.g.: Bacillus subtilis with the kcat = 70 [1/s]) OR
- Applying the Vmax for the T. thermophile (Errafiy, Soukri , 2012)
More plausible is the first application as the organism, to which the data referred, is closer related to E.coli and the data has been taken for the wild-type enzyme. Furthermore, B. subtilis has been cultivated under standard conditions and the activity of the enzyme hasn’t been influenced by any additional environmental factor.
References
- Errafiy N, Soukri A (2012). Purification and partial characterization of glyceraldehyde-3-phosphate dehydrogenase from the ciliate Tetrahymena thermophila. Acta Biochimica et Biophysica Sinica, Volume 44(6), 527-534.
 "
"