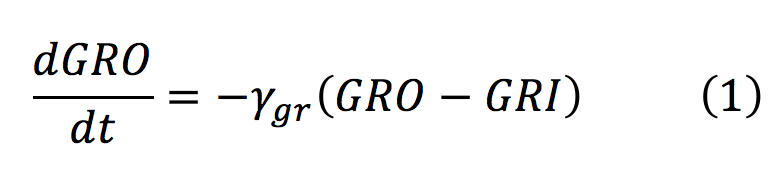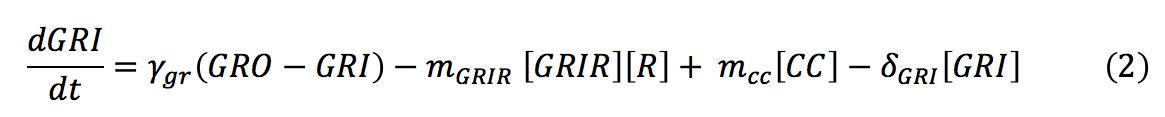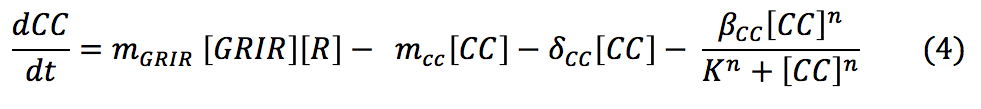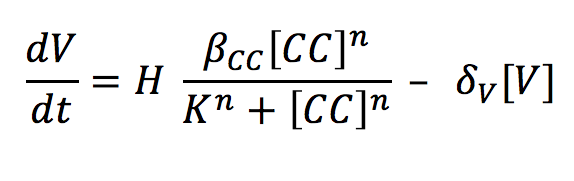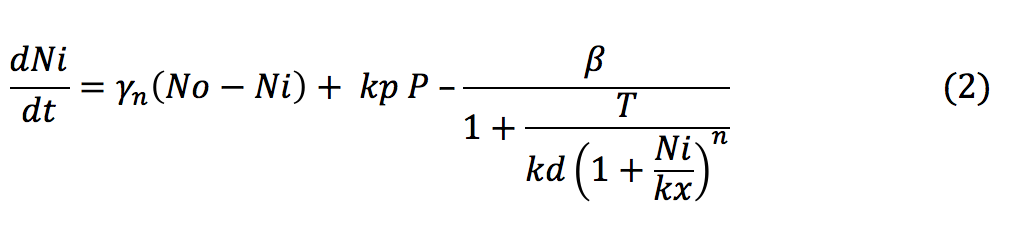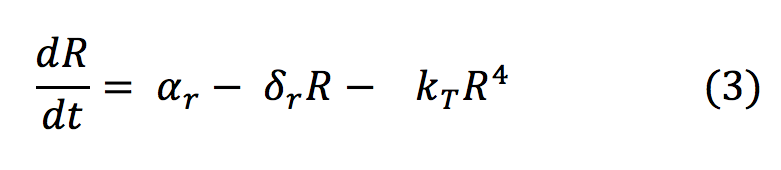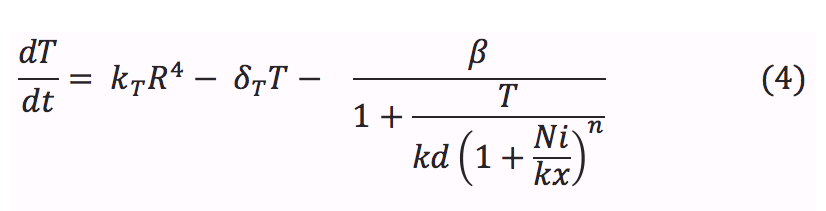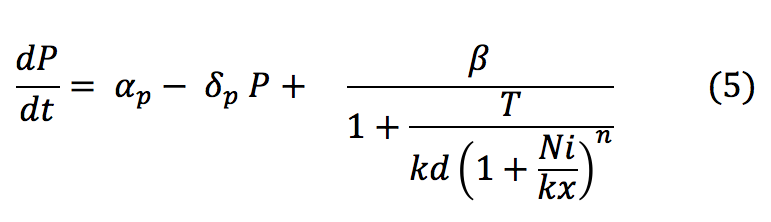Team:Colombia Uniandes/Modeling
From 2013.igem.org
Differential Equations
We use a mathematical model to check if the design works before starting to work at the laboratory. To develop this model the first step is to create a deterministic model, which is based on differential equations. This type of model describes the mean behavior for each of the substances in the synthetic circuit over time. However, it does not take into account the probabilities involved in each of the events described, the population interactions and the noise of the system.
The deterministic model is based on the law of mass conservation and expresses the inputs and outputs of the system with expressions that use the law of mass action and known models like Hill's or Michaelis Menten.
Below are shown and explained the differential equations that allowed us to work in both projects.
Glucocorticoid Sensor
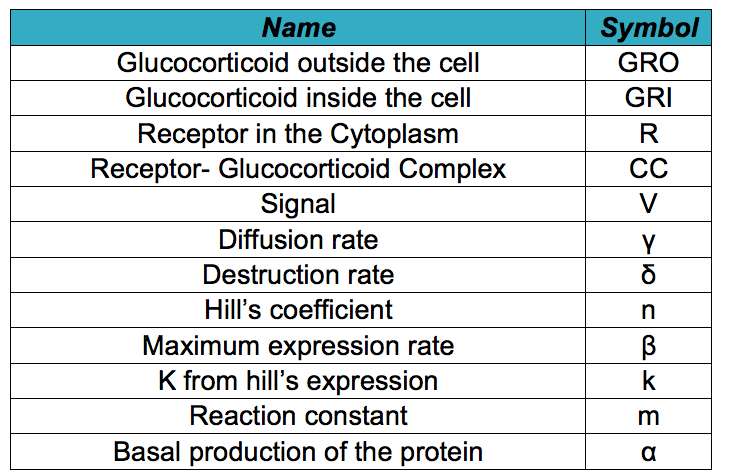
Below it is shown differential equations for the glucocorticoid sensor based on the circuit shown in the project description. However, before describing the modeled system, it is necessary to know all the symbols that will be used in the documents and the simulation. The following table contains all the substances involved in the processes and the constants required for the simulation.
- GRO: Animals release glucocorticoids under stress situations such as famine, injuries, anxiety or fear. This hormone is the one we will detect as an indicator of stressful conditions for the animal. This differential equation expresses the concentration of glucocorticoids outside the cell, which only depends on their diffusion rate into the cell.
- GRI: The amount of glucocorticoids inside the yeast cell depends on the degradation rate of the hormone, the complex formation and the import rate of the glucocorticoids inside the cell, which depends on the diffusion rate of the hormone in relation with the concentration of the hormone outside and inside the cell.
- R: The receptor's production in the cytoplasm is regulated by a constitutive promoter. This protein binds to the glucocorticoid and forms a dimer. So its concentration on the cell depends on the basal production, the complex production and the degradation rate.
- CC: Once the glucocorticoid binds to the receptor, the dimer is made. Its concentration in the cell will depend on the amount of glucocorticoid available in the cytosol, the amount of receptors produced and the affinity between the ligand and the protein. We also consider the degradation rate of the complex and its binding to the signal promoter enhancer.
- V: The complex binds to an enhancer and this union unfolds the DNA and allows the RNA polymerase to produce our visible indicator (mCherry). The concentration of this signal will depend on the binding of the complex and the enhancer and its degradation rate.
Nickel removal system
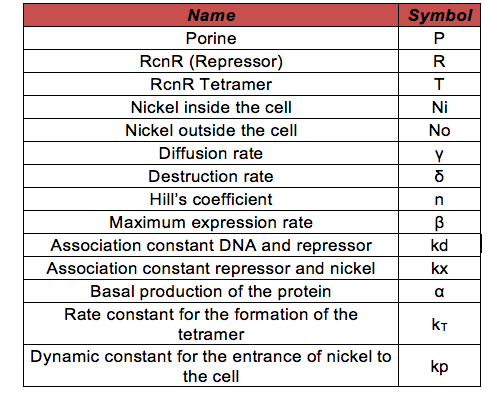
As in the Glucocorticoid Sensor project the differential equations were based on the circuit shown in the project description . Before explaining the equations ,it is necessary to know all the symbols that will be use in the documents and the simulation. The following table contains all the substances involved in the processes and the constants required for the simulation.
- No: We are going to work with contaminated water that is going to have Nickel ions in the media outside our bacteria. The change in concentration of theses ions only depends in the rate at which they are taken by the cells. It may enter by diffusion or by the porines.
- Ni: The amount of nickel inside the bacteria depends on the rate at which it enters to the cell by diffusion and by the porines. Also it is affected by the union of the ions with the repressor.
- R: The repressor is produced at a constant rate by a constitutive promoter. Then binds to three other molecules to form a tetrameric protein
- T: The tretamer is formed when four repressor subunits bind together, it binds to the DNA in order to block the production of the porine and when there is nickel inside the cell each of the repressor subunits coordinates a nickel atom unbinding the repressor from DNA and enabling transcription downstream.
- P: The Porine is produced at a low basal when the tretameric repressor is binded to the promoter. Once the repressor binds to the nickel it is released and allows the production of the porine.
As you can see there are many constants that define the system, so the next step in the mathematical model is to fin those parameters
 "
"