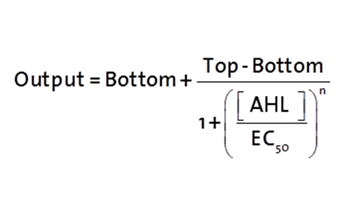Team:ETH Zurich/Experiments 5
From 2013.igem.org
| Line 5: | Line 5: | ||
[[File:Gfp_od_sigmoid1.png|350px|right|thumb|<b>Figure 1: Sensivity curve of the N-3-oxo-C6-l-homoserine lactone </b> The OHHL concentration range is plotted against the standarized fluorescence (au)]] | [[File:Gfp_od_sigmoid1.png|350px|right|thumb|<b>Figure 1: Sensivity curve of the N-3-oxo-C6-l-homoserine lactone </b> The OHHL concentration range is plotted against the standarized fluorescence (au)]] | ||
| - | <p><br><br>In order to select mutated pLuxR promoters we need to know about the sensivity of the wild type construct. The test range was inspired form the paper about [http://www.ncbi.nlm.nih.gov/pubmed/18760602 Evaluation of a focused library of N-Acryl L-Homoserine lactone reveals a new set of potent quorum sensing modulators].The paper shows sensivities of the pLuxR promoter to different sets of OHHL molecules in Vibrio fisheri. We need to adjust the ranges for our <i>E.coli DH5alpha</i> strain in a second run and finally get our sensivity curve over the linear range of [0 nM],[0.25 nM],[0.5 nM],[1 nM],[2 nM],[3 nM],[4 nM],[5 nM],[ | + | <p><br><br>In order to select mutated pLuxR promoters we need to know about the sensivity of the wild type construct. The test range was inspired form the paper about [http://www.ncbi.nlm.nih.gov/pubmed/18760602 Evaluation of a focused library of N-Acryl L-Homoserine lactone reveals a new set of potent quorum sensing modulators].The paper shows sensivities of the pLuxR promoter to different sets of OHHL molecules in Vibrio fisheri. We need to adjust the ranges for our <i>E.coli DH5alpha</i> strain in a second run and finally get our sensivity curve over the linear range of [0 nM],[0.25 nM],[0.5 nM],[1 nM],[2 nM],[3 nM],[4 nM],[5 nM],[10 nM],[20 nM],[30 nM],[40 nM]and[50 nM].<br><br> The experimental set-up includes a 96-well plate filled with 180 uL LB media , 10uL of receiver cells and 10 uL of different OHHL concentrations, everything in triplicates. (+ blank and negative control). The experiment runs for 16 hours in order to monitor the evolution of fluorescence and later choose the steady state to create the sensivity curve.The data are analyzed with GraphPad Prism 6.0.<br></p> |
<br clear="all"/> | <br clear="all"/> | ||
| Line 22: | Line 22: | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | <b>Liquid culture</b><br><br>To gain precise data out of our fluorescence analysis, to better characterize the different promoters, we use the single analysis to obtain high throughtput data.<br>The protocol was the following :<br>From overnight cultures we inoculate | + | <b>Liquid culture</b><br><br>To gain precise data out of our fluorescence analysis, to better characterize the different promoters, we use the single analysis to obtain high throughtput data.<br>The protocol was the following :<br>From overnight cultures we inoculate 50 mL Falcon™ tubes containing 5mL LB media, 5 uL OHHL and the antibiotic. We evaluate a range of 10 different concentrations from <br>10<sup>-4</sup>nM OHHL to 10<sup>5</sup>nM OHHL by a 10 fold increase, according to the results of the plate reader analysis. The measurements were done in duplicates.</p> |
<br clear="all"/> | <br clear="all"/> | ||
[[File:C6.png|left|630px|thumb|<b>Figure 2: Sensitivity curve of the Wild Type in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System. The first graph on the left is the fluorescence(au) against the concentration (nM). The second graph shows the fluorescence(au)over the whole range OHHL concentrations.]] | [[File:C6.png|left|630px|thumb|<b>Figure 2: Sensitivity curve of the Wild Type in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System. The first graph on the left is the fluorescence(au) against the concentration (nM). The second graph shows the fluorescence(au)over the whole range OHHL concentrations.]] | ||
| Line 28: | Line 28: | ||
[[File:G1.png|left|455px|thumb|<b>Figure 3: Sensitivity curve of the PluxR1 (G1) mutated promoter in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System. The first graph on the left is the fluorescence(au) against the concentration (nM). The second graph shows the fluorescence(au) over the whole range OHHL concentrations.]] | [[File:G1.png|left|455px|thumb|<b>Figure 3: Sensitivity curve of the PluxR1 (G1) mutated promoter in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System. The first graph on the left is the fluorescence(au) against the concentration (nM). The second graph shows the fluorescence(au) over the whole range OHHL concentrations.]] | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | <p>The first graph (on the left)of <i>Figure 2.</i> is the | + | <p>The first graph (on the left)of <i>Figure 2.</i> is the sensitivity curve of the J08955-GFP construct with the wild type R0062 promoter. The EC<sub>50</sub> indicates the half maximal effective concentratio and is 0.02 nM for the wild type. We had a few background noise during the fluorescence measurements of the wild type construct, that's why the R<sup>2</sup>=0.8.<br>The second graph (on the right)of <i>Figure 2.</i> is the fluorescence in function of the <b>side scatter</b> over the whole concentration range of OHHL. The <b>side scatter</b> is a measurement of refracted light that occurs at each interface in the cell. The refracted light is collected and amplified in a photomultiplier tube. The <b>SSC</b> gives information about the cellular granularity and complexity (e.g. dead cells).<br>In the end we note that the EC<sub>50</sub> (0.02nM) from single cell analysis is 100 times lower than the EC<sub>50</sub> (2 nM) calculated out of the Tecan Infinite M200 PRO™ plate reader measurements<br><br>The first graph (on the left)of <i>Figure 3.</i> shows the sensivity curve of the isolated pLuxR1 (or G1 according to the deepwell plate identification). The EC<sub>50</sub>=6'250nM is remarquably high andwell correlating data according to R<sup>2</sup>=0.97. The second graph (on the right)of <i>Figure 3.</i> is the fluorescence in function of the <b>side scatter</b>over the whole concentration range of OHHL</p> |
<br clear="all"/> | <br clear="all"/> | ||
[[File:Comparison.png|left|400px|thumb|<b>Figure 4. Comparison of the Wild Type and PluxR1 mutant sensitivity curves in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System]] | [[File:Comparison.png|left|400px|thumb|<b>Figure 4. Comparison of the Wild Type and PluxR1 mutant sensitivity curves in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System]] | ||
| - | <p>The direct plot of both sensivity curves shows the clearly shifted sensivity of the pLuxR1 promoter in comparison to the wild type R0062 promoter. The EC<sub>50</sub> shit from 0.02nM for the wild type to 6' | + | <p>The direct plot of both sensivity curves shows the clearly shifted sensivity of the pLuxR1 promoter in comparison to the wild type R0062 promoter. The EC<sub>50</sub> shit from 0.02nM for the wild type to 6'250 nM for the pLuxR1 promoter is equal to a 312'500 fold increase.<br> This promoter will be used in our final set-up to only respond to high concentrations of OHHL and therefore be activated, when two sender cells are surrounding the receiver cell, and expressing the [https://2013.igem.org/Team:ETH_Zurich/Experiments_7 AES] hydrolase responsible for the colorimetric conversion of the specific substrate to a visible purple output. |
<br clear="all"/> | <br clear="all"/> | ||
<br> | <br> | ||
Revision as of 15:26, 29 September 2013
Contents |
Native N-3-Oxo-Hexanoyl-l-Homoserine Lactone tests using the Tecan Infinite M200 PRO™ plate reader
In order to select mutated pLuxR promoters we need to know about the sensivity of the wild type construct. The test range was inspired form the paper about [http://www.ncbi.nlm.nih.gov/pubmed/18760602 Evaluation of a focused library of N-Acryl L-Homoserine lactone reveals a new set of potent quorum sensing modulators].The paper shows sensivities of the pLuxR promoter to different sets of OHHL molecules in Vibrio fisheri. We need to adjust the ranges for our E.coli DH5alpha strain in a second run and finally get our sensivity curve over the linear range of [0 nM],[0.25 nM],[0.5 nM],[1 nM],[2 nM],[3 nM],[4 nM],[5 nM],[10 nM],[20 nM],[30 nM],[40 nM]and[50 nM].
The experimental set-up includes a 96-well plate filled with 180 uL LB media , 10uL of receiver cells and 10 uL of different OHHL concentrations, everything in triplicates. (+ blank and negative control). The experiment runs for 16 hours in order to monitor the evolution of fluorescence and later choose the steady state to create the sensivity curve.The data are analyzed with GraphPad Prism 6.0.
PluxR random mutagenesis PCR
The pLuxR R0062 wild type promoter was mutated using site directed mutagenesis on specific sites according to the paper [put paper here]
PluxR promoter with different sensitivity analyzed using the single cell analyzer BD LSRFortessa™ Flow Cytometer System
The fitting of the following graphs was perfomed using the previous equation.the equation consists of :
Output = sfGFP levels [au] (“eGFP level”)
Top = maximal sfGFP level [au] (eGFP level at “full induction”)
Bottom = minimal sfGFP level [au] (“leakiness”)
n = Hill coefficient (“cooperativity”)
EC50 = Half-maximal effective concentration (“sensitivity”)
Liquid culture
To gain precise data out of our fluorescence analysis, to better characterize the different promoters, we use the single analysis to obtain high throughtput data.
The protocol was the following :
From overnight cultures we inoculate 50 mL Falcon™ tubes containing 5mL LB media, 5 uL OHHL and the antibiotic. We evaluate a range of 10 different concentrations from
10-4nM OHHL to 105nM OHHL by a 10 fold increase, according to the results of the plate reader analysis. The measurements were done in duplicates.</p>

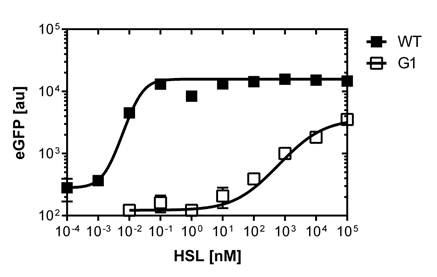
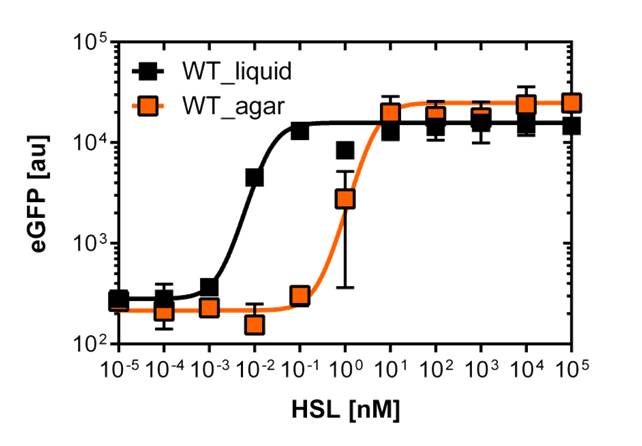
The first graph (on the left)of Figure 2. is the sensitivity curve of the J08955-GFP construct with the wild type R0062 promoter. The EC50 indicates the half maximal effective concentratio and is 0.02 nM for the wild type. We had a few background noise during the fluorescence measurements of the wild type construct, that's why the R2=0.8.
The second graph (on the right)of Figure 2. is the fluorescence in function of the side scatter over the whole concentration range of OHHL. The side scatter is a measurement of refracted light that occurs at each interface in the cell. The refracted light is collected and amplified in a photomultiplier tube. The SSC gives information about the cellular granularity and complexity (e.g. dead cells).
In the end we note that the EC50 (0.02nM) from single cell analysis is 100 times lower than the EC50 (2 nM) calculated out of the Tecan Infinite M200 PRO™ plate reader measurements
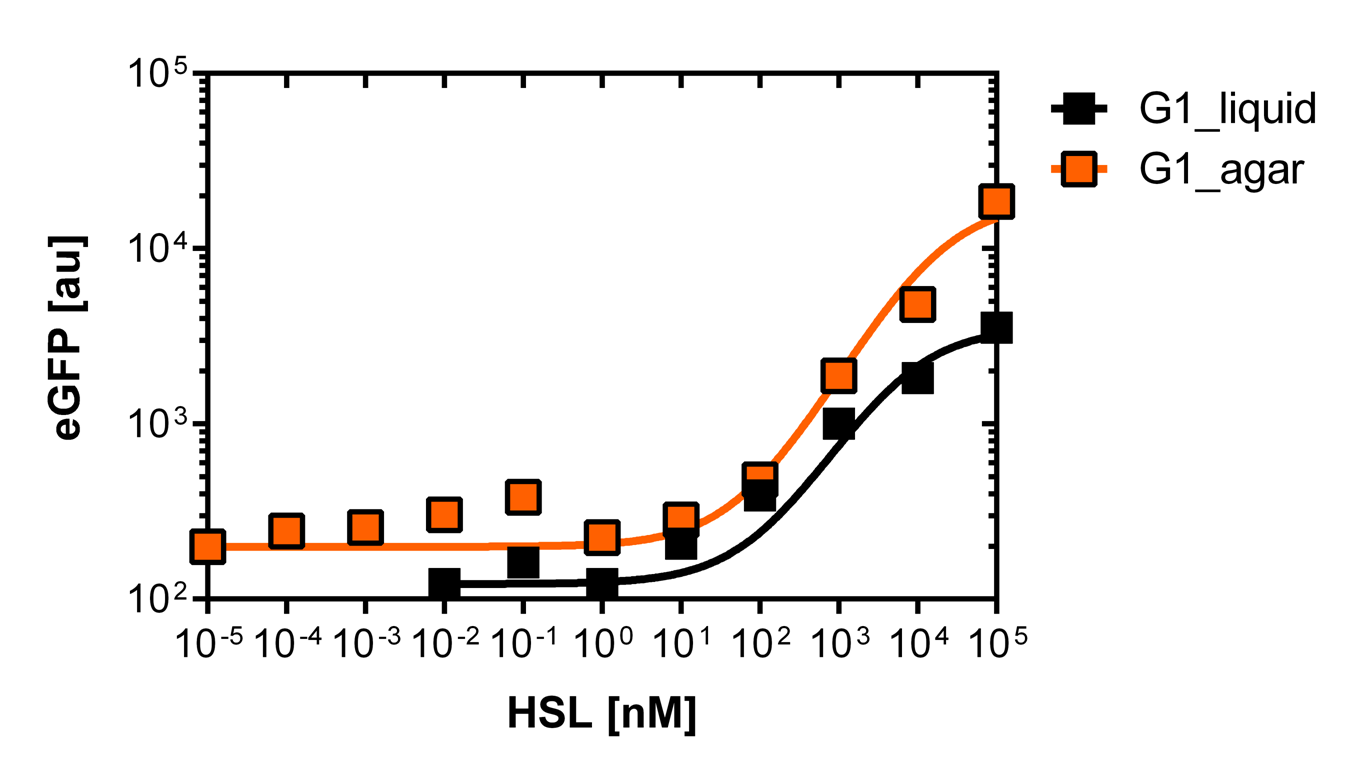
The first graph (on the left)of Figure 3. shows the sensivity curve of the isolated pLuxR1 (or G1 according to the deepwell plate identification). The EC50=6'250nM is remarquably high andwell correlating data according to R2=0.97. The second graph (on the right)of Figure 3. is the fluorescence in function of the side scatterover the whole concentration range of OHHL
The direct plot of both sensivity curves shows the clearly shifted sensivity of the pLuxR1 promoter in comparison to the wild type R0062 promoter. The EC50 shit from 0.02nM for the wild type to 6'250 nM for the pLuxR1 promoter is equal to a 312'500 fold increase.
This promoter will be used in our final set-up to only respond to high concentrations of OHHL and therefore be activated, when two sender cells are surrounding the receiver cell, and expressing the AES hydrolase responsible for the colorimetric conversion of the specific substrate to a visible purple output.
On Agar plates
For the real colisweeper game we need to know about the behavior of the promoters on agar plates.
We set-up an experiment which consists in pooring agar plates containing different OHHL concentration going form 10-5nM to 105nM by 10 fold increase. The wild type and the pLuxR1 promoter were streaked onto those plates to analyze the sensivity to OHHL of the wild type and the promoter on an agar plate. After 15hours of incubation at 37 degree celsius some colonies were picked and resuspended in Phosphate Buffered Solution (PBS) for the single cell analysis , using again the BD LSRFortessa™ Flow Cytometer System.
 "
"