Team:Bielefeld-Germany/Project/Phenazine
From 2013.igem.org
| (25 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<html> | <html> | ||
<style> | <style> | ||
| - | |||
| - | |||
#globalwrapper ul {padding-left:40px; padding-right:40px;} | #globalwrapper ul {padding-left:40px; padding-right:40px;} | ||
| Line 27: | Line 25: | ||
#globalwrapper .bigbutton p{padding-left:5px; padding-right:5px; padding-top:2px;} | #globalwrapper .bigbutton p{padding-left:5px; padding-right:5px; padding-top:2px;} | ||
| - | .bigbutton{width: | + | .bigbutton{width:130px; height:50px; line-height:50px; font-size:1.2em; margin-right:10px; display:table;} |
.bigbutton a{display:block; height:100%;} | .bigbutton a{display:block; height:100%;} | ||
| Line 49: | Line 47: | ||
<html> | <html> | ||
<h1>Phenazine</h1> | <h1>Phenazine</h1> | ||
| - | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left: | + | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left:20px; clear:both;"> |
<div class="bigbutton"> | <div class="bigbutton"> | ||
| - | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Abstract">Projects Overview</a></div> | + | <p><a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Abstract">Projects Overview</a></p></div> |
| + | <div class="bigbutton"> | ||
| + | <p><a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Mediators">Mediators Overview</a></p></div> | ||
<div class="bigbutton"> | <div class="bigbutton"> | ||
<a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Phenazine#Theory">Theory</a></div> | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Phenazine#Theory">Theory</a></div> | ||
<div class="bigbutton"> | <div class="bigbutton"> | ||
| - | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Phenazine#Genetic_Approach">Genetic Approach</a></div> | + | <p><a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Phenazine#Genetic_Approach">Genetic<br> Approach</a></p></div> |
<div class="bigbutton"> | <div class="bigbutton"> | ||
<a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Phenazine#Results">Results</a></div> | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Project/Phenazine#Results">Results</a></div> | ||
| Line 65: | Line 65: | ||
==Overview== | ==Overview== | ||
| - | + | [[File:IGEM Bielefeld 2013Phenazine-3D-balls-B.png|300px|thumb|left| '''Figure 1:''' Molecular structure of phenazine.]] | |
| - | [[ | + | |
| - | + | ||
<p align="justify"> | <p align="justify"> | ||
| - | + | Phenazines are water-soluble secondary metabolites secreted by some bacterial species. Almost all their properties can be attributed to their ability to undergo redox transformations. Some soil-born bacteria commonly produce phenazine-1-carboxylic acid ([http://www.ebi.ac.uk/chebi/chebiOntology.do?chebiId=CHEBI:62412 PCA])) against fungal root disease. (Niknejad Kazempour, [http://www.agriculturaits.czu.cz/pdf_files/vol_42_4_pdf/kazempour.pdf 2009]) Bacteria are also able to use this substance as a mediator for electron shuttling, which leads to an efficiency gain if a phenazine-producing strain is present in a MFC. | |
| + | <br> | ||
| + | Problems arise with the different phenazine producing host strains. Most of them are classified as pathogens and therefore unsuitable for application in our lab and MFC. | ||
| + | Hence, all experiments carried out in this section were soluble done with ''Pseudomonas fluorescences'' which is classified as risk group 1. | ||
| + | <br> | ||
| + | Initially PCA overproduction seemed to be a promising approach to achieve higher electron transfers. Because of the extremely complicated cloning procedure as well as the good advice of the consulted MFC expert [https://2013.igem.org/Team:Bielefeld-Germany/Human_Practice/Experts Dr. Falk Harnisch], we abandoned this sub-project in an early stage and searched for alternatives. We concentrated on an overexpression of riboflavin, which serves as an electrone shuttle itself and of the glycerol dehydrohenase ''gldA'', which increases NADH concentration. (see sections [https://2013.igem.org/Team:Bielefeld-Germany/Project/Riboflavine Riboflavin] and [https://2013.igem.org/Team:Bielefeld-Germany/Project/GldA ''gldA''] on our Wiki for more information). | ||
| + | Still, quite a lot of work was invested into investigation of phenazine functions and synthesis in bacteria, of so we present a short theoretical overview of this topic. | ||
| + | The only entry in the labjournal on phenazine is found [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/July here] in Week 13. | ||
</p> | </p> | ||
| - | + | <br> | |
| - | + | ||
==Theory== | ==Theory== | ||
| Line 80: | Line 84: | ||
Phenazines comprise a large group of heterocyclic nitrogen-containing substances, having a tricyclic ring in its core. They are substituted at different points around their rings, which alters water solubility and other properties. After discovery of naturally occurring phenazines, more than 6000 were chemically synthesized. (Mavrodi, [http://www.ncbi.nlm.nih.gov/pubmed/16719720 2006]). [http://en.wikipedia.org/wiki/Neutral_red Neutral red], a well-known pH-indicator, also belongs to this class. | Phenazines comprise a large group of heterocyclic nitrogen-containing substances, having a tricyclic ring in its core. They are substituted at different points around their rings, which alters water solubility and other properties. After discovery of naturally occurring phenazines, more than 6000 were chemically synthesized. (Mavrodi, [http://www.ncbi.nlm.nih.gov/pubmed/16719720 2006]). [http://en.wikipedia.org/wiki/Neutral_red Neutral red], a well-known pH-indicator, also belongs to this class. | ||
<br><br> | <br><br> | ||
| - | Phenazine derivatives were one of the first secondary bacterial metabolites that were consequently studied since their discovery in 1859. The first substance of this family, a blue pigment of ''Pseudomonas | + | Phenazine derivatives were one of the first secondary bacterial metabolites that were consequently studied since their discovery in 1859. The first substance of this family, a blue pigment of ''Pseudomonas aeruginosa'', was called [http://en.wikipedia.org/wiki/Pyocyanine «pyocyanine»] (short PYO). It has antimicrobial properties that raised further interest in these substances. |
| - | Phenazines are produced by many bacterial | + | Phenazines are produced by many bacterial species, but they are mostly studied in representatives of fluorescent pseudomonads. A single bacterial strain usually produces two or more species-specific phenazines, except ''Pseudomonas fluorescens'' 2-79 that is known to produce only PCA, a broad-spectrum antibiotic, which is a casual agent active against a variety of root pathogens (Mavrodi et al., [http://www.ncbi.nlm.nih.gov/pubmed/11591691 2001]). |
<br><br> | <br><br> | ||
| - | + | Biological phenazines are known for their broad-spectrum of antibiotical, antiparasaticand even anti-malaria properties (Handelsmann et al., [http://www.plantcell.org/content/8/10/1855.long 1996]). | |
| - | Consequently it was shown | + | Consequently, it was shown that almost all properties of phenazines could be attributed to their ability to undergo redox transformations (Muller, [http://www.ncbi.nlm.nih.gov/pubmed/8541351 1995]). This discovery brought up the idea, that these molecules can be used as an electron shuttle in a MFC. Indeed, it was shown that external addition of phenazines, both PCA and PCN ([http://www.ebi.ac.uk/chebi/searchId.do?chebiId=62240 phenazine-1-carboxamide]) alike, causes a significant raise of current flow (The Hai Pham, [http://www.ncbi.nlm.nih.gov/pubmed/18688612 2008]). Surprisingly, a mixed culture of phenazine-secreting and phenazine non-secreting strains showed similar results, proving that phenazine can be used for redox cycling not only by its producer (Hernandez et al. [http://www.ncbi.nlm.nih.gov/pubmed/14766572 2004]). |
| - | + | ||
<br><br> | <br><br> | ||
| - | + | Many strains of ''Pseudomonas fluorescens'', common soil inhabitants, are known to produce the phenazine compound PCA. The phenazine synthesis pathway was studied in detail. The seven-gene cluster [http://www.ncbi.nlm.nih.gov/nuccore/L48616.1 ''phzABCDEFG''], 8505 bp long, was firstly identified in ''Pseudomonas fluorescens'' 2-79.</p> | |
| - | + | <br> | |
| + | <p align="justify">Research on phenazine diversity and genetic evolution, (Mavrodi, [http://www.ncbi.nlm.nih.gov/pubmed/20008172 2010]) stated, that many species of fluorescent pseudomonas have the corresponding genes and are able to produce phenazines, so we expected that orthologs are found in the ''Pseudomonas fluorescens'' 50090 that we obtained from [http://www.dsmz.de DSMZ]. | ||
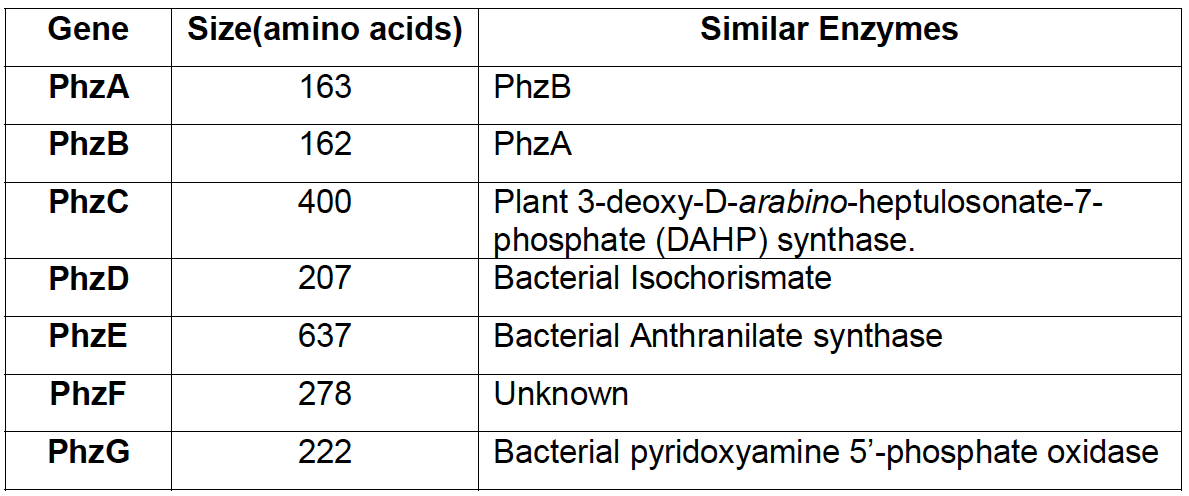
| + | [[File:IGEM Bielefeld 2013 phz genes table.png|thumb|left|400px|'''Figure 2''': Comparison of sequences between enzymes in phenazine cluster and other proteins with known function, according to research of [http://www.ncbi.nlm.nih.gov/pubmed/11562236 MacDonald et al. 2001.]]] | ||
| + | </p> | ||
<br><br> | <br><br> | ||
==Genetic Approach== | ==Genetic Approach== | ||
| - | [[File:IGEM Bielefeld 2013 Phenazines genes.png|thumb|center|500px|'''Figure | + | [[File:IGEM Bielefeld 2013 Phenazines genes.png|thumb|center|500px|'''Figure 3:''' Phenazine synthesis cluster, comprising an autoinducer synthase (''phzI'') gene, positive regulator protein (''phzR'') and pathway-coding [http://www.ncbi.nlm.nih.gov/nuccore/L48616.1 ''phzABCDEFG''] genes.]] |
| - | + | ||
<br> | <br> | ||
| + | |||
<p align="justify"> | <p align="justify"> | ||
| - | + | The coding cluster revealed seven forbidden restriction sites, making a cloning in accordance with the iGEM-rules a steep challenge. Unfortunately, amplification of the cluster was never successful. We varied different parameters and tried different enzymes, but we always saw a false priming and multiple amplificates. So we decided to look for applicable alternatives. Riboflavin, a natural mediator seemed an alternative of choice and our decision was rewarded by successful results. | |
| - | + | </p> | |
| - | + | ||
| - | We varied different parameters and tried different enzymes | + | |
| - | + | ||
| - | Riboflavin, a natural mediator | + | |
| - | + | ||
==Results== | ==Results== | ||
| - | + | <p align="justify"> | |
| - | + | There are no practical results in this section. The project was set aside after unsuccessful attempts to amplify the phenazine-coding fragment from ''Pseudomonas fluorescens'' sp. The only entry in the labjournal on phenazine is found [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/July here] in Week 13. | |
| + | </p> | ||
==References== | ==References== | ||
| - | * Mavrodi D.V, Tomashow L.S. (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. [http://www.ncbi.nlm.nih.gov/pubmed/16719720 | + | * Mavrodi D.V, Tomashow L.S. (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. [http://www.ncbi.nlm.nih.gov/pubmed/16719720 ''Annual review of Phytopathology 44:417– 445''] |
| - | * Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS (2001) Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. [http://www.ncbi.nlm.nih.gov/pubmed/11591691 | + | * Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS (2001) Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. [http://www.ncbi.nlm.nih.gov/pubmed/11591691 ''J Bacteriol 183:6454–6465''] |
| - | * Handelsman, J. and E. V. Stabb (1996) Biocontrol of soilborne plant pathogens. [http://www.plantcell.org/content/8/10/1855.long | + | * Handelsman, J. and E. V. Stabb (1996) Biocontrol of soilborne plant pathogens. [http://www.plantcell.org/content/8/10/1855.long ''Plant Cell 8(10): 1855-69.''] |
| - | * Vining L.C. (1990). Functions of secondary metabolites. [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.44.100190.002143 | + | * Vining L.C. (1990). Functions of secondary metabolites. [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.44.100190.002143 ''Annu Rev Microbiol: 44:395-427''] |
| - | * Muller M. Scavenging of neutrophil-derived superoxide anion by 1-hydroxyphenazine, a phenazine derivative associated with chronic Pseudomonas aeruginosa infection: relevance to cystic fibrosis. (1995) [http://www.ncbi.nlm.nih.gov/pubmed/8541351 | + | * Muller M. Scavenging of neutrophil-derived superoxide anion by 1-hydroxyphenazine, a phenazine derivative associated with chronic Pseudomonas aeruginosa infection: relevance to cystic fibrosis. (1995) [http://www.ncbi.nlm.nih.gov/pubmed/8541351 ''Biochim Biophys Acta: 1272(3):185-9.''] |
| - | *Pham T.H. (1998) Use of Pseudomonas species producing phenazine-based metabolites in the anodes of microbial fuel cells to improve electricity generation. [http://www.ncbi.nlm.nih.gov/pubmed/18688612 | + | *Pham T.H. (1998) Use of Pseudomonas species producing phenazine-based metabolites in the anodes of microbial fuel cells to improve electricity generation. [http://www.ncbi.nlm.nih.gov/pubmed/18688612 ''Appl Microbiol Biotechnology: 80(6):985-93.''] |
| - | * Hernandes M.E (2004) Phenazines and other redox-active antibiotics promote microbial mineral reduction [http://www.ncbi.nlm.nih.gov/pubmed/14766572 | + | * Hernandes M.E (2004) Phenazines and other redox-active antibiotics promote microbial mineral reduction [http://www.ncbi.nlm.nih.gov/pubmed/14766572 ''Appl Environ Microbiol.:70(2):921-8.''] |
| - | * McDonald M, Mavrodi DV, Thomashow LS, Floss HG. (2001). Phenazine biosynthesis in Pseudomonas fluorescens: branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. [http://www.ncbi.nlm.nih.gov/pubmed/11562236 | + | * McDonald M, Mavrodi DV, Thomashow LS, Floss HG. (2001). Phenazine biosynthesis in Pseudomonas fluorescens: branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. [http://www.ncbi.nlm.nih.gov/pubmed/11562236 ''J. Am. Chem. Soc. 123:9459–60''] |
| - | + | ||
| - | + | ||
| + | * Mavrodi D.V. (2010) Diversity and evolution of the phenazine biosynthesis pathway. [http://www.ncbi.nlm.nih.gov/pubmed/20008172 ''Appl Environ Microbiol.:866-79''.] | ||
| + | * Niknejad Kazempour M., Anvary M. (2009) CLONING OF PHENAZINE CARBOXYLIC ACID GENES OF ''FUSARIU MONILIFORME'' ANTAGONISTS BACTERIA IN ''ESHERICHIA COLI'' DH5α [http://www.agriculturaits.czu.cz/pdf_files/vol_42_4_pdf/kazempour.pdf ''AGRICULTURA TROPICA ET SUBTROPICA VOL. 42 (4) 157-161''] | ||
| Line 138: | Line 141: | ||
</div> | </div> | ||
| + | <div id="asdf"> | ||
| + | <html> | ||
| + | <div id="nav2" style="width:210px; padding-bottom:5px; padding-left:15px;"> | ||
| + | <div class="navbutton" id="home" style="float:left; padding-left:55px; margin-left:10px; padding-top:0px;"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany" title="Jump to Frontpage"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/f/f6/Bielefeld-Germany2013-ButtonHome.png" height="15px"> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="navbutton" id="top" style="float:left; padding-left:10px; margin-left:10px; padding-top:0px;"> | ||
| + | <a href="#" title="Jump to top"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/a/ab/Bielefeld-Germany2013-Up_orange_new.png" height="15px"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
| - | + | <div id="rightcol" style="width:210px; height:100%; overflow-y:auto; box-shadow:0px 0px 2px 0px grey;" padding:0px 20px;> | |
| - | <div id="rightcol" style="width:210px; height: | + | |
__TOC__ | __TOC__ | ||
| - | + | </div> | |
| + | |||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 22:34, 26 October 2013
Phenazine
Overview
Phenazines are water-soluble secondary metabolites secreted by some bacterial species. Almost all their properties can be attributed to their ability to undergo redox transformations. Some soil-born bacteria commonly produce phenazine-1-carboxylic acid ([http://www.ebi.ac.uk/chebi/chebiOntology.do?chebiId=CHEBI:62412 PCA])) against fungal root disease. (Niknejad Kazempour, [http://www.agriculturaits.czu.cz/pdf_files/vol_42_4_pdf/kazempour.pdf 2009]) Bacteria are also able to use this substance as a mediator for electron shuttling, which leads to an efficiency gain if a phenazine-producing strain is present in a MFC.
Problems arise with the different phenazine producing host strains. Most of them are classified as pathogens and therefore unsuitable for application in our lab and MFC.
Hence, all experiments carried out in this section were soluble done with Pseudomonas fluorescences which is classified as risk group 1.
Initially PCA overproduction seemed to be a promising approach to achieve higher electron transfers. Because of the extremely complicated cloning procedure as well as the good advice of the consulted MFC expert Dr. Falk Harnisch, we abandoned this sub-project in an early stage and searched for alternatives. We concentrated on an overexpression of riboflavin, which serves as an electrone shuttle itself and of the glycerol dehydrohenase gldA, which increases NADH concentration. (see sections Riboflavin and gldA on our Wiki for more information).
Still, quite a lot of work was invested into investigation of phenazine functions and synthesis in bacteria, of so we present a short theoretical overview of this topic.
The only entry in the labjournal on phenazine is found here in Week 13.
Theory
Phenazines comprise a large group of heterocyclic nitrogen-containing substances, having a tricyclic ring in its core. They are substituted at different points around their rings, which alters water solubility and other properties. After discovery of naturally occurring phenazines, more than 6000 were chemically synthesized. (Mavrodi, [http://www.ncbi.nlm.nih.gov/pubmed/16719720 2006]). [http://en.wikipedia.org/wiki/Neutral_red Neutral red], a well-known pH-indicator, also belongs to this class.
Phenazine derivatives were one of the first secondary bacterial metabolites that were consequently studied since their discovery in 1859. The first substance of this family, a blue pigment of Pseudomonas aeruginosa, was called [http://en.wikipedia.org/wiki/Pyocyanine «pyocyanine»] (short PYO). It has antimicrobial properties that raised further interest in these substances.
Phenazines are produced by many bacterial species, but they are mostly studied in representatives of fluorescent pseudomonads. A single bacterial strain usually produces two or more species-specific phenazines, except Pseudomonas fluorescens 2-79 that is known to produce only PCA, a broad-spectrum antibiotic, which is a casual agent active against a variety of root pathogens (Mavrodi et al., [http://www.ncbi.nlm.nih.gov/pubmed/11591691 2001]).
Biological phenazines are known for their broad-spectrum of antibiotical, antiparasaticand even anti-malaria properties (Handelsmann et al., [http://www.plantcell.org/content/8/10/1855.long 1996]).
Consequently, it was shown that almost all properties of phenazines could be attributed to their ability to undergo redox transformations (Muller, [http://www.ncbi.nlm.nih.gov/pubmed/8541351 1995]). This discovery brought up the idea, that these molecules can be used as an electron shuttle in a MFC. Indeed, it was shown that external addition of phenazines, both PCA and PCN ([http://www.ebi.ac.uk/chebi/searchId.do?chebiId=62240 phenazine-1-carboxamide]) alike, causes a significant raise of current flow (The Hai Pham, [http://www.ncbi.nlm.nih.gov/pubmed/18688612 2008]). Surprisingly, a mixed culture of phenazine-secreting and phenazine non-secreting strains showed similar results, proving that phenazine can be used for redox cycling not only by its producer (Hernandez et al. [http://www.ncbi.nlm.nih.gov/pubmed/14766572 2004]).
Many strains of Pseudomonas fluorescens, common soil inhabitants, are known to produce the phenazine compound PCA. The phenazine synthesis pathway was studied in detail. The seven-gene cluster [http://www.ncbi.nlm.nih.gov/nuccore/L48616.1 phzABCDEFG], 8505 bp long, was firstly identified in Pseudomonas fluorescens 2-79.
Research on phenazine diversity and genetic evolution, (Mavrodi, [http://www.ncbi.nlm.nih.gov/pubmed/20008172 2010]) stated, that many species of fluorescent pseudomonas have the corresponding genes and are able to produce phenazines, so we expected that orthologs are found in the Pseudomonas fluorescens 50090 that we obtained from [http://www.dsmz.de DSMZ].
Genetic Approach
The coding cluster revealed seven forbidden restriction sites, making a cloning in accordance with the iGEM-rules a steep challenge. Unfortunately, amplification of the cluster was never successful. We varied different parameters and tried different enzymes, but we always saw a false priming and multiple amplificates. So we decided to look for applicable alternatives. Riboflavin, a natural mediator seemed an alternative of choice and our decision was rewarded by successful results.
Results
There are no practical results in this section. The project was set aside after unsuccessful attempts to amplify the phenazine-coding fragment from Pseudomonas fluorescens sp. The only entry in the labjournal on phenazine is found here in Week 13.
References
- Mavrodi D.V, Tomashow L.S. (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. [http://www.ncbi.nlm.nih.gov/pubmed/16719720 Annual review of Phytopathology 44:417– 445]
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS (2001) Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. [http://www.ncbi.nlm.nih.gov/pubmed/11591691 J Bacteriol 183:6454–6465]
- Handelsman, J. and E. V. Stabb (1996) Biocontrol of soilborne plant pathogens. [http://www.plantcell.org/content/8/10/1855.long Plant Cell 8(10): 1855-69.]
- Vining L.C. (1990). Functions of secondary metabolites. [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.44.100190.002143 Annu Rev Microbiol: 44:395-427]
- Muller M. Scavenging of neutrophil-derived superoxide anion by 1-hydroxyphenazine, a phenazine derivative associated with chronic Pseudomonas aeruginosa infection: relevance to cystic fibrosis. (1995) [http://www.ncbi.nlm.nih.gov/pubmed/8541351 Biochim Biophys Acta: 1272(3):185-9.]
- Pham T.H. (1998) Use of Pseudomonas species producing phenazine-based metabolites in the anodes of microbial fuel cells to improve electricity generation. [http://www.ncbi.nlm.nih.gov/pubmed/18688612 Appl Microbiol Biotechnology: 80(6):985-93.]
- Hernandes M.E (2004) Phenazines and other redox-active antibiotics promote microbial mineral reduction [http://www.ncbi.nlm.nih.gov/pubmed/14766572 Appl Environ Microbiol.:70(2):921-8.]
- McDonald M, Mavrodi DV, Thomashow LS, Floss HG. (2001). Phenazine biosynthesis in Pseudomonas fluorescens: branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. [http://www.ncbi.nlm.nih.gov/pubmed/11562236 J. Am. Chem. Soc. 123:9459–60]
- Mavrodi D.V. (2010) Diversity and evolution of the phenazine biosynthesis pathway. [http://www.ncbi.nlm.nih.gov/pubmed/20008172 Appl Environ Microbiol.:866-79.]
- Niknejad Kazempour M., Anvary M. (2009) CLONING OF PHENAZINE CARBOXYLIC ACID GENES OF FUSARIU MONILIFORME ANTAGONISTS BACTERIA IN ESHERICHIA COLI DH5α [http://www.agriculturaits.czu.cz/pdf_files/vol_42_4_pdf/kazempour.pdf AGRICULTURA TROPICA ET SUBTROPICA VOL. 42 (4) 157-161]
 "
"