Team:SJTU-BioX-Shanghai/Project/Regulator/Integrating
From 2013.igem.org
(Difference between revisions)
(→Gathering Quantitative Data -- sgRNA targeting luciferase) |
|||
| (3 intermediate revisions not shown) | |||
| Line 58: | Line 58: | ||
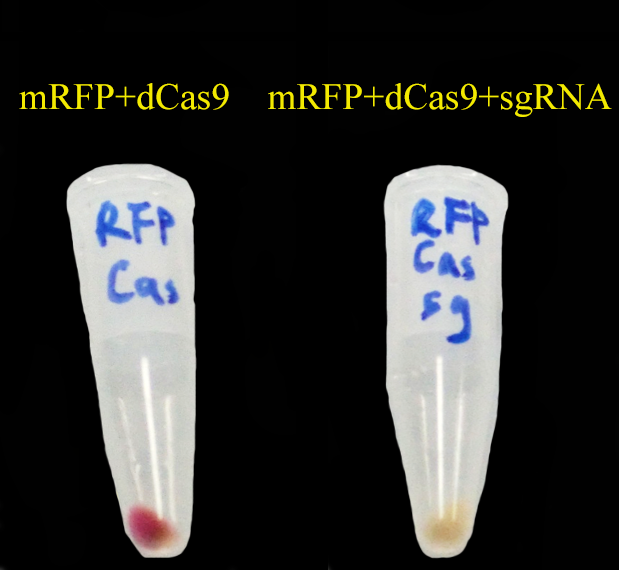
As expected, '''mRFP repressed by CRISPRi (dCas9 and gR4mRFP) is not as red as the control.''' | As expected, '''mRFP repressed by CRISPRi (dCas9 and gR4mRFP) is not as red as the control.''' | ||
<br><br> | <br><br> | ||
| - | The sequence of gR4mRPF is given in our part BBa_K1026003 . This sequence comes from the following literature: QI, LEI S., LARSON, MATTHEW H., GILBERT, LUKE A., DOUDNA, JENNIFER A., WEISSMAN, JONATHAN S., ARKIN, ADAM P. & LIM, WENDELL A. 2013. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell, 152, 1173-1183. | + | The sequence of gR4mRPF is given in our part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1026003 BBa_K1026003]. This sequence comes from the following literature: QI, LEI S., LARSON, MATTHEW H., GILBERT, LUKE A., DOUDNA, JENNIFER A., WEISSMAN, JONATHAN S., ARKIN, ADAM P. & LIM, WENDELL A. 2013. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell, 152, 1173-1183. |
<br><br> | <br><br> | ||
And <font size=4>'''CAUTION'''</font> that: | And <font size=4>'''CAUTION'''</font> that: | ||
* There shall '''not''' be extra nucleotides between promoter and sgRNA itself. | * There shall '''not''' be extra nucleotides between promoter and sgRNA itself. | ||
gRNA shall be exactly transcribed out. Any extra nucleotides would potentially jeopardize the normal function of the gRNA. Therefore, '''BioBrick scars are not allowed here'''. Instead of cut-and-ligation, inverse PCR or In-Fusion is preferred for the assembly task. | gRNA shall be exactly transcribed out. Any extra nucleotides would potentially jeopardize the normal function of the gRNA. Therefore, '''BioBrick scars are not allowed here'''. Instead of cut-and-ligation, inverse PCR or In-Fusion is preferred for the assembly task. | ||
| - | Our part '''BBa_K1026003''' can be an example of no-scar assembly. | + | Our part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1026003 '''BBa_K1026003'''] can be an example of no-scar assembly. |
* A reliable terminator is applied here to '''ensure the termination of sgRNA'''. | * A reliable terminator is applied here to '''ensure the termination of sgRNA'''. | ||
<br> | <br> | ||
| Line 93: | Line 93: | ||
<br> | <br> | ||
Since we do not know the sequence of a working sgRNA that targets luciferase, so we designed two gR4mRFP that complements the coding sequence of luciferase gene ourselves according to those design criteria: one is '''closer to the start codon (the proximal one)''', whereas the other is relatively further (the '''distal one'''). | Since we do not know the sequence of a working sgRNA that targets luciferase, so we designed two gR4mRFP that complements the coding sequence of luciferase gene ourselves according to those design criteria: one is '''closer to the start codon (the proximal one)''', whereas the other is relatively further (the '''distal one'''). | ||
| - | However it is necessary for us to test these sgRNA. So we again inserted these two sgRNAs into a constitutive operon. | + | However it is necessary for us to test these sgRNA. So we again inserted these two sgRNAs into a constitutive operon. We choose the proximal one for later use. |
| - | + | ||
<br><br> | <br><br> | ||
<font size=4>Determining the '''Working Curve''' of our Metabolic Gear Box</font size=4> | <font size=4>Determining the '''Working Curve''' of our Metabolic Gear Box</font size=4> | ||
| Line 104: | Line 103: | ||
| - | {{Template: | + | {{Template:13SJTU_footer}} |
Latest revision as of 18:10, 17 October 2013
 "
"