Team:SJTU-BioX-Shanghai/Prospect
From 2013.igem.org
(→CRISPRi-on) |
(→Absolutely Automatic) |
||
| Line 56: | Line 56: | ||
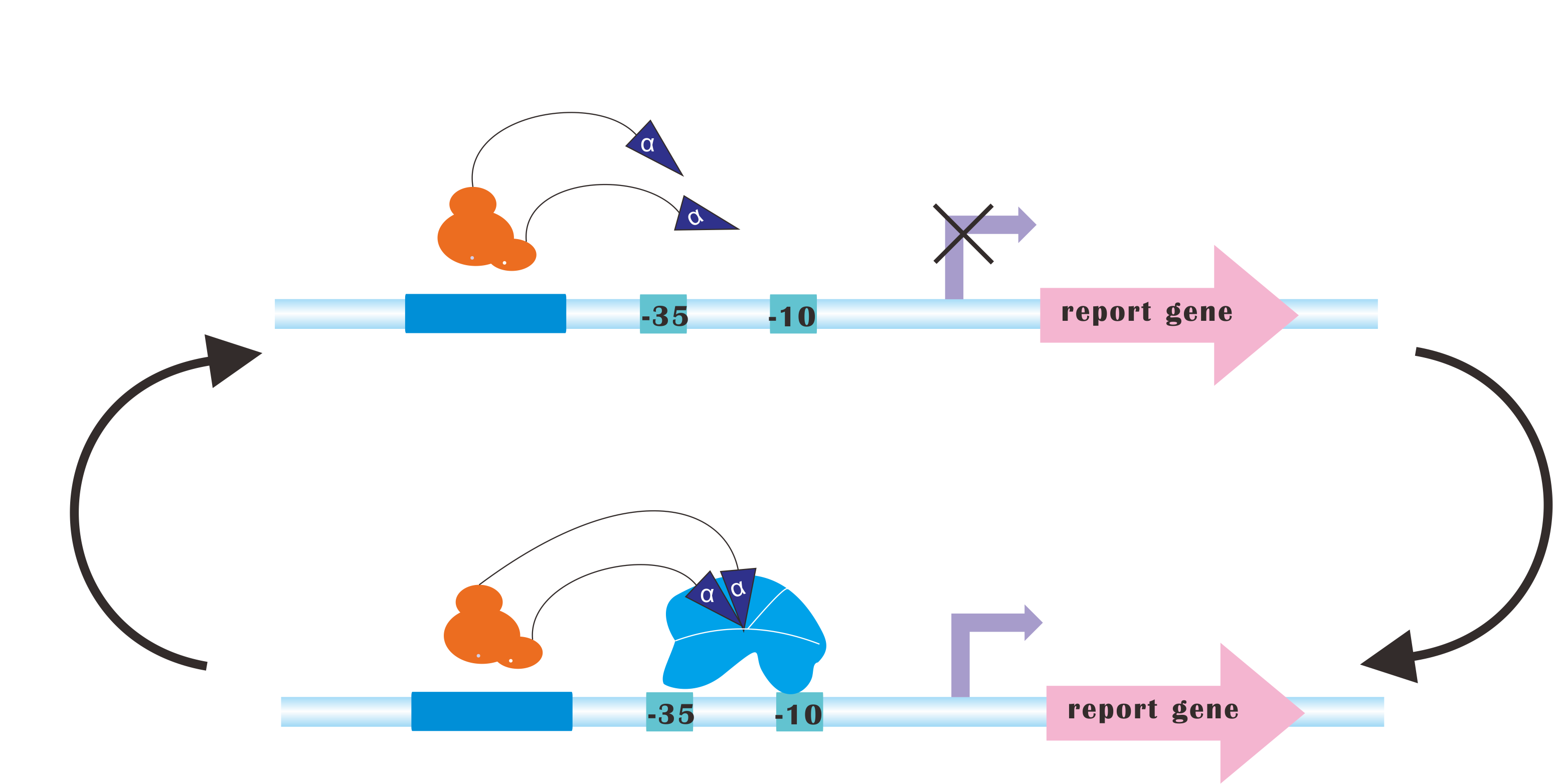
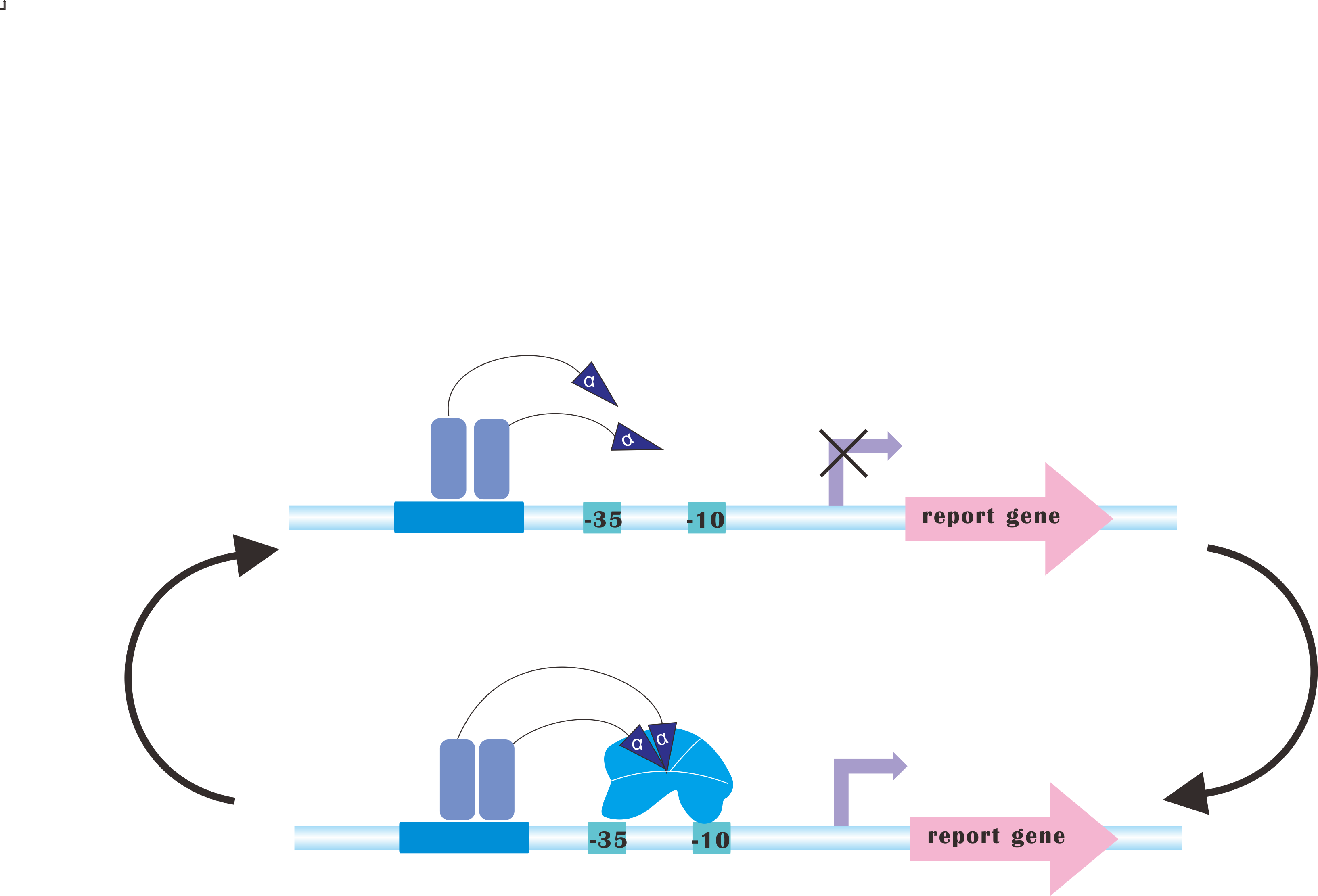
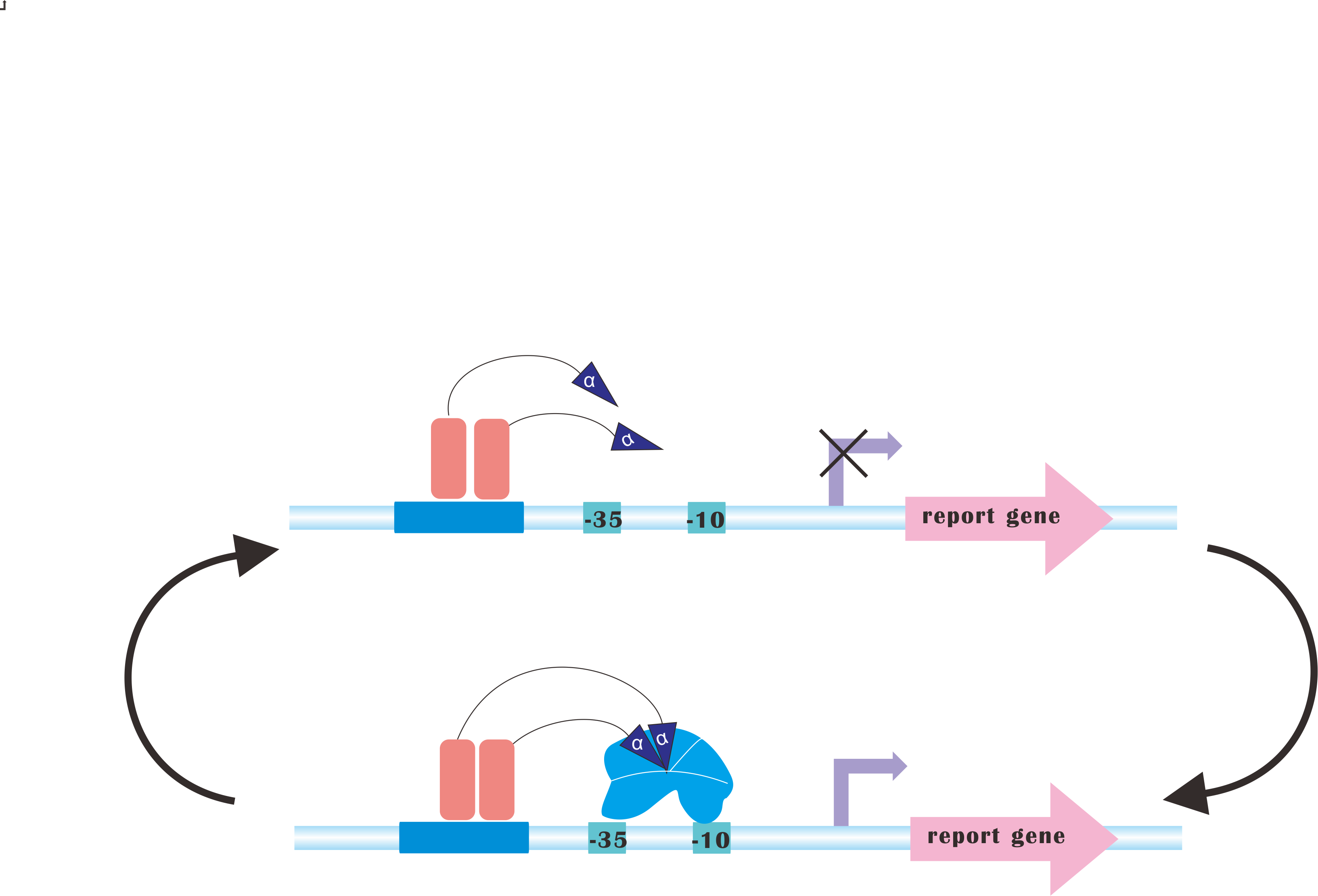
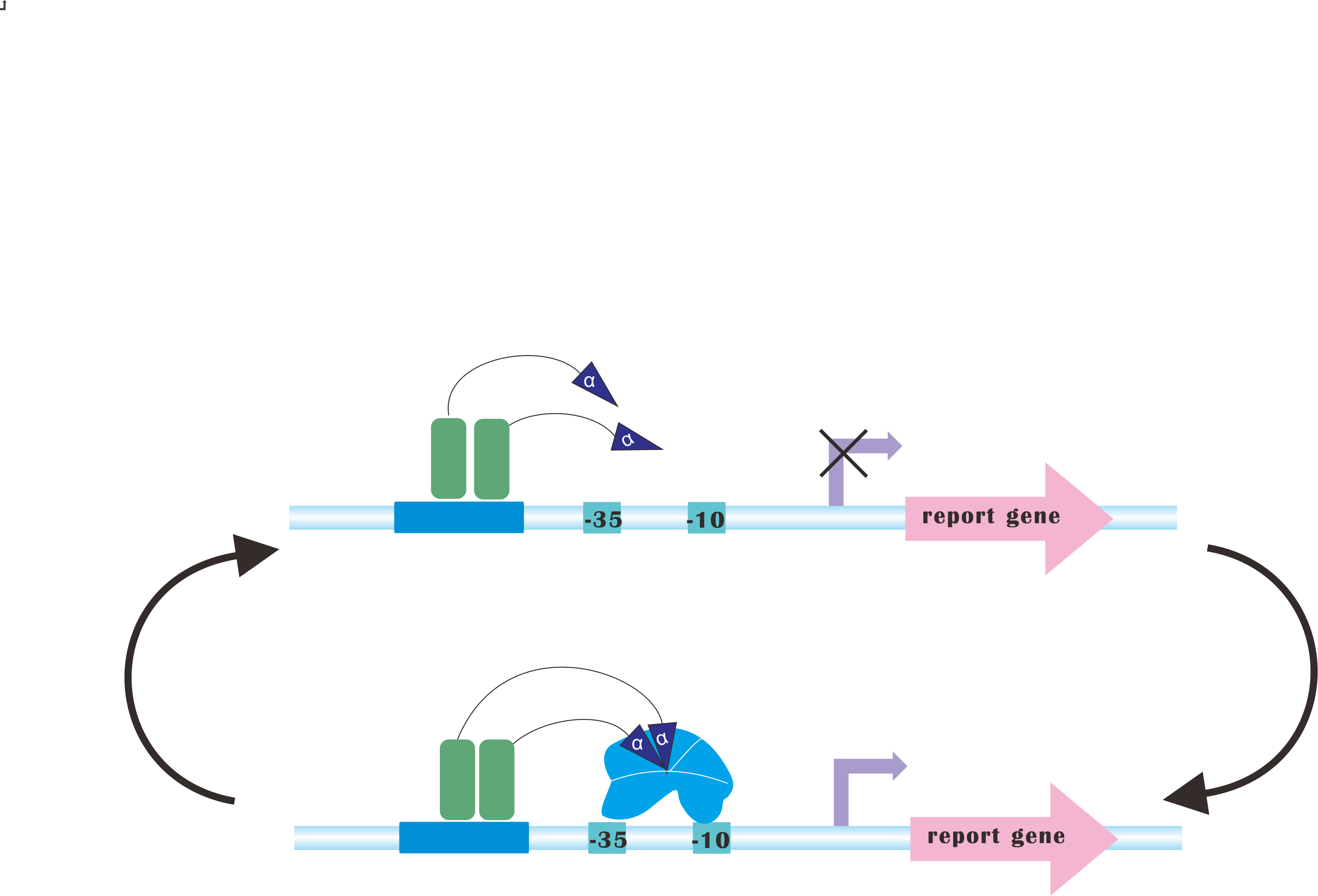
At present, our platform only analyzes metabolic fluxes (namely, SBML files), conduct flux balance analysis, calculating optimal expression levels of those genes and send commands to the luminous device. But after the regulation process, you may still need to re-adjust those parameters according to the producing rate of desired product(s). | At present, our platform only analyzes metabolic fluxes (namely, SBML files), conduct flux balance analysis, calculating optimal expression levels of those genes and send commands to the luminous device. But after the regulation process, you may still need to re-adjust those parameters according to the producing rate of desired product(s). | ||
But how about we add a feedback path! And the system automatically iterate to sample the FBA space, finding an optimal, or at least a quasi-optimal for you. It is hard, but we would get a try. :) | But how about we add a feedback path! And the system automatically iterate to sample the FBA space, finding an optimal, or at least a quasi-optimal for you. It is hard, but we would get a try. :) | ||
| + | <br><br> | ||
| + | |||
| + | <html> | ||
| + | <h1 style="color:grey;">References</h1> | ||
| + | <p style="color:grey;"> | ||
| + | <br> | ||
| + | CAMSUND, D., LINDBLAD, P. & JARAMILLO, A. 2011. Genetically engineered light sensors for control of bacterial gene expression. Biotechnol J, 6, 826-36. | ||
| + | <br/><br/> | ||
| + | CHENG, A. W., WANG, H., YANG, H., SHI, L., KATZ, Y., THEUNISSEN, T. W., RANGARAJAN, S., SHIVALILA, C. S., DADON, D. B. & JAENISCH, R. 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Research, 23, 1163-1171. | ||
| + | </p></html> | ||
Latest revision as of 05:21, 18 October 2013
|
| ||
|
 "
"