Team:ETH Zurich/pre proc
From 2013.igem.org
| (21 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
<b>Natural Lux signalling system</b> | <b>Natural Lux signalling system</b> | ||
<br> | <br> | ||
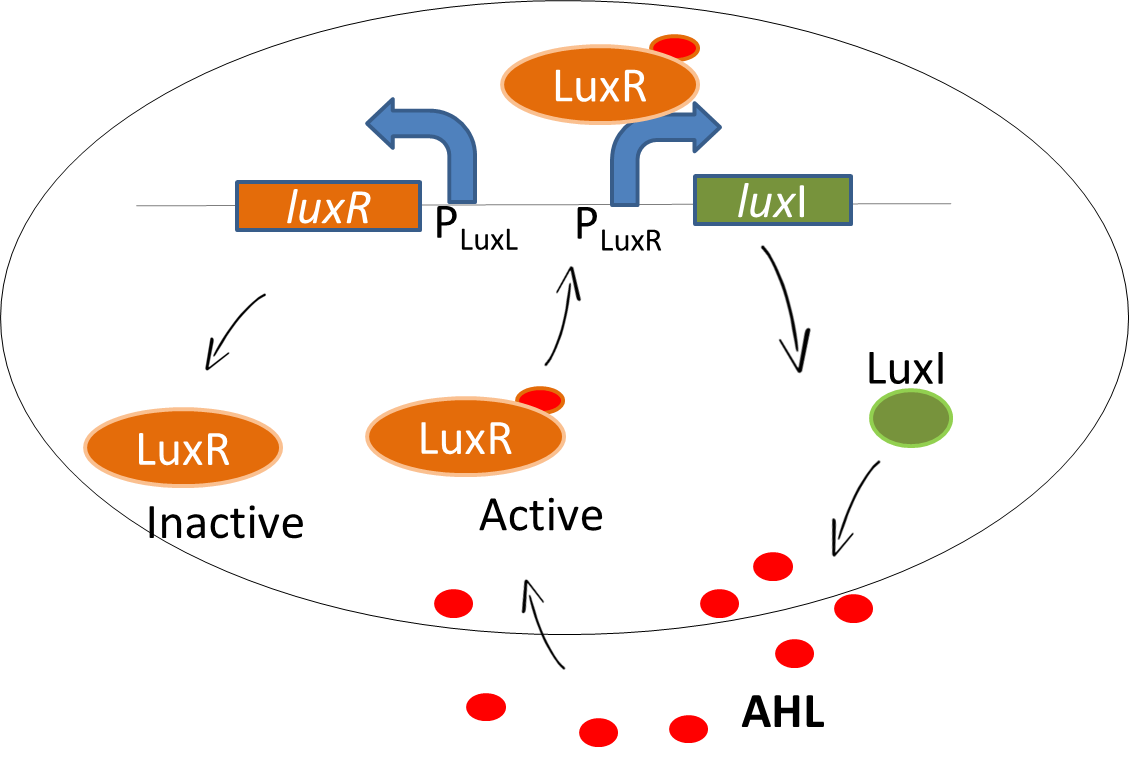
| - | <p align="justify">As our system is based on a sender-receiver module, we use the lux signalling system in our bio-game. In <i> | + | <p align="justify">As our system is based on a sender-receiver module, we use the well-known quorum sensing mechanism of the lux signalling system in our bio-game. In <i>Vibrio fischeri</i>, the <i>luxI</i> gene encodes for the enzyme that catalyses the production of the signaling molecule N-acyl-homoserine lactone, <b>AHL</b>. The AHL molecule is capable of <b>diffusion</b> in and out of cells. Meanwhile, the LuxR protein is produced which is driven by promoter P<sub>luxL</sub>. When unbound, this protein is in the inactive state. The binding of AHL to LuxR protein converts inactive LuxR to an <b>active LuxR-AHL complex</b>. The complex binds to the P<sub>LuxR</sub> promoter and drives the gene expression upstream of the operon [http://www.fasebj.org/content/7/11/1016 (1)] that leads to more production of AHL. Thereby, the available LuxR is bound with AHL which downregulates the P<sub>LuxL</sub> promoter.</p> |
| - | + | ||
<br> | <br> | ||
| Line 15: | Line 14: | ||
| - | [[File: | + | [[File:I love iGEM 13.png|700px|center|Thumb|<b>Figure 2. Biological circuit of mine and non-mine in Colisweeper </b>|]] |
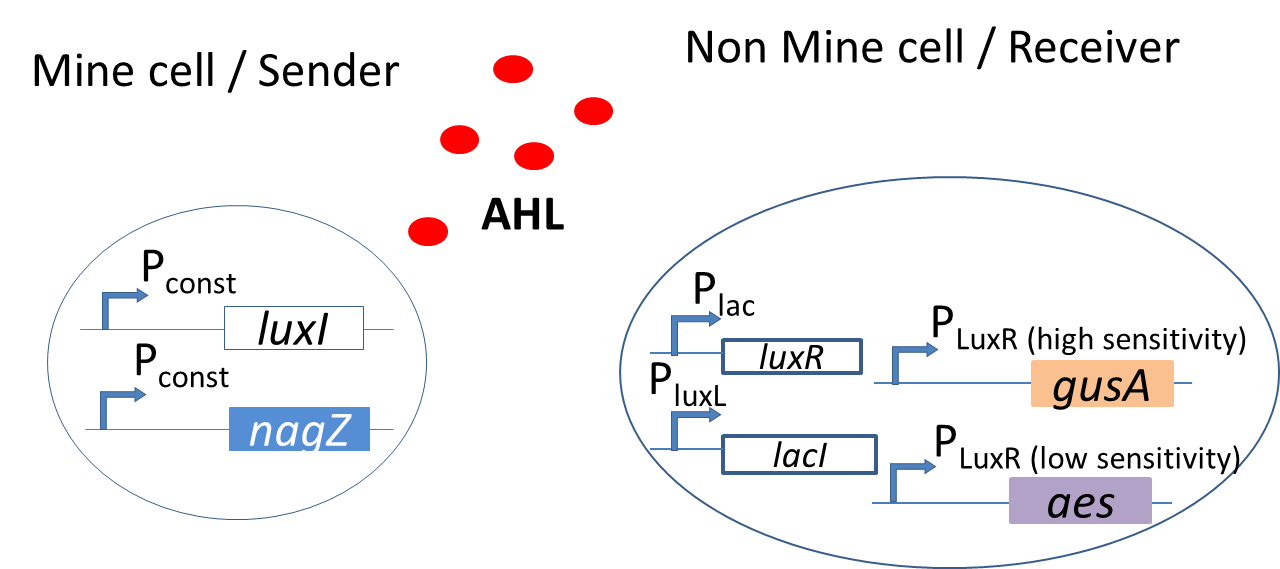
| - | <p align="justify">In the bio-game, the mines are the sender cells and the non-mines are the receiver cells. The mines constitutively express LuxI under [http://parts.igem.org/Part:BBa_J23110 promoter (Part BBa_J23110)] and hence [https://2013.igem.org/Team:ETH_Zurich/Experiments_2 AHL]. This molecule then diffuses through the agar and can be processed by the non-mines. | + | <p align="justify">In the bio-game, the mines are the sender cells and the non-mines are the receiver cells. The mines constitutively express LuxI under [http://parts.igem.org/Part:BBa_J23110 promoter (Part BBa_J23110)] and hence produce [https://2013.igem.org/Team:ETH_Zurich/Experiments_2 AHL]. This molecule then diffuses through the agar and can be processed by the non-mines. |
| - | The non-mines produce | + | The non-mines produce LuxR under [http://parts.igem.org/Part:BBa_J09855 promoter (Part BBa_J09855)] and so the incoming AHL will bind to the LuxR protein to form an active complex AHL-LuxR. The non-mines also contain [https://2013.igem.org/Team:ETH_Zurich/Processing_2 P<sub>LuxR</sub> promoters] that are tuned to different AHL sensitivities. The AHL-LuxR complex will bind to the promoters and drive the expression of the reporter [https://2013.igem.org/Team:ETH_Zurich/Experiments_7 hydrolases.] |
Depending on the amount of AHL that is processed, different expression of output colors give the player the cue for the next move in the bio-game. </p> | Depending on the amount of AHL that is processed, different expression of output colors give the player the cue for the next move in the bio-game. </p> | ||
<br> | <br> | ||
| - | <h1>Diffusion of | + | <h1>Diffusion of signalling molecule AHL from mine to non-mine</h1> |
<br> | <br> | ||
| - | <p align="justify">In order to study the communication between the mines and non-mines via the signalling molecule AHL, diffusion tests were performed. It was vital to know the exact diffusion pattern of AHL from the mine to have an idea of the placement of the colonies on the agar-mine field. By studying the <b> diffusion time and distance</b> of AHL around the sender, we were able to confirm the <b>honey-comb pattern</b> as the design of the mine grid. With this design, colonies are placed on the edges of each hexagon in the honey-comb pattern. See results of these experiments [https://2013.igem.org/Team:ETH_Zurich/Experiments_2 here] </p> | + | <p align="justify">In order to study the communication between the mines and non-mines via the signalling molecule AHL, diffusion tests were performed. It was vital to know the exact diffusion pattern of AHL from the mine to have an idea of the placement of the colonies on the agar-mine field. By studying the <b> diffusion time and distance</b> of AHL around the sender, we were able to confirm the <b>honey-comb pattern</b> as the design of the mine grid. With this design, colonies are placed on the edges of each hexagon in the honey-comb pattern. See results of these experiments [https://2013.igem.org/Team:ETH_Zurich/Experiments_2 here]. </p> |
| + | <h1>References</h1> | ||
| + | 1. Meighen, E.A.; FASEB J. 7: 1016-1022 (Aug 1993) | ||
Latest revision as of 01:59, 29 October 2013
Contents |
LuxR signaling system
Natural Lux signalling system
As our system is based on a sender-receiver module, we use the well-known quorum sensing mechanism of the lux signalling system in our bio-game. In Vibrio fischeri, the luxI gene encodes for the enzyme that catalyses the production of the signaling molecule N-acyl-homoserine lactone, AHL. The AHL molecule is capable of diffusion in and out of cells. Meanwhile, the LuxR protein is produced which is driven by promoter PluxL. When unbound, this protein is in the inactive state. The binding of AHL to LuxR protein converts inactive LuxR to an active LuxR-AHL complex. The complex binds to the PLuxR promoter and drives the gene expression upstream of the operon [http://www.fasebj.org/content/7/11/1016 (1)] that leads to more production of AHL. Thereby, the available LuxR is bound with AHL which downregulates the PLuxL promoter.
Exploiting the lux signalling system in Colisweeper
In the bio-game, the mines are the sender cells and the non-mines are the receiver cells. The mines constitutively express LuxI under [http://parts.igem.org/Part:BBa_J23110 promoter (Part BBa_J23110)] and hence produce AHL. This molecule then diffuses through the agar and can be processed by the non-mines. The non-mines produce LuxR under [http://parts.igem.org/Part:BBa_J09855 promoter (Part BBa_J09855)] and so the incoming AHL will bind to the LuxR protein to form an active complex AHL-LuxR. The non-mines also contain PLuxR promoters that are tuned to different AHL sensitivities. The AHL-LuxR complex will bind to the promoters and drive the expression of the reporter hydrolases. Depending on the amount of AHL that is processed, different expression of output colors give the player the cue for the next move in the bio-game.
Diffusion of signalling molecule AHL from mine to non-mine
In order to study the communication between the mines and non-mines via the signalling molecule AHL, diffusion tests were performed. It was vital to know the exact diffusion pattern of AHL from the mine to have an idea of the placement of the colonies on the agar-mine field. By studying the diffusion time and distance of AHL around the sender, we were able to confirm the honey-comb pattern as the design of the mine grid. With this design, colonies are placed on the edges of each hexagon in the honey-comb pattern. See results of these experiments here.
References
1. Meighen, E.A.; FASEB J. 7: 1016-1022 (Aug 1993)
 "
"