Team:Colombia Uniandes/Parts
From 2013.igem.org
Trafalmejo (Talk | contribs) |
(→Glucocorticoid sensor) |
||
| (54 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Http://2013.igem.org/Team:Colombia Uniandes/Bootstrap}} | {{Http://2013.igem.org/Team:Colombia Uniandes/Bootstrap}} | ||
| + | <html> | ||
| + | <div class="container"> | ||
| + | <div class="span11"> | ||
| - | + | <h2><center>Parts</center></h2> | |
| - | + | <div class="accordion" id="accordion2"> | |
| - | + | <div class="accordion-group"> | |
| - | + | <div class="accordion-heading"> | |
| - | + | <a class="accordion-toggle" data-toggle="collapse" data-parent="#accordion2" href="#collapseOne"> | |
| - | + | Glucocorticoid sensor | |
| - | + | </a> | |
| - | + | </div> | |
| - | + | <div id="collapseOne" class="accordion-body collapse"> | |
| - | + | <div class="accordion-inner"> | |
| - | + | </html> | |
| - | + | ==Glucocorticoid sensor== | |
| - | + | ||
| - | + | Here are the parts sent to the registry: | |
| - | + | ||
| - | + | [[File:partsGC.jpg|700px|center|]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
{| border="1" align="center" | {| border="1" align="center" | ||
| - | |+''' | + | |+'''Name table''' |
| - | | style="text-align: center;" | | + | | style="text-align: center;" |Part Name |
| - | | style=" | + | | style="text-align: center;" |Registry Number |
|- | |- | ||
| - | | style="text-align: center;" | | + | | style="text-align: center;" |E1 |
| - | | style=" | + | | style="text-align: center;" |BBa_K1144001 |
|- | |- | ||
| - | | style="text-align: center;" | | + | | style="text-align: center;" |E2 |
| - | | style=" | + | | style="text-align: center;" |BBa_K1144002 |
| - | |- | + | |- |
| - | | style="text-align: center;" | | + | | style="text-align: center;" |E3 |
| - | | style=" | + | | style="text-align: center;" |BBa_K1144003 |
| + | |- | ||
| + | | style="text-align: center;" |E4 | ||
| + | | style="text-align: center;" |BBa_K1144004 | ||
| + | |- | ||
| + | | style="text-align: center;" |E1GF | ||
| + | | style="text-align: center;" |BBa_K1144005 | ||
| + | |- | ||
| + | | style="text-align: center;" |E2GF | ||
| + | | style="text-align: center;" |BBa_K1144006 | ||
| + | |- | ||
| + | | style="text-align: center;" |E3GF | ||
| + | | style="text-align: center;" |BBa_K1144007 | ||
| + | |- | ||
| + | | style="text-align: center;" |E4GF | ||
| + | | style="text-align: center;" |BBa_K1144008 | ||
| + | |||
|- | |- | ||
| - | |||
| - | |||
|} | |} | ||
| + | You can see the confirmation gels of the parts below: | ||
| - | |||
| - | + | [[File:Gel1GC.jpg|410px|thumb|left|''' PCR confirmation: E1GF-E1GF(2)-E2GF-E2GF(2)-WM-E3GF-E3GF(2)-E4GF-E4GF(2)''']] [[File:Gel2GC.jpg|400px|thumb|center|'''Co-transformation: WM-E1GF2-E2GF-E3GF1-E3GF2-E4GF-WM- E3GF3-E4GF2-E2GF2 | |
| - | | | + | ''']] |
| - | + | [[File:GEL3.jpg|400px|thumb|center|'''PCR confirmation of the parts cloned in the pSB1C3 backbone using the specifically designed primers 15 and 31 (see primers)''']] | |
| - | | | + | |
| - | + | ||
| - | + | ==='''Characterization of reporters'''=== | |
| - | | | + | |
| - | + | To assess the strength of our promoters when induced with Dexamethasone, we performed a fluorometric assay using mCherry as our reporter.Since we don't have our transactivating protein ready yet, we co-transformed our parts into the ''E. coli'' DH10B strain with the pAT7002 vector (Aoyama and Chua, 1997), which contains a well characterized Glucocorticoid Responsive Element that also uses the GAL4 DNA binding domain. | |
| - | + | ||
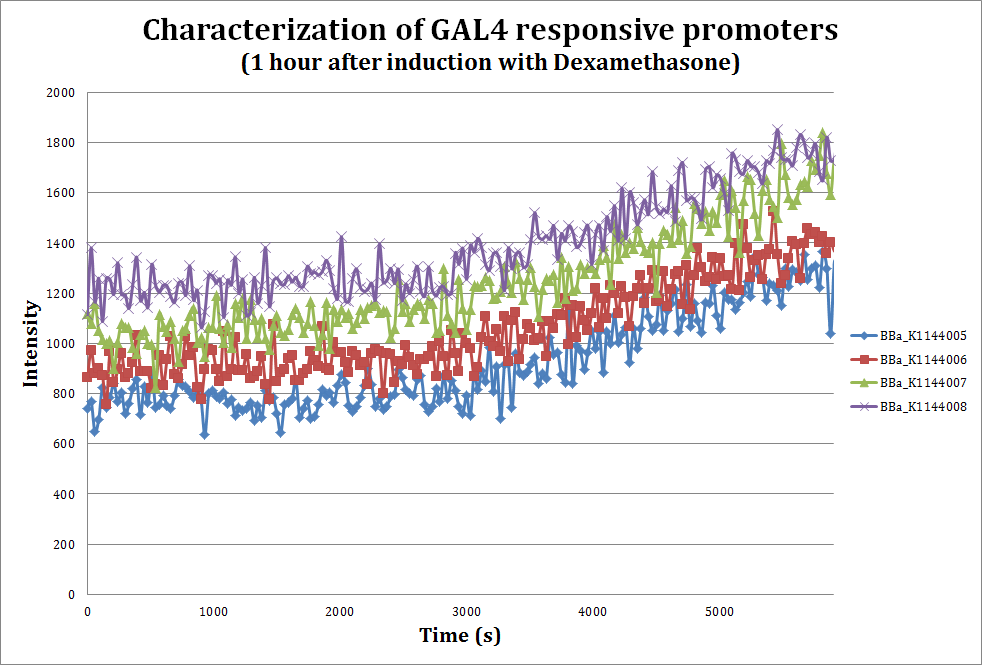
| - | + | [[File:Response1hr.jpg|700px|thumb|center|''' Unsaturated curve where we see how the mCherry reporter appears after the addition of 10 uM dexamethasone, a glucocorticoid. Emmision intensity was measured at 607nm after excitation at 586nm''']] | |
| - | |- | + | |
| - | + | ||
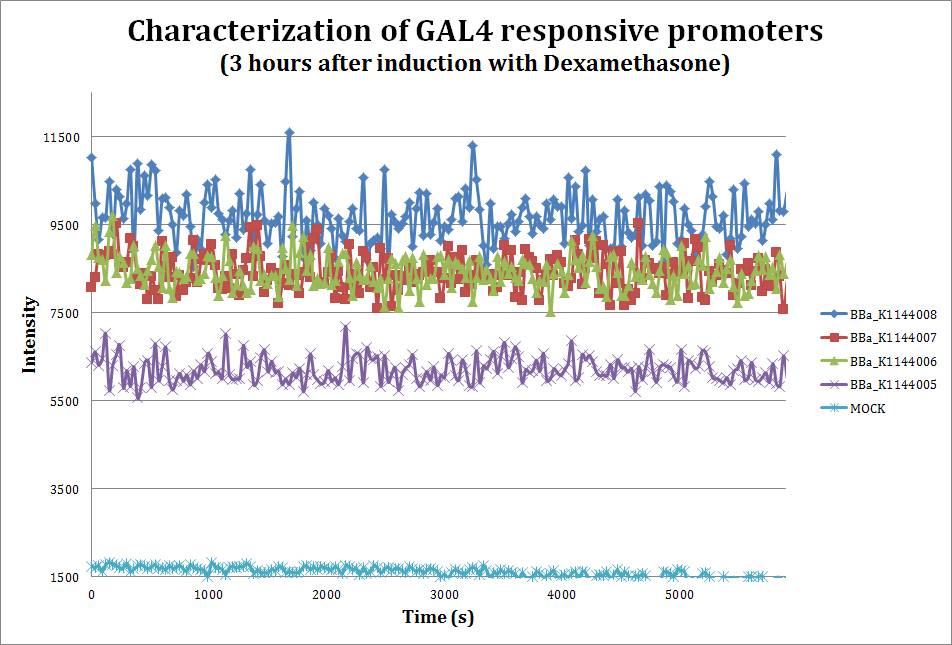
| - | + | [[File:Response3hr.jpg|700px|thumb|center|''' Saturated curve for fluorescence. After three hours of the addition of 10 uM dexamethasone the reporter (mCherry) reaches its highest signal. We can see the different strengths depending on the number of UAS boxes. Emmision intensity was measured at 607nm after excitation at 586nm''']] | |
| - | + | ||
| - | + | ||
| - | + | We also decided to visually inspect our induced transformants. Here two of the images taken using a epifluorescent microscopy with a TRITC filter. | |
| - | |- | + | |
| - | + | ||
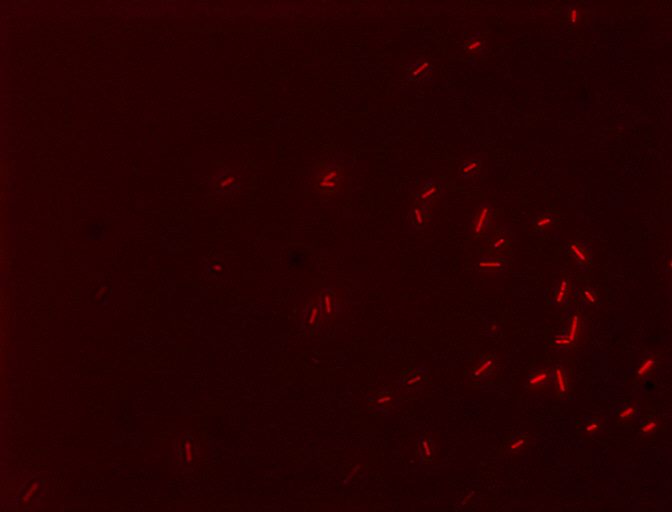
| + | [[File:E2GF.jpg|370px|thumb|left|''' Cells with the E2GF part (BBa_K1144006) showing their fluorescent reporter, mCherry, after induction with 10 uM dexamethasone! The filter used was TRITC''']] | ||
| + | |||
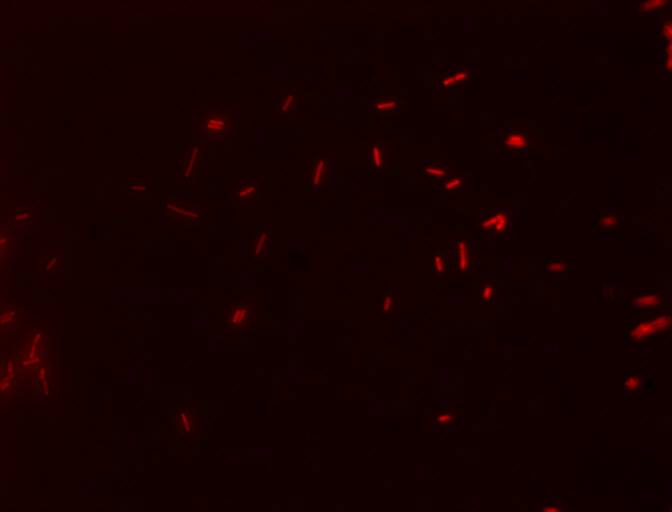
| + | [[File:E4GF.jpg|370px|thumb|right|''' Cells with the E4GF part (BBa_K1144008) showing their fluorescent reporter, mCherry, after induction with 10 uM dexamethasone! The filter used was TRITC''']] | ||
| + | |||
| + | |||
| + | ====References==== | ||
| + | Aoyama, T. & Chua N. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. ''The Plant Journal, 11''(3): 605-612. | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="accordion-group"> | ||
| + | <div class="accordion-heading"> | ||
| + | <a class="accordion-toggle" data-toggle="collapse" data-parent="#accordion2" href="#collapseTwo"> | ||
| + | Nickel Removal System | ||
| + | </a> | ||
| + | </div> | ||
| + | <div id="collapseTwo" class="accordion-body collapse"> | ||
| + | <div class="accordion-inner"> | ||
| + | </html> | ||
| + | |||
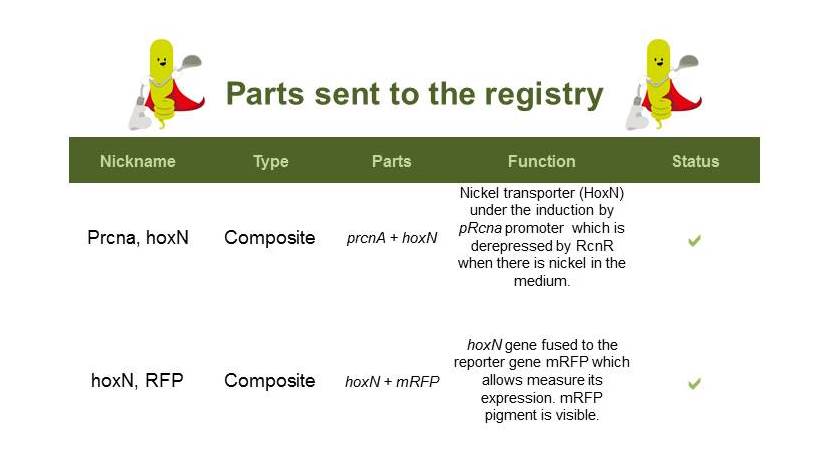
| + | ==Nickel Removal System== | ||
| + | |||
| + | Here are the parts sent to the registry: | ||
| + | |||
| + | [[File:partsNi.jpg|700px|center|]] | ||
| + | |||
| + | [[File:Prcna_hoxN.jpg|400px|thumb|center|Prcna-HoxN and HoxN-RFP]] | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| - | + | </html> | |
Latest revision as of 03:40, 28 September 2013
Parts
Contents |
Glucocorticoid sensor
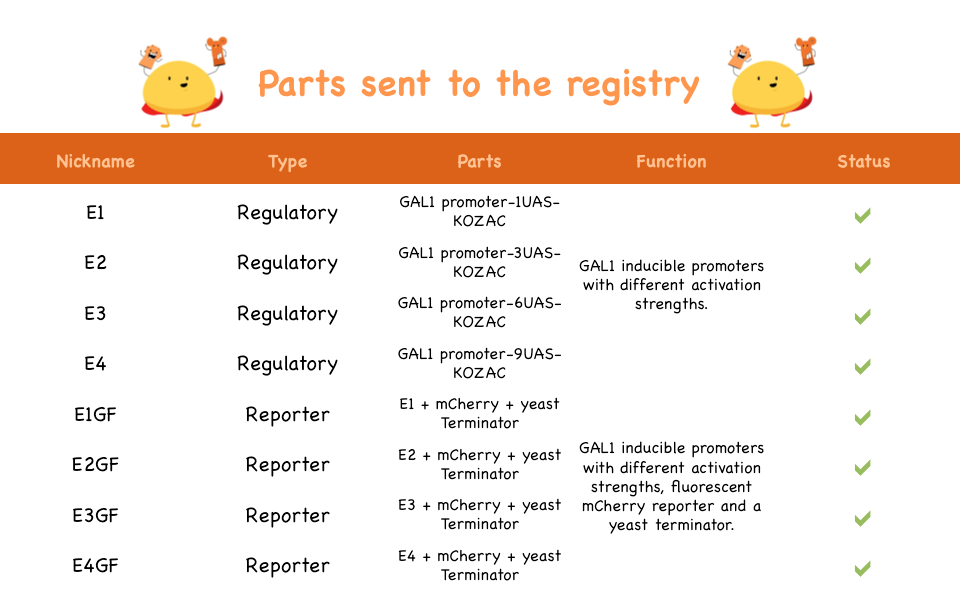
Here are the parts sent to the registry:
| Part Name | Registry Number |
| E1 | BBa_K1144001 |
| E2 | BBa_K1144002 |
| E3 | BBa_K1144003 |
| E4 | BBa_K1144004 |
| E1GF | BBa_K1144005 |
| E2GF | BBa_K1144006 |
| E3GF | BBa_K1144007 |
| E4GF | BBa_K1144008 |
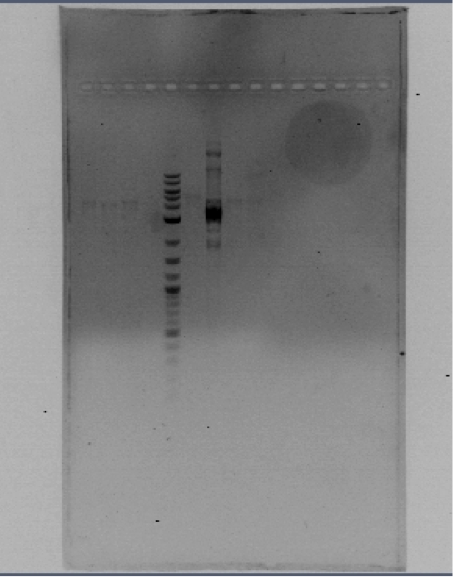
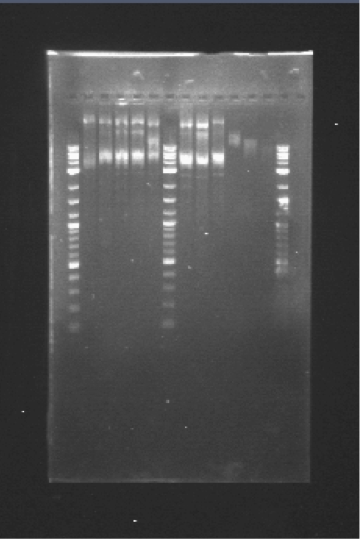
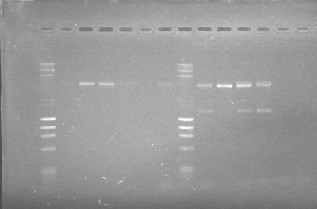
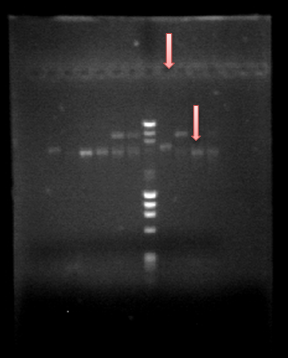
You can see the confirmation gels of the parts below:
Characterization of reporters
To assess the strength of our promoters when induced with Dexamethasone, we performed a fluorometric assay using mCherry as our reporter.Since we don't have our transactivating protein ready yet, we co-transformed our parts into the E. coli DH10B strain with the pAT7002 vector (Aoyama and Chua, 1997), which contains a well characterized Glucocorticoid Responsive Element that also uses the GAL4 DNA binding domain.
We also decided to visually inspect our induced transformants. Here two of the images taken using a epifluorescent microscopy with a TRITC filter.
References
Aoyama, T. & Chua N. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. The Plant Journal, 11(3): 605-612.
 "
"