Team:SJTU-BioX-Shanghai/Parts
From 2013.igem.org
| Line 1: | Line 1: | ||
| - | + | {{Template:13SJTU_header}} | |
| + | {{Template:13SJTU_nav}} | ||
| + | <table><tr> | ||
| + | <td valign="top" width="200"> | ||
| + | {{Template:12SJTU_floatnav_head}} | ||
<html> | <html> | ||
| - | < | + | <script type="text/javascript"> |
| - | < | + | // <![CDATA[ |
| - | + | var myMenu; | |
| - | + | window.onload = function() { | |
| - | + | myMenu = new SDMenu("my_menu"); | |
| - | + | myMenu.oneSmOnly = true; | |
| - | + | myMenu.remember = false; | |
| - | + | myMenu.init(); | |
| - | + | myMenu.expandMenu(myMenu.submenus[0]); | |
| - | + | ||
| - | </ | + | }; |
| + | // ]]> | ||
| + | </script> | ||
</html> | </html> | ||
| + | {{Template:13SJTU_floatnav_parts}} | ||
| - | + | {{Template:12SJTU_floatnav_foot}} | |
| - | + | <td valign="top" width="750"> | |
| - | ! | + | __NOTOC__ |
| - | + | <!----------------------------------------------------从这里开始写wiki---------------------------------> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | =Overview= | ||
| - | + | This year we made the components of CRISPRi system into parts and submitted to the iGEM Registry. With continuously increasing interest in work relevant to CRISPRi system, we believe these 2 parts will provide lots of convenience to users. Since in our project we hope that light could be the only parameter that we directly adjust, we did some improvements to the pre-existing parts: inserting them into a constant expressing operon. | |
| - | + | ||
| - | + | All of our parts can be divided into 2 categories, the CRISPRi system and the light sensing system. | |
| + | |||
| + | =CRISPRi= | ||
| + | |||
| + | Original Components: dCas9 protein(BBa_K1026001) and sgRNA(BBa_K1026003) | ||
| + | |||
| + | Constitutive Expressing Components: dCas9 protein(BBa_K1026000) and sgRNA(BBa_K1026002) | ||
| + | |||
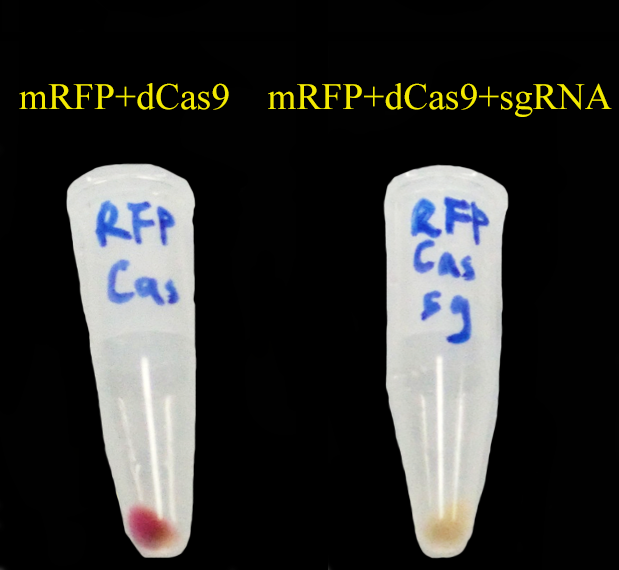
| + | We have tested CRISPRi system though the following experiment: After successful transformation, the control group expressed dCas9 and mRFP constitutively while the case group expressed dCas9, sgRNA and mRFP. The color of these two groups should be different due to the knock down effect of CRISPRi system. | ||
| + | |||
| + | The result is in consistent with our expectation: | ||
| + | |||
| + | [[File:2013_iGEM_SJTU_CRISPRi_control.png|350px]] | ||
| + | |||
| + | Additionally, the base-pairing region can be changed according to different people's needs. Inverted PCR will provide an easy way to replace this region and make it a universal and convenient regulating tool. | ||
| + | |||
| + | We also built the part that combines a promoter regulated by light sensor with sgRNA(BBa_K1026004), if used together with light sensing system, the red light will negatively control the expression amount of sgRNA, thus affecting the reporter genes or functional genes. | ||
| + | |||
| + | =Light Sensor= | ||
| + | |||
| + | After careful discussion and selection, we chose three light-controlled gene expression systems induced by red, green, and blue respectively. Main parts of these 3 sensing systems all have been used by 2011_Uppsala Team. | ||
| + | |||
| + | From the perspective of convenience, we improved several of these parts: | ||
| + | |||
| + | Constitutively Expressed Red Light Sensor Operon(BBa_K1026005) | ||
| + | |||
| + | Constitutively Expressed Blue Light Sensor Operon(BBa_K1026011) | ||
| + | |||
| + | Constitutively Expressed pcyA Operon(BBa_K1026014) | ||
<groupparts>iGEM013 SJTU-BioX-Shanghai</groupparts> | <groupparts>iGEM013 SJTU-BioX-Shanghai</groupparts> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!----------------------------------------------------到这里结束---------------------------------------> | ||
| + | </td> | ||
| + | </tr></table> | ||
| + | |||
| + | |||
| + | |||
| + | {{Template:13SJTU_footer}} | ||
Revision as of 22:58, 27 September 2013
|
| ||
|
 "
"