Team:ETH Zurich/Infoproc
From 2013.igem.org
| Line 10: | Line 10: | ||
If we located bacterial colonies in the place of squares like in the traditional game, the colonies placed in the corners would be further away compared to the colonies along the edges. We decided on a hexagonal honeycomb-like pattern, where colonies are placed at all edges of the hexagon except from the center. In this set-up the number of mine colonies restricted is to only one, two or three that can surround a non-mine colony. This also facilitates the selection of suitable reporter enzymes. To plate the field, liquid cultures of mine and non-mine cells are grown to an OD<sub>600</sub> of 0.5, then, using a pipette, 1.5μl of liquid culture are placed according to the grid. | If we located bacterial colonies in the place of squares like in the traditional game, the colonies placed in the corners would be further away compared to the colonies along the edges. We decided on a hexagonal honeycomb-like pattern, where colonies are placed at all edges of the hexagon except from the center. In this set-up the number of mine colonies restricted is to only one, two or three that can surround a non-mine colony. This also facilitates the selection of suitable reporter enzymes. To plate the field, liquid cultures of mine and non-mine cells are grown to an OD<sub>600</sub> of 0.5, then, using a pipette, 1.5μl of liquid culture are placed according to the grid. | ||
<br> </p> | <br> </p> | ||
| + | |||
<br clear="all" /> | <br clear="all" /> | ||
| + | {{:Team:ETH_Zurich/templates/footer}} | ||
Revision as of 10:52, 1 October 2013
Information processing
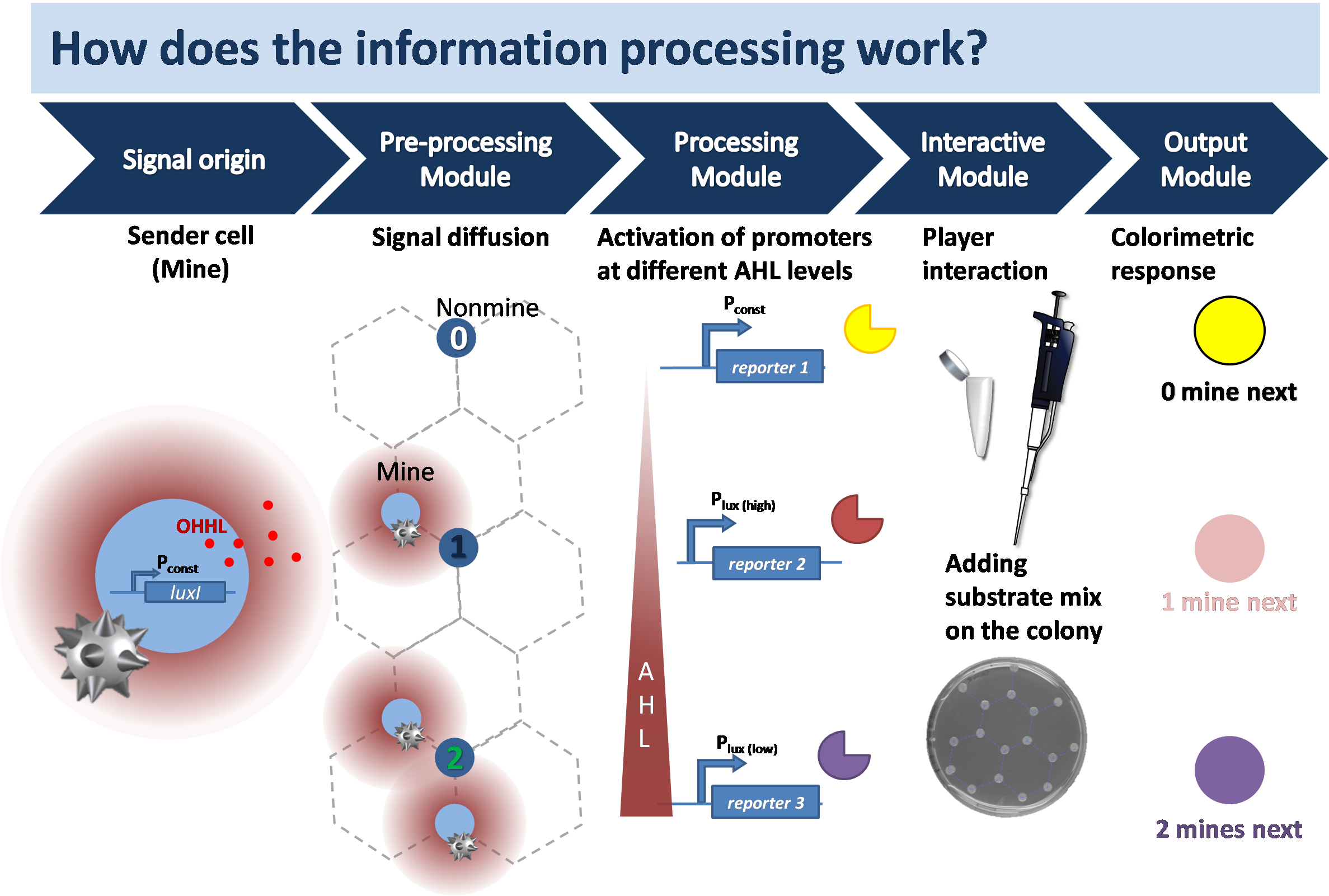
Each minesweeper square in the computer game corresponds to a bacterial colony on the agar mine-field. We have two bacterial strains: 1. the mine strain with the sender cells and 2. the non-mine strain with the receiver cells. The cells communicate through diffusion of OHHL and change color to give the player information in order to logically carry out the next move in the game. The colonies remain white as seen on the agar plate until any substrate is added. The left and the right click of the mouse is simulated with the addition of the multi-substrate mix and the single substrate.
The biology is explained here. The sender colony secretes the quorum sensing molecule 3-oxo-N-hexanoyl-L-homoserine lactone
(OHHL) that diffuses through the agar to the surrounding cells. The receiver cells passively take up the OHHL by diffusion where the signaling molecule forms a complex with the inactive LuxR to activate it. The information is translated via mutated pLuxR promoters of different OHHL affinities which leads to the secretion of different hydrolases. Within minutes after the addition of substrate a change in color indicates the identity of the played colony and number of surrounding mine colonies.
If we located bacterial colonies in the place of squares like in the traditional game, the colonies placed in the corners would be further away compared to the colonies along the edges. We decided on a hexagonal honeycomb-like pattern, where colonies are placed at all edges of the hexagon except from the center. In this set-up the number of mine colonies restricted is to only one, two or three that can surround a non-mine colony. This also facilitates the selection of suitable reporter enzymes. To plate the field, liquid cultures of mine and non-mine cells are grown to an OD600 of 0.5, then, using a pipette, 1.5μl of liquid culture are placed according to the grid.
 "
"