Team:ETH Zurich/Experiments 3
From 2013.igem.org
| Line 22: | Line 22: | ||
[[File:AesReaction.png|thumb|center|800px| <b>Figure 2.</b> Acetyl esterase catalyzed hydrolysis reaction of magenta butyrate.]]<br clear="all"/> | [[File:AesReaction.png|thumb|center|800px| <b>Figure 2.</b> Acetyl esterase catalyzed hydrolysis reaction of magenta butyrate.]]<br clear="all"/> | ||
<br><b>Enzyme kinetics</b> | <br><b>Enzyme kinetics</b> | ||
| - | [[File:Aes_fluorescent.png|thumb|right| | + | [[File:Aes_fluorescent.png|thumb|right|200px|<b>Figure 3.</b> Cell lysate of <i>Escherichia coli</i> overexpressing acetyl esterase after reaction with 4-MU-butyrate.]] |
| - | + | For the kinetics assay, the ''aes'' encoded protein was tested with the fluorescent substrate 4-MU-butyrate. The picture on the right was taken with a common single lens reflex camera mounted on a dark hood at λ<sub>Ex</sub> 365 nm. | |
| - | [[File:AES_reaction.png|thumb|center| | + | |
| - | To characterize the | + | [[File:AES_reaction.png|thumb|center|400px|<b>Figure 4. Enzymatic reaction of Aes with 4-MU-butyrate.</b>]]<br> |
| + | To characterize the enzymes, we conducted fluorometric assays to obtain K<sub>m</sub> values. Bacterial cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm. | ||
The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™. | The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™. | ||
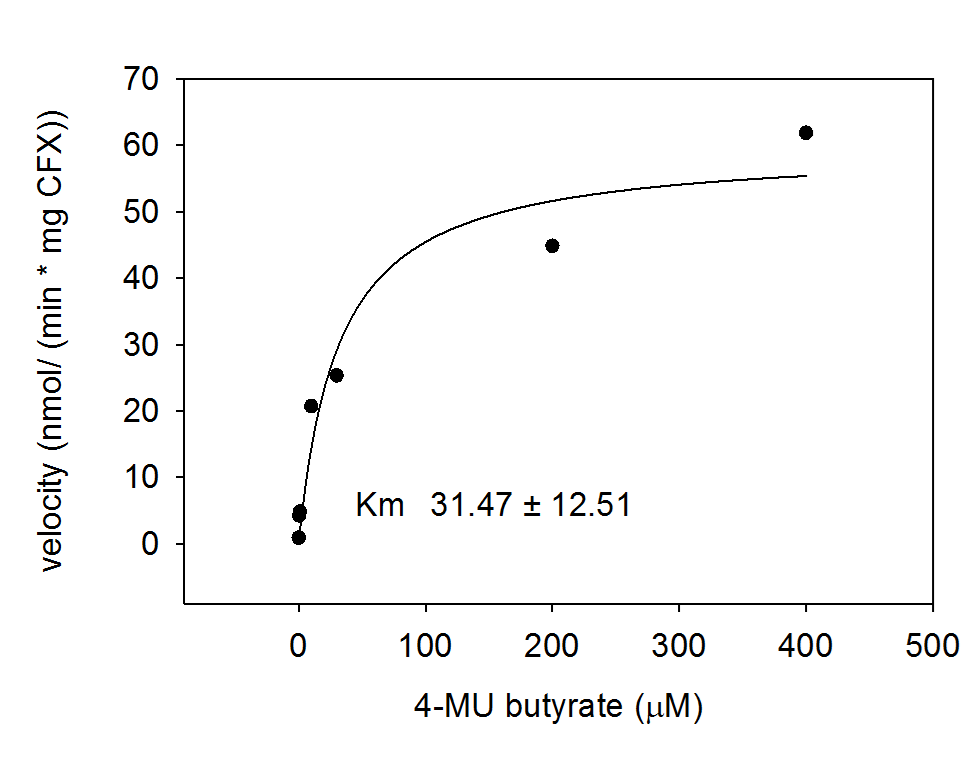
| - | + | [[File:Hydrolase AES.png|thumb|center|400px|<b>Figure 5.</b> Michaelis-Menten-Kinetics of cell lysate from <i>Escherichia coli</i> overexpressing acetyl esterase: plots velocity versus substrate concentration (10 μL, 30 μL, 100 μL, 200 μL, 400 μL) in 20 mM Tris buffer of pH 8. A kinetic value for K<sub>m</sub> obtained by fitting the raw data to standard the Michaelis Menten equation; K<sub>m</sub> = 31.5 ± 12.5 μM. All assays were carried out in triplicates, results are presented as means.]] | |
| - | [[File:Hydrolase AES.png|thumb|center| | + | |
<br clear="all"/> | <br clear="all"/> | ||
| Line 48: | Line 48: | ||
<br><b>Enzyme kinetics</b> | <br><b>Enzyme kinetics</b> | ||
| - | [[File:PhoA_fluorescent.png|thumb|right| | + | [[File:PhoA_fluorescent.png|thumb|right|200px|<b>Figure 3.</b> Cell lysate from <i>Escherichia coli</i> overexpressing PhoA-HIS (left) and PhoA (right) after reaction with 4-MU-phosphate.]] |
For the kinetics assay, we tested both the ''Citrobacter'' ''phoA'' and ''phoA''-HIS encoded proteins with the fluorescent substrate 4-MU-phosphate. The picture on the right was taken with a common single lens reflex camera mounted on a dark hood at λ<sub>Ex</sub> 365 nm. | For the kinetics assay, we tested both the ''Citrobacter'' ''phoA'' and ''phoA''-HIS encoded proteins with the fluorescent substrate 4-MU-phosphate. The picture on the right was taken with a common single lens reflex camera mounted on a dark hood at λ<sub>Ex</sub> 365 nm. | ||
[[File:phoa_fluorescent_reaction.png|frame|center|<b>Figure 4. Enzymatic reaction of PhoA with 4-MU-phosphate.</b>]] | [[File:phoa_fluorescent_reaction.png|frame|center|<b>Figure 4. Enzymatic reaction of PhoA with 4-MU-phosphate.</b>]] | ||
| Line 59: | Line 59: | ||
| - | <h1> | + | <h1>β-Galactosidase (LacZ)</h1> |
| - | |||
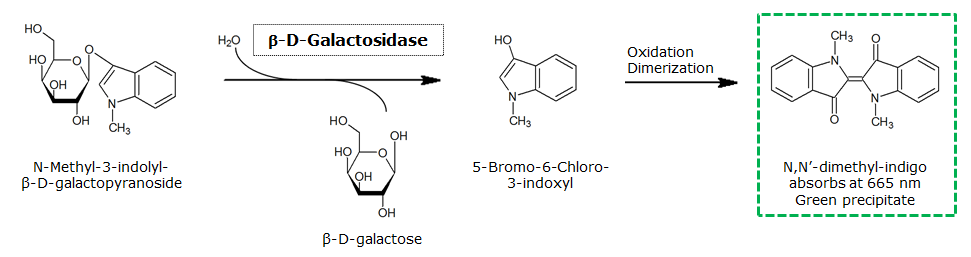
[[File:LacZ and GreenGal.jpeg|400px]] | [[File:LacZ and GreenGal.jpeg|400px]] | ||
[[File:LacZReaction.png|center]] | [[File:LacZReaction.png|center]] | ||
| - | |||
| - | |||
| - | |||
<br clear="all"/> | <br clear="all"/> | ||
| - | <h1> | + | <h1>β-Glucuronidase (GusA)</h1> |
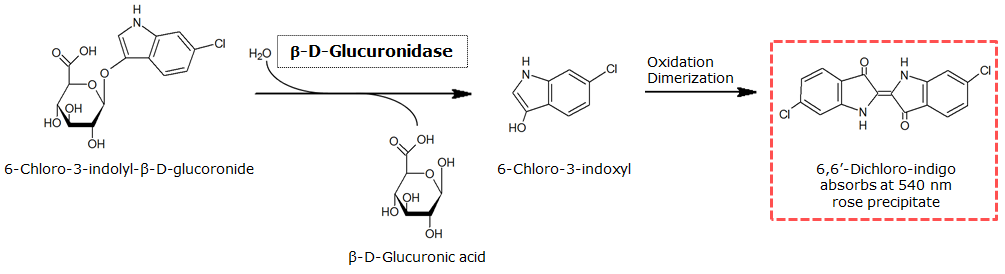
[[File:GusA_colored.png|frame|right|<b>Figure 1.</b> Liquid culture from <i>Escherichia coli</i> overexpressing β-glucuronidase after reacting with Salmon-β-glucuronide.]] | [[File:GusA_colored.png|frame|right|<b>Figure 1.</b> Liquid culture from <i>Escherichia coli</i> overexpressing β-glucuronidase after reacting with Salmon-β-glucuronide.]] | ||
To test the functionality of β-glucuronidase, a liquid culture of <i>Escherichia coli</i> overexpressing β-glucuronidase was grown until an OD<sub>600</sub> of 0.4 - 0.6 was reached before addition of Salmon-β-glucuronide, a substrate for β-glucuronidase which produces a rose color after cleavage. The final concentration of the substrate in liquid culture was 1 mM and the full conversion of this substrate was reached by overnight incubation at 37°C. <br clear="all"/> | To test the functionality of β-glucuronidase, a liquid culture of <i>Escherichia coli</i> overexpressing β-glucuronidase was grown until an OD<sub>600</sub> of 0.4 - 0.6 was reached before addition of Salmon-β-glucuronide, a substrate for β-glucuronidase which produces a rose color after cleavage. The final concentration of the substrate in liquid culture was 1 mM and the full conversion of this substrate was reached by overnight incubation at 37°C. <br clear="all"/> | ||
| Line 84: | Line 80: | ||
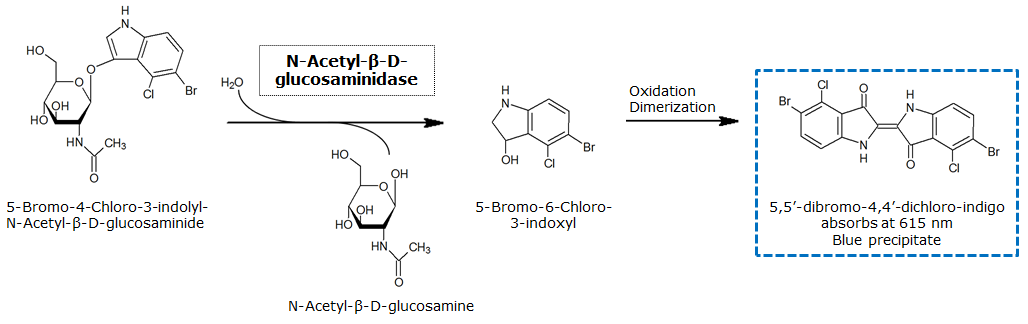
| - | <h1>β-N-Acetylglucosaminidase(NagZ)</h1> | + | <h1>β-N-Acetylglucosaminidase (NagZ)</h1> |
| - | |||
[[File:NagZReaction.png|center]] | [[File:NagZReaction.png|center]] | ||
| - | |||
| - | |||
| - | |||
<br clear="all"/> | <br clear="all"/> | ||
Revision as of 02:29, 5 October 2013
Contents |
Enzyme-substrate reactions

To generate visible output by adding substrates to colonies in Colisweeper, we made use of orthogonal enzyme-substrate reactions. A set of chromogenic substrates was chosen to produce different colors depending on the abundant hydrolases and thereby to uncover the identity of each colony to the player.
Chromogenic substrates incorporate a chromophore whose absorbance properties change after the enzyme reaction, and the color signal produced then is directly related to the enzyme-catalyzed reaction.
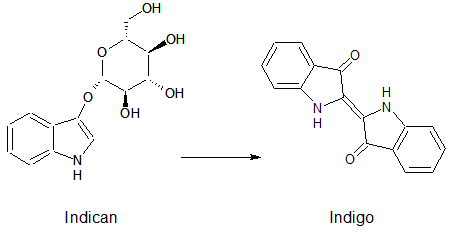
Indican is a chromogenic glycoside hydrolase substrate which belongs to a family of natural glycosides found in plants. Cleavage of the glycosidic bond forms an unstable hydroxyindole intermediate, which dimerizes by oxidation to form indigo as a blue precipitate:
Numerous enzyme substrates have been designed following this natural product example, giving rise to many colored phenols that are used to detect enzyme activities.
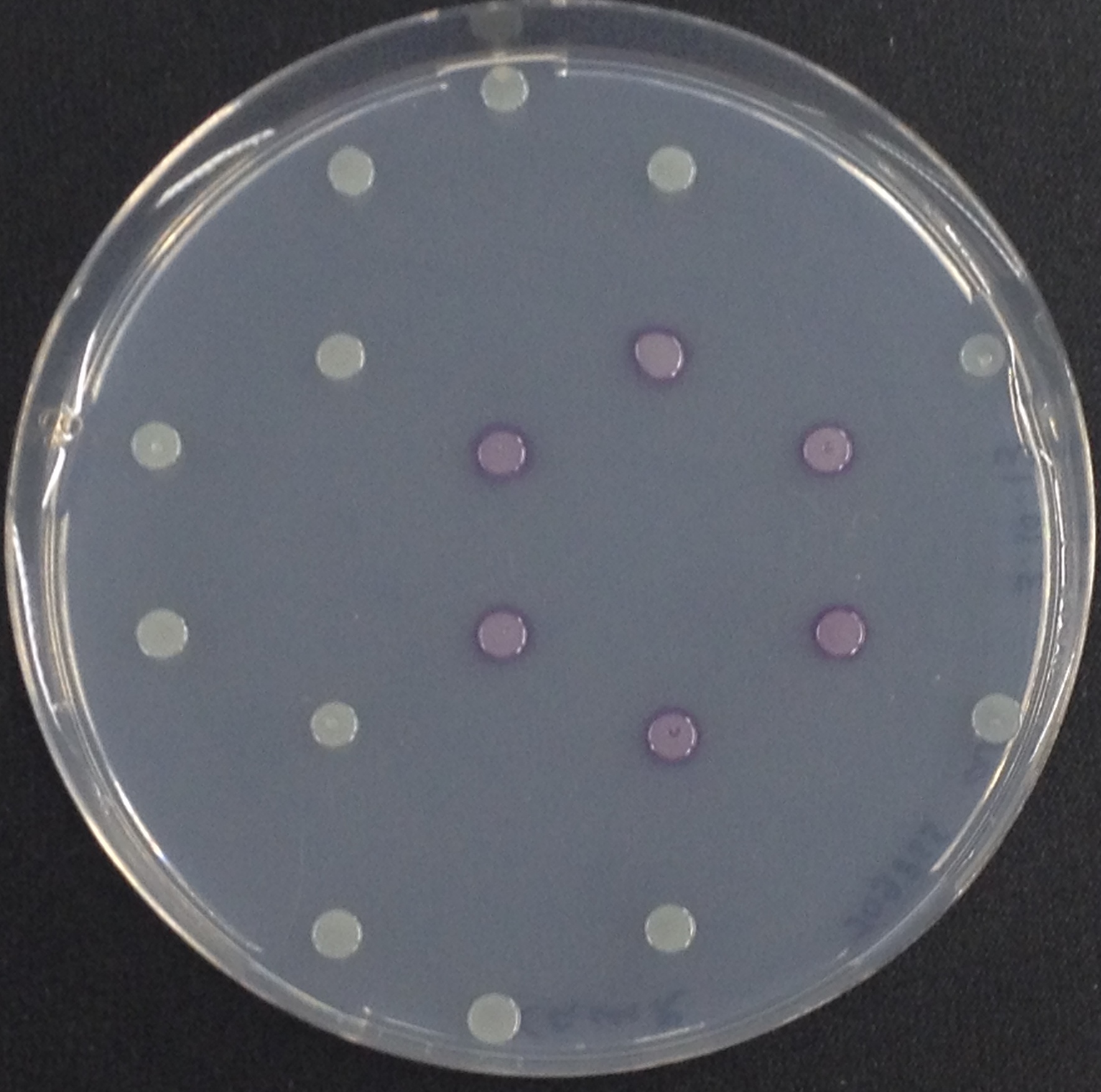
As shown in Figure 1, some of the hydrolases used in the Colisweeper reporter system can catalyze hydrolysis of various substrates, with different chromophores that give rise to a wide range of colors. This variety of substrates and color outputs enables change of positions and function of these enzymes. Other possible substrates that can be used for the enzymes of the Colisweeper reporter system can be found in the Hydrolases section.
In Colisweeper, the set of enzyme-substrate pairs are chosen as illustrated in the following table:
Acetyl esterase (Aes)
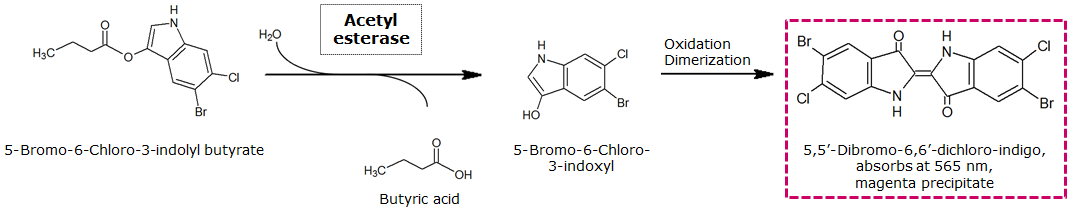
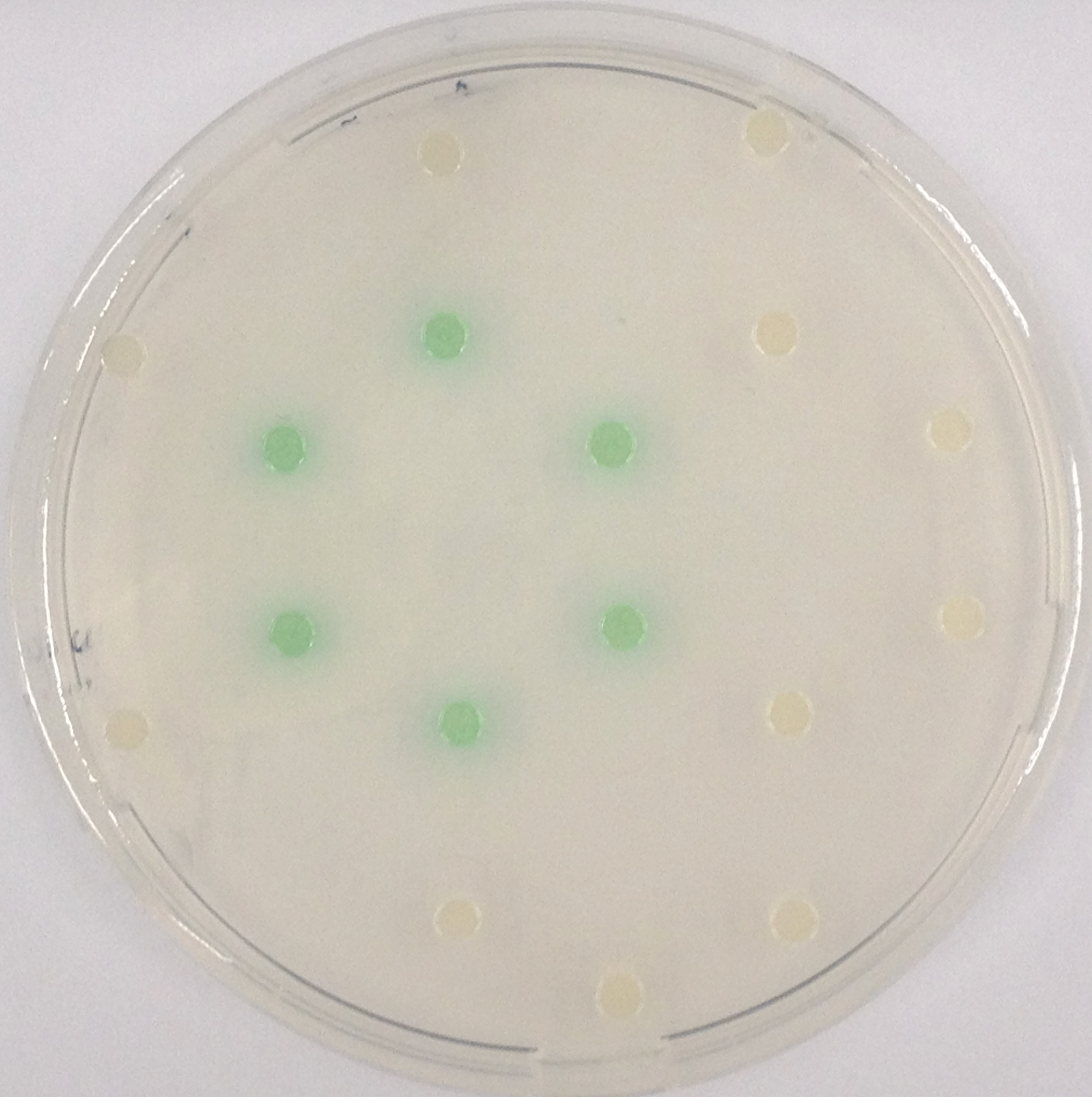
To assess color development after reaction of the enzyme with the chromogenic substrate, a liquid culture of our triple knockout Escherichia coli strain overexpressing Aes was grown until an OD600 of 0.4 - 0.6 was reached before addition of magenta butyrate to a final concentration of 250 µM. To study the color development in the actual Colisweeper game setup, colonies were plated by pipetting 1.5 µl of triple knockout Escherichia coli liquid culture (OD600 of 0.4 - 0.6) on an M9 agar plate. Addition of 1.5 µl of 20 mM magenta butyrate onto each colony results in color generation visible after a few minutes at room temperature.
Enzyme kinetics
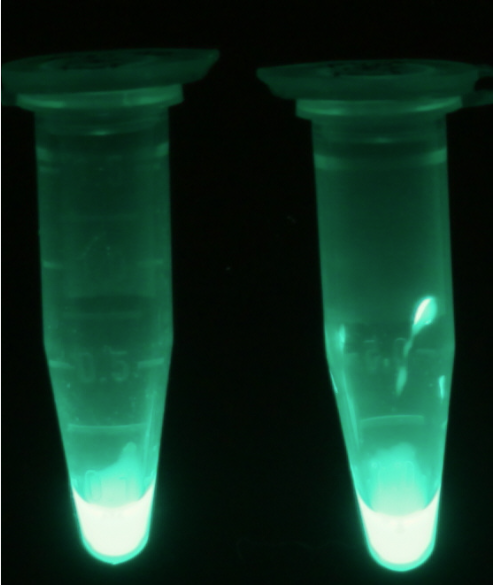
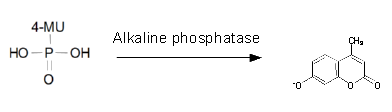
For the kinetics assay, the aes encoded protein was tested with the fluorescent substrate 4-MU-butyrate. The picture on the right was taken with a common single lens reflex camera mounted on a dark hood at λEx 365 nm.
To characterize the enzymes, we conducted fluorometric assays to obtain Km values. Bacterial cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λEx 365 nm and λEm 445 nm. The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™.
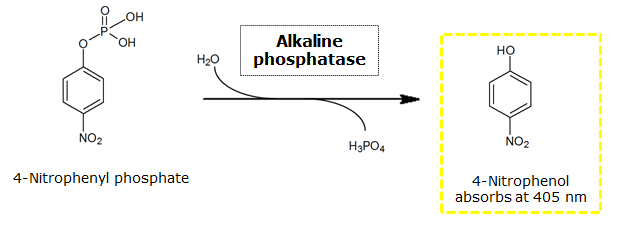
Alkaline phosphatase(PhoA)
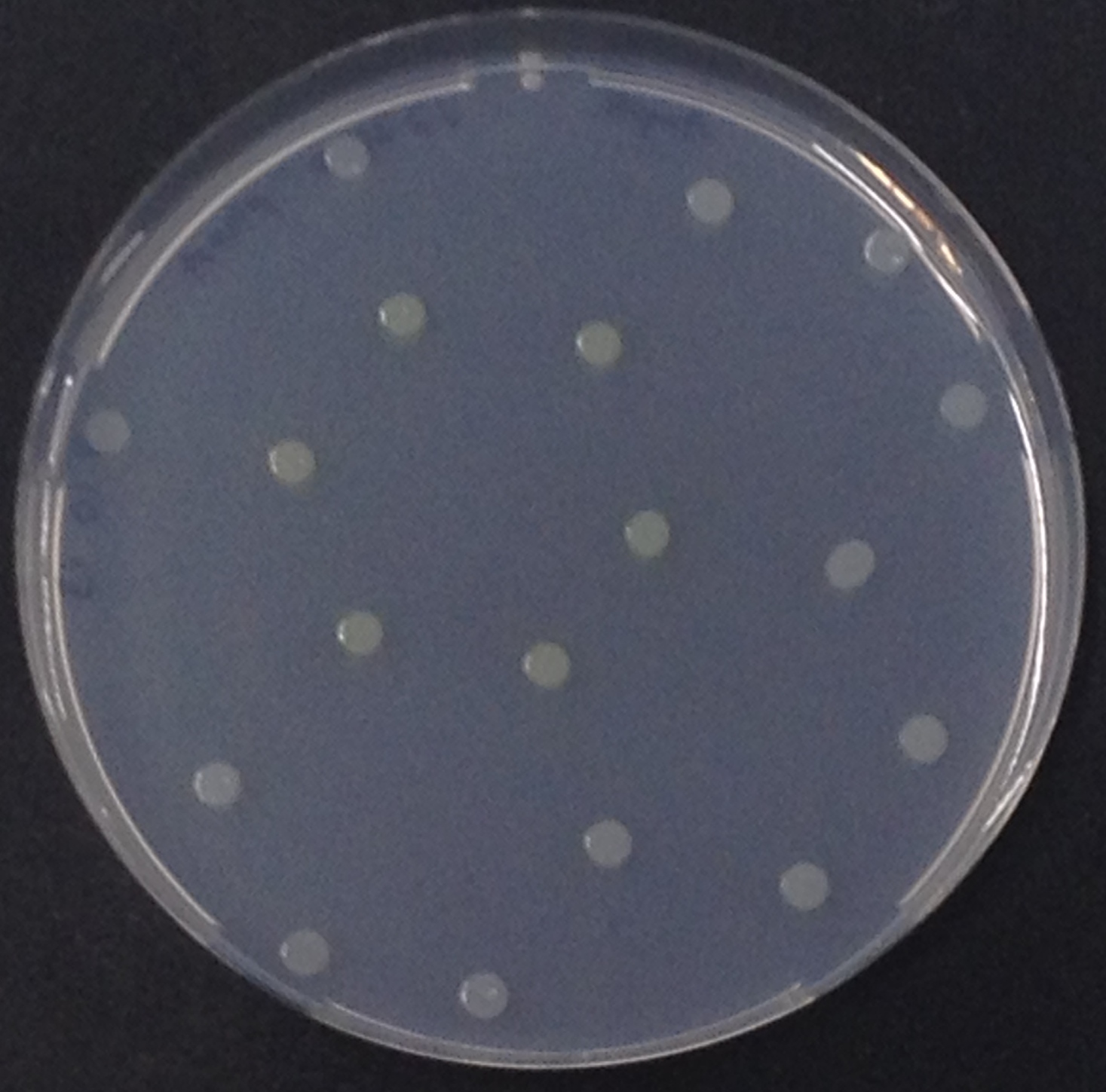
To test the functionality of this alkaline phosphatase, liquid culture of Escherichia coli overexpressing this hydrolase was grown until an OD600 of 0.4 - 0.6 was reached before addition of p-nitrophenyl phosphate (pNPP) to a final concentration of 5 µM. This substrate gives rise to yellow color after hydrolysis by the alkaline phosphatase. In the Colisweeper game, substrates are pipetted onto colonies. Figure Bla shows color development on colonies five minutes after addition of 1.5 µl of a 0.5 M pNPP solution.
Characterization of PhoA-HIS tagged protein
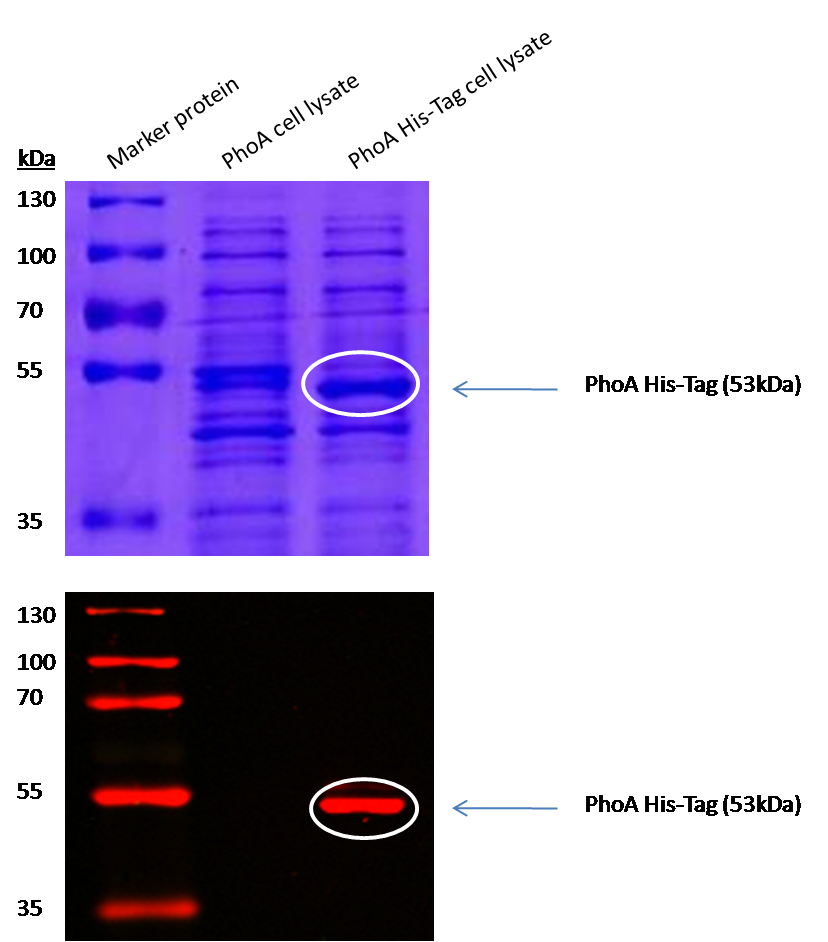
Enzyme kinetics
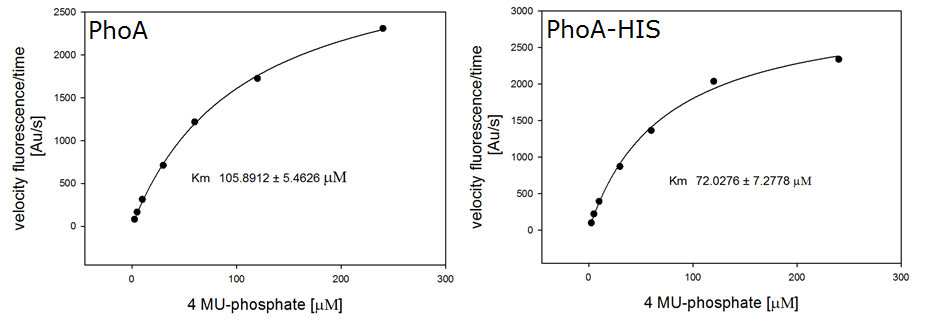
For the kinetics assay, we tested both the Citrobacter phoA and phoA-HIS encoded proteins with the fluorescent substrate 4-MU-phosphate. The picture on the right was taken with a common single lens reflex camera mounted on a dark hood at λEx 365 nm.
To characterize the enzymes, we conducted fluorometric assays to obtain Km values. Bacterial cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λEx 365 nm and λEm 445 nm. The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:
β-Galactosidase (LacZ)
β-Glucuronidase (GusA)
To test the functionality of β-glucuronidase, a liquid culture of Escherichia coli overexpressing β-glucuronidase was grown until an OD600 of 0.4 - 0.6 was reached before addition of Salmon-β-glucuronide, a substrate for β-glucuronidase which produces a rose color after cleavage. The final concentration of the substrate in liquid culture was 1 mM and the full conversion of this substrate was reached by overnight incubation at 37°C.
Enzyme kinetics
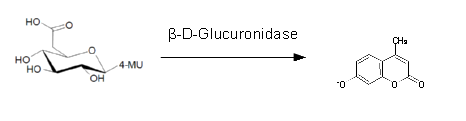
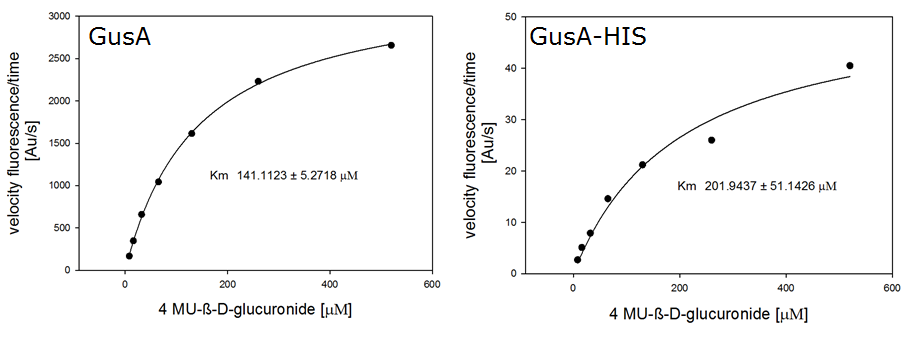
For the kinetics assay, we tested both gusA and gusA-HIS encoded proteins with the fluorescent substrate 4-MU-β-D-Glucuronide. The picture on the right was taken with a common single lens reflex camera mounted on a dark hood at λEx 365 nm.
To characterize the enzymes, we conducted fluorometric assays to obtain Km values. Bacterial cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λEx 365 nm and λEm 445 nm. The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:
β-N-Acetylglucosaminidase (NagZ)
 "
"