Team:ETH Zurich/Experiments 3
From 2013.igem.org
| Line 90: | Line 90: | ||
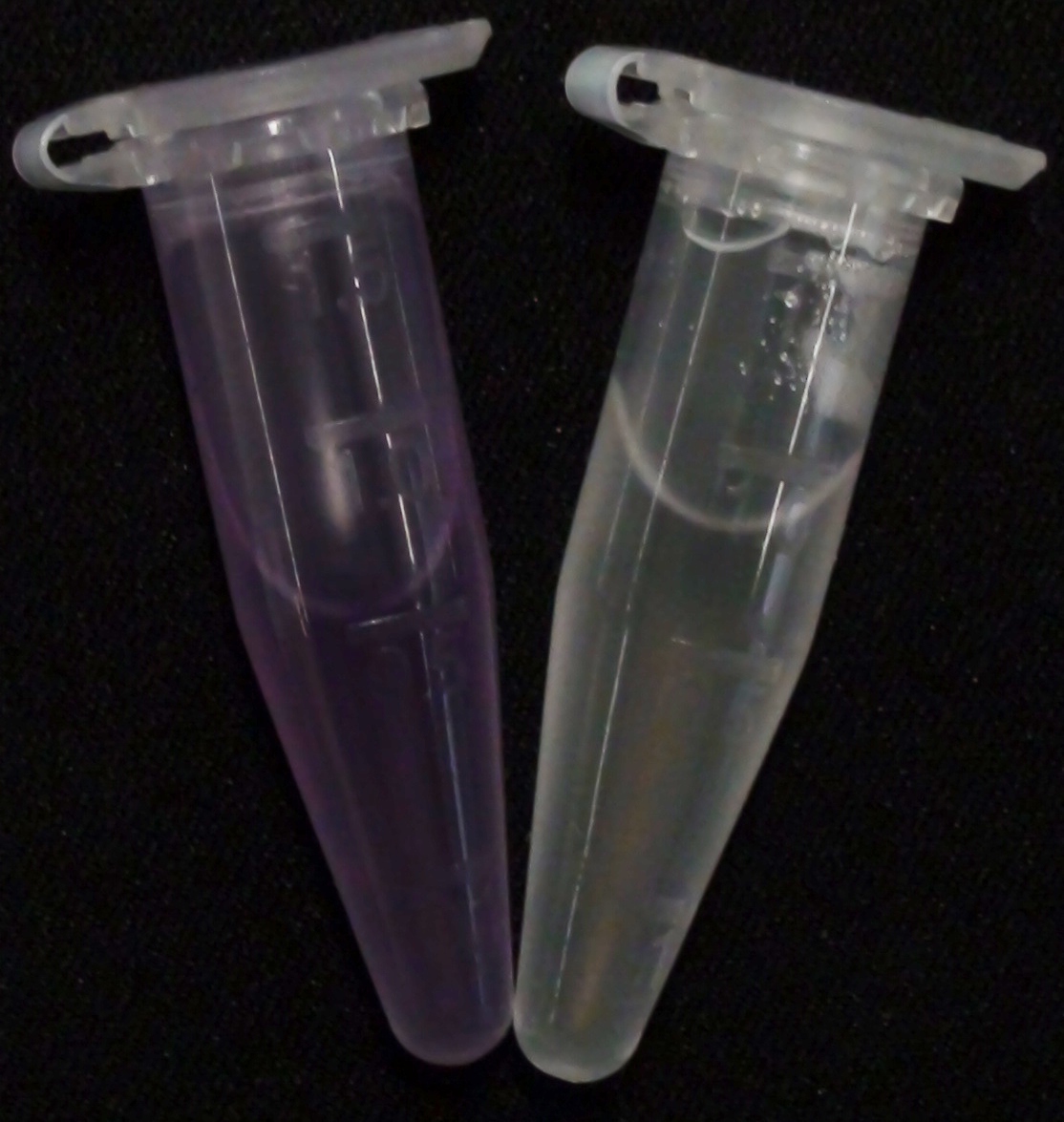
[[File:NagZ pNPGluNAc.JPG|thumb|right|200px|<b>Figure 23.</b> Liquid culture of the triple knockout <i>Escherichia coli</i> strain overexpressing NagZ; after reaction with pNP-GluNAc.]] | [[File:NagZ pNPGluNAc.JPG|thumb|right|200px|<b>Figure 23.</b> Liquid culture of the triple knockout <i>Escherichia coli</i> strain overexpressing NagZ; after reaction with pNP-GluNAc.]] | ||
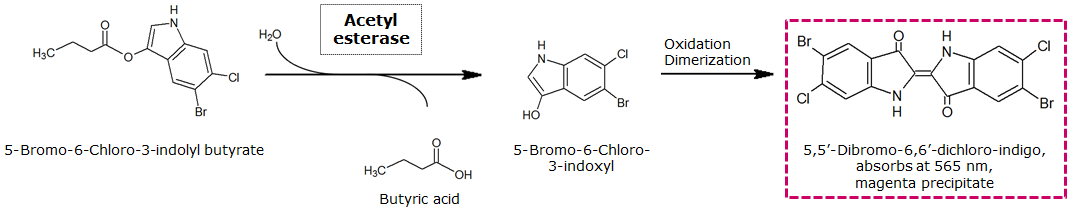
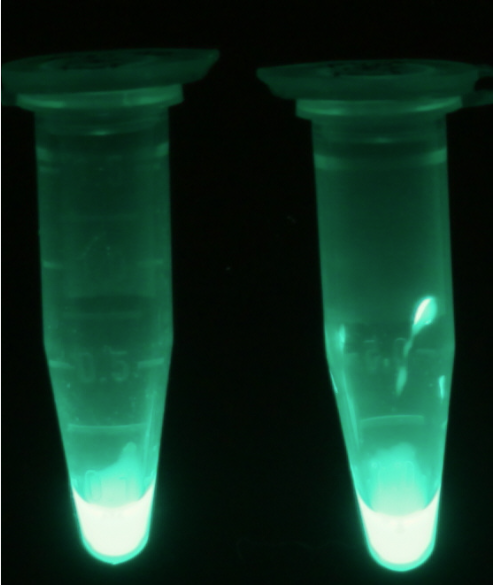
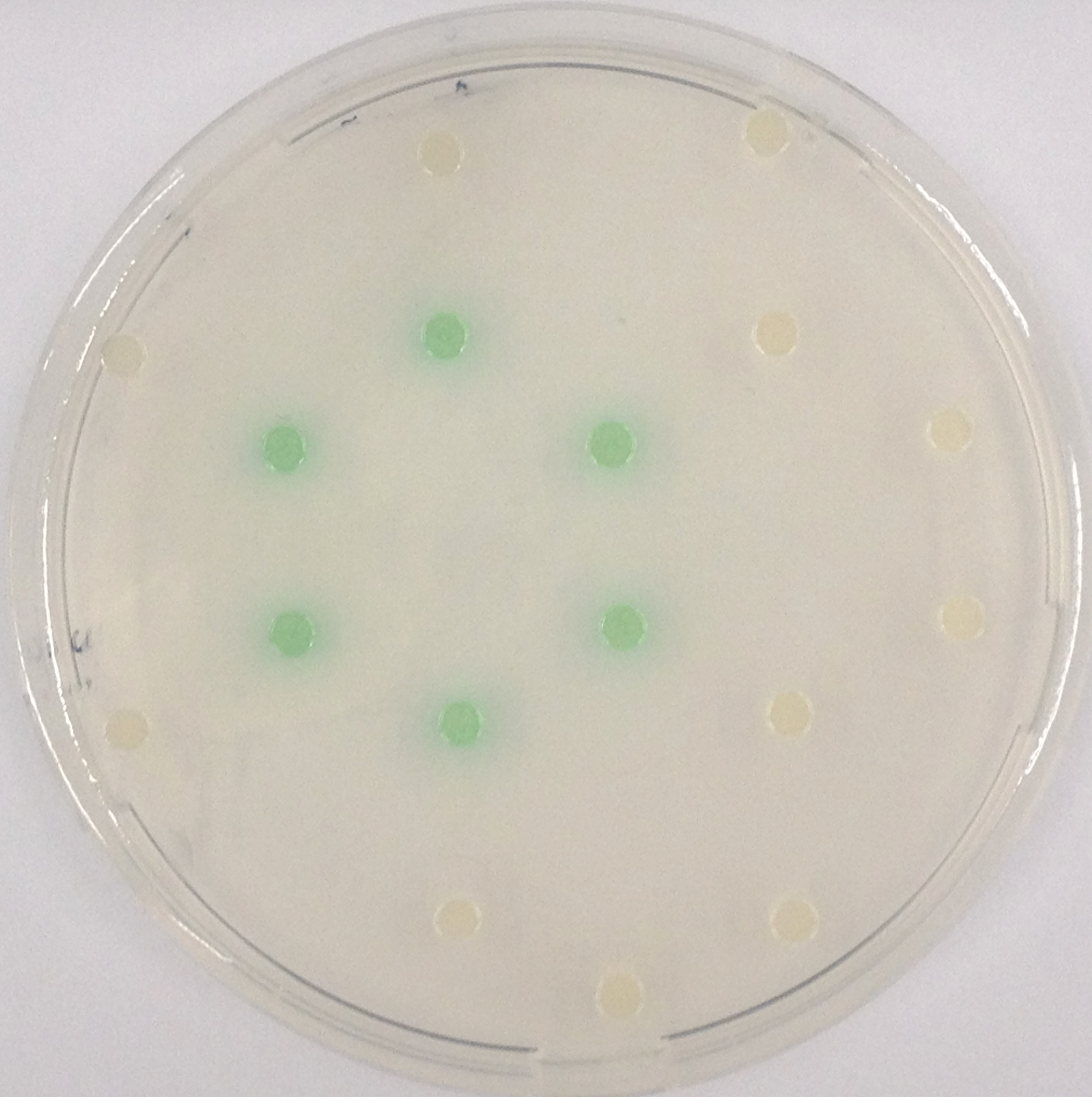
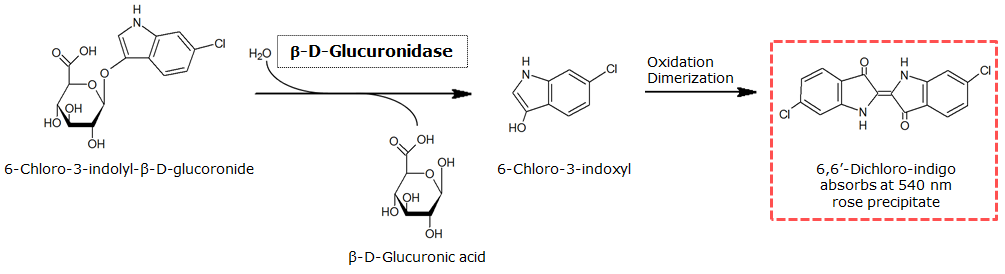
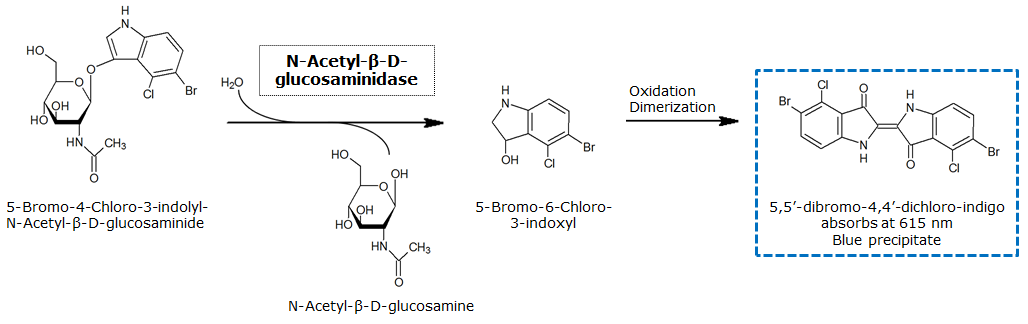
<p align="justify"><b>Chromogenic assay</b><br>NagZ was proposed to be constitutively expressed in the mine colonies, processing its blue color-yielding substrate 5-Bromo-4-Chloro-3-indolyl-β-N-acetylglucosaminide (X-GluNAc), as shown in Figure 25. However, we could not initially manage to get color from the reaction with this enzyme-substrate pair. Using p-nitrophenyl-β-N-acetylglucosaminide (pNP-GluNAc) at an end concentration of 0.01 mM in an ''Escherichia coli'' liquid culture with an OD<sub>600</sub> of 0.4 - 0.6, development of yellow color could be observed (Figure 23), confirming that NagZ is expressed in our mine cells.<br> | <p align="justify"><b>Chromogenic assay</b><br>NagZ was proposed to be constitutively expressed in the mine colonies, processing its blue color-yielding substrate 5-Bromo-4-Chloro-3-indolyl-β-N-acetylglucosaminide (X-GluNAc), as shown in Figure 25. However, we could not initially manage to get color from the reaction with this enzyme-substrate pair. Using p-nitrophenyl-β-N-acetylglucosaminide (pNP-GluNAc) at an end concentration of 0.01 mM in an ''Escherichia coli'' liquid culture with an OD<sub>600</sub> of 0.4 - 0.6, development of yellow color could be observed (Figure 23), confirming that NagZ is expressed in our mine cells.<br> | ||
| - | According to Orenga et al. [https://2013.igem.org/Team:ETH_Zurich/Experiments_3#hydrolase_reference 1], the substrate incorporating the indoxyl chromophore is less preferentially hydrolyzed by bacterial NagZ in comparison to yeast NagZ, and more preferentially hydrolyzes the alizarine-based substrate they applied. To increase hydrolysis of the indoxyl-based substrate by our NagZ, we added the substrate to a liquid culture supplemented with BSA. The end concentration in liquid culture of both X-GluNAc and 5-Bromo-6-Chloro-3-indolyl-β-N-acetylglucosaminide (Magenta GluNAc) were at 1 mM (Figure 23). | + | According to Orenga et al. ([https://2013.igem.org/Team:ETH_Zurich/Experiments_3#hydrolase_reference 1]), the substrate incorporating the indoxyl chromophore is less preferentially hydrolyzed by bacterial NagZ in comparison to yeast NagZ, and more preferentially hydrolyzes the alizarine-based substrate they applied. To increase hydrolysis of the indoxyl-based substrate by our NagZ, we added the substrate to a liquid culture supplemented with BSA, which enhanced hydrolysis dramatically. The end concentration in liquid culture of both X-GluNAc and 5-Bromo-6-Chloro-3-indolyl-β-N-acetylglucosaminide (Magenta GluNAc), a magenta color-yielding substrate for NagZ that we found by chance :), were at 1 mM (Figure 23). |
<br></p> | <br></p> | ||
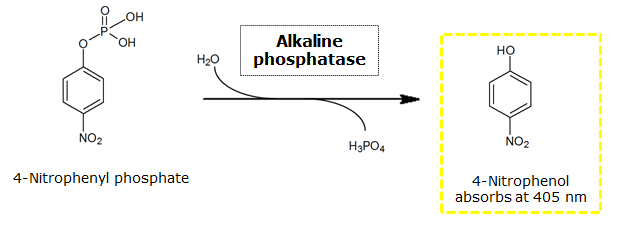
[[File:PNP-GluNAc.png|thumb|center|400px| <b>Figure 24.</b> β-N-Acetylglucosaminidase catalyzed hydrolysis of the chromogenic substrate pNP-GluNAc.]]<br> | [[File:PNP-GluNAc.png|thumb|center|400px| <b>Figure 24.</b> β-N-Acetylglucosaminidase catalyzed hydrolysis of the chromogenic substrate pNP-GluNAc.]]<br> | ||
Revision as of 16:15, 26 October 2013
Contents[hide] |
Enzyme-substrate reactions
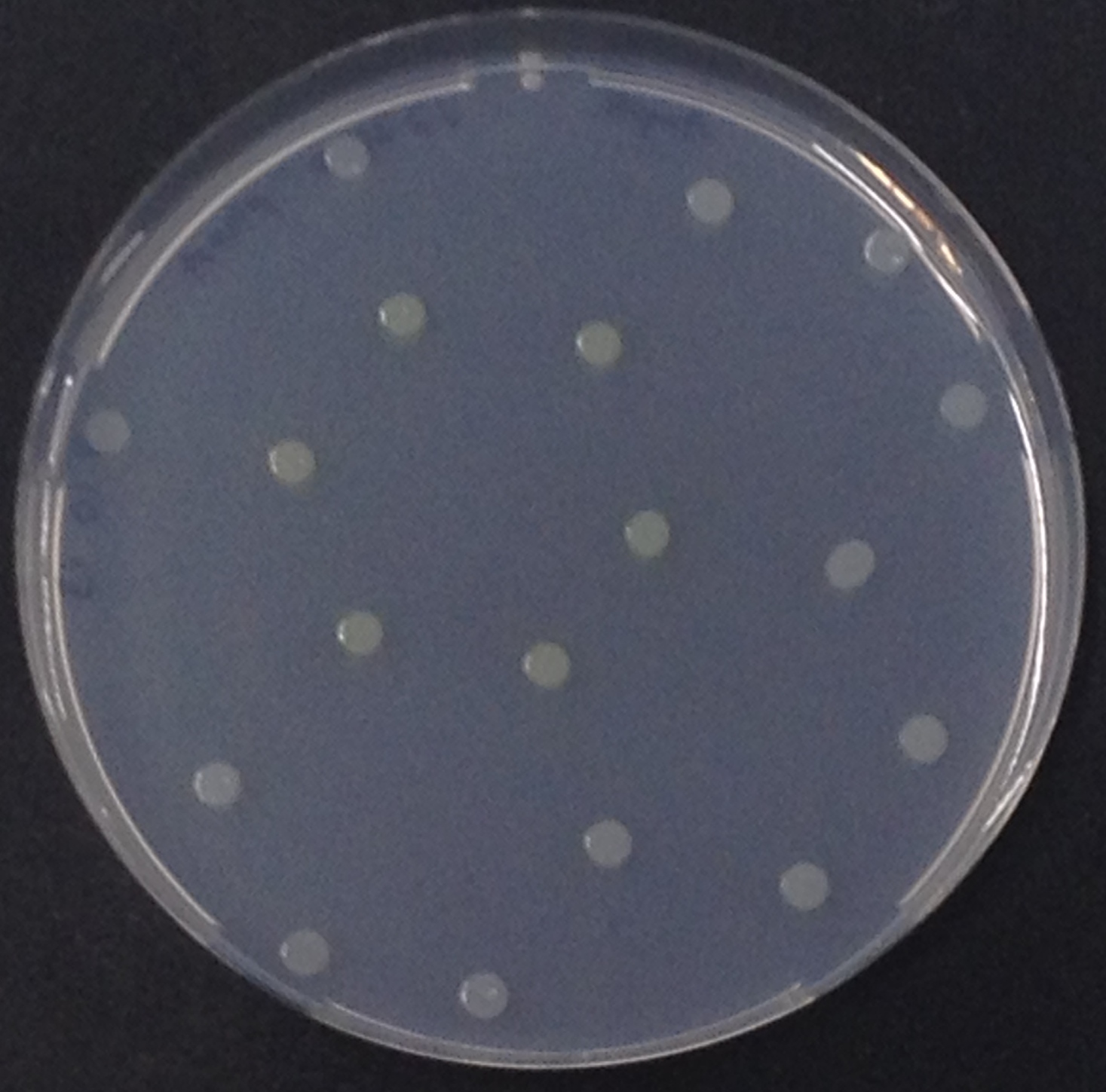
To generate visible output by adding substrates to colonies in Colisweeper, we made use of orthogonal enzyme-substrate reactions. A set of chromogenic substrates was chosen to produce different colors depending on the abundant hydrolases and thereby to uncover the identity of each colony to the player.
The set of enzyme-substrate pairs chosen for the Colisweeper project, and characterizations of their reactions, are described below. Information on other possible substrates that can be used for the enzymes of the Colisweeper reporter system can be found in the reporter system section.
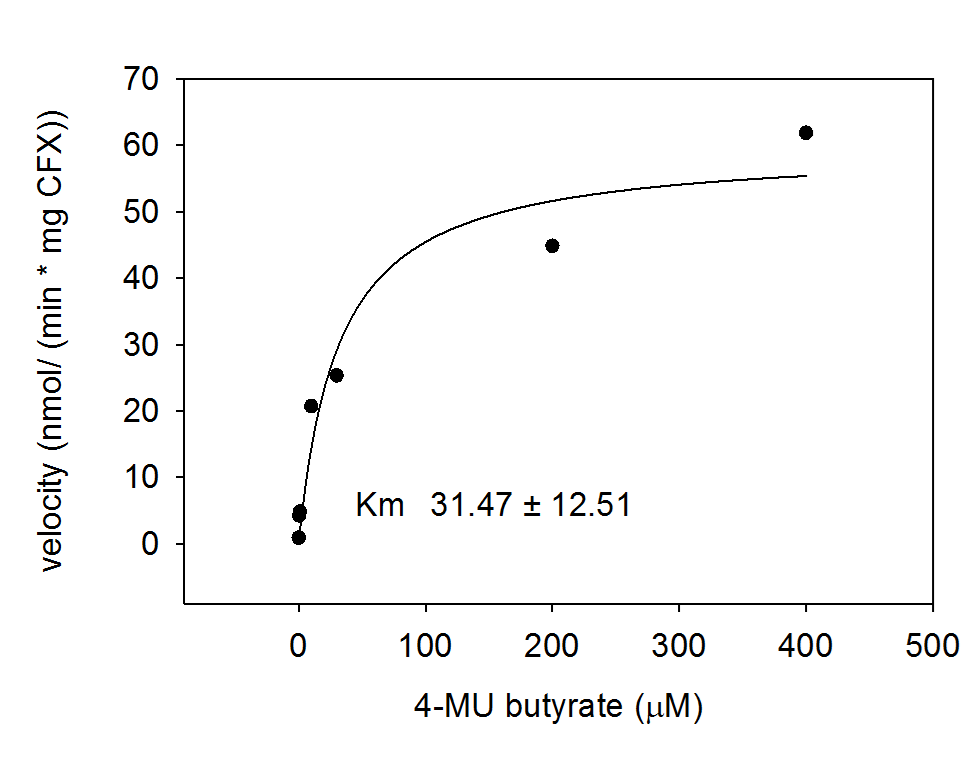
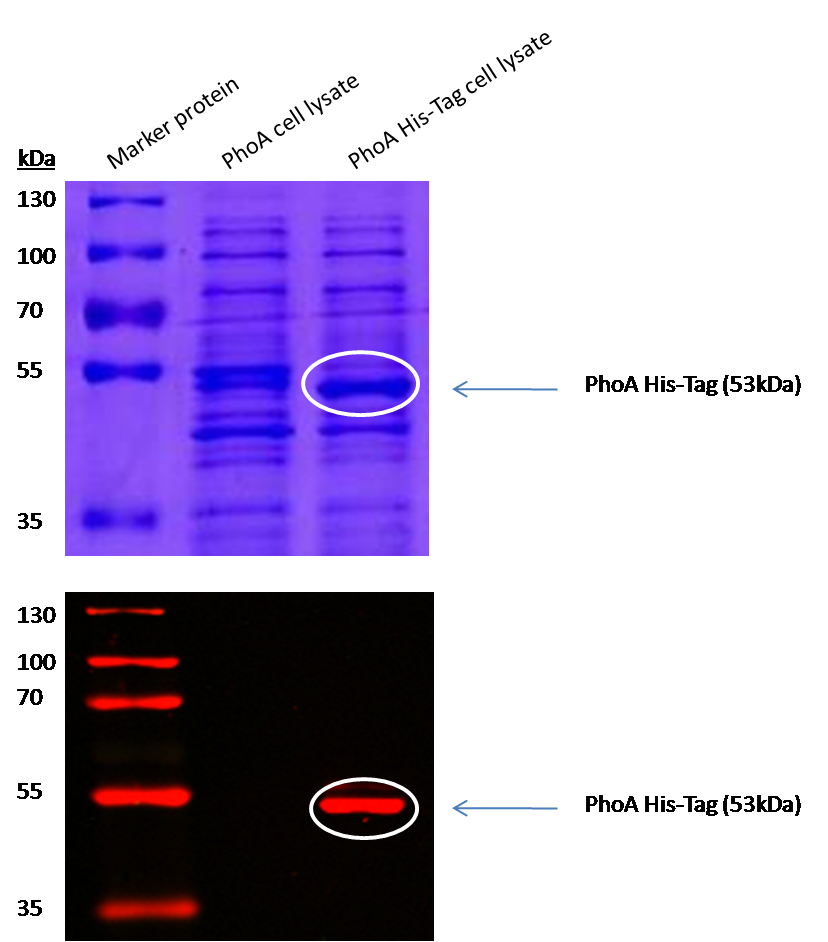
To characterize the hydrolases used in Colisweeper, we conducted substrate tests in liquid cultures as well as on colonies, enzyme kinetics and detection of HIS-tagged protein by Western Blot. Additionally, crosstalk assays and color overlay tests (multiple enzyme-substrate reactions together) have been performed for this project. An overview of chromogenic substrates used can be found in the Materials section.
Expand the boxes to see the characterization of the hydrolases
Acetyl esterase (Aes)
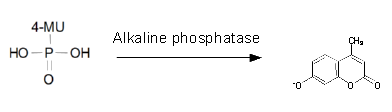
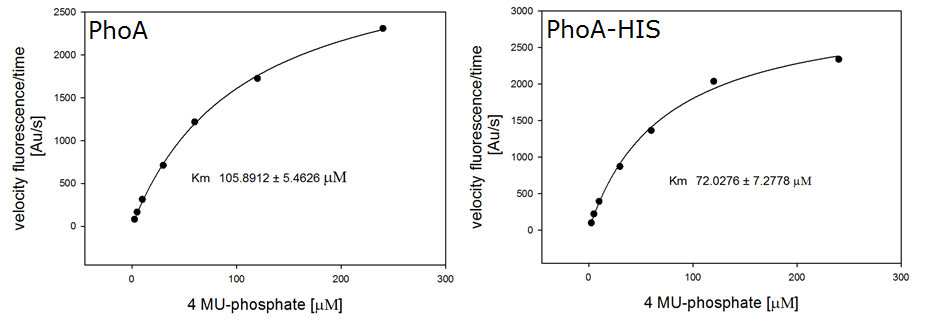
Alkaline phosphatase (PhoA)
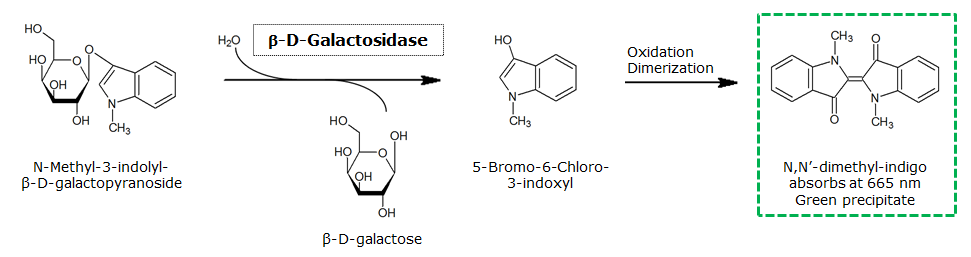
β-Galactosidase (LacZ)
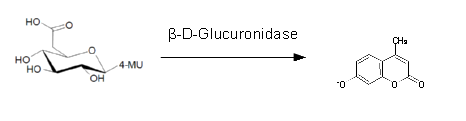
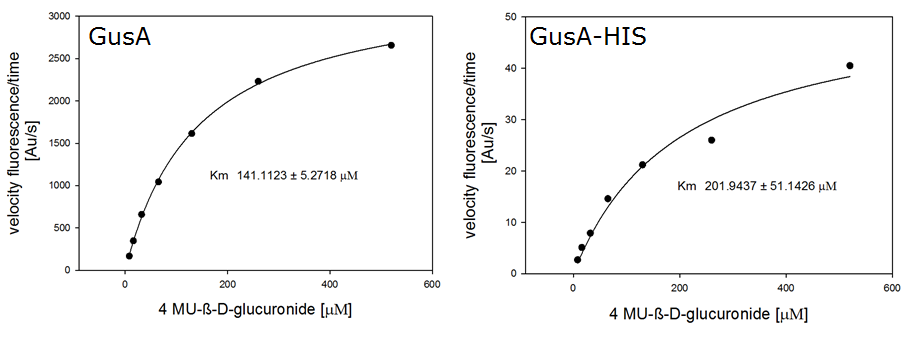
β-Glucuronidase (GusA)
β-N-Acetylglucosaminidase (NagZ)
Crosstalk
To ensure orthogonality between the enzyme-substrate reactions used in Colisweeper, a crosstalk test has been done to ensure that all overexpressed enzymes specifically cleave their assigned substrate.
Color overlay
To play Colisweeper, colors for each colony identity have to be clearly distinguishable by eye. Because we are using high-pass filters to differentially process certain AHL levels, we had to make sure that a mix of cleaved product would show distinguishable colors.
References
(1) Orenga S, Roger-Dalbert C, James A, Perry J, US Patent 20090017481 (2009).
Absorption: Biosynth
 "
"