Team:Bielefeld-Germany/Results
From 2013.igem.org
| Line 82: | Line 82: | ||
==MFC== | ==MFC== | ||
| + | |||
| + | |||
*<p align="justify">After extensive optimization, we were able to build a well-functioning Microbial Fuel Cell and establish a protocol for measuring the power output of our electricity generating cultures. We built a stack of five Fuel Cells which was successfully used to power different LEDs and the motor of a small fan. Furthermore, a 3D model was designed, which can be printed out using a 3D printer. This model was made available for download on our wiki, so anyone interested can build a fuel cell of their own.[[Team:Bielefeld-Germany/Project/MFC#Results| Read more]].</p> | *<p align="justify">After extensive optimization, we were able to build a well-functioning Microbial Fuel Cell and establish a protocol for measuring the power output of our electricity generating cultures. We built a stack of five Fuel Cells which was successfully used to power different LEDs and the motor of a small fan. Furthermore, a 3D model was designed, which can be printed out using a 3D printer. This model was made available for download on our wiki, so anyone interested can build a fuel cell of their own.[[Team:Bielefeld-Germany/Project/MFC#Results| Read more]].</p> | ||
| Line 92: | Line 94: | ||
==Riboflavin== | ==Riboflavin== | ||
| + | |||
| + | |||
*<p align="justify">Riboflavin possesses the ability to be a potent redox mediator. By turning the ''rib''-gene cluster from ''Shewanella oneidensis'' into a BioBrick and subsequently cloning it into the desired chassis ''Escherichia coli'', the iGEM Team Bielefeld was able to significantly raise the amount of riboflavin produced by ''E. coli''. <br> This means that the transformation of ''E. coli'' with <bbpart>BBa_K1172303</bbpart> and <bbpart>BBa_K1172306</bbpart>, respectively, represents a viable option when considering genetic optimization of microorganisms intended for usage in Microbial Fuel Cells (MFC).[[Team:Bielefeld-Germany/Project/Riboflavine#Results| Read more]].</p> | *<p align="justify">Riboflavin possesses the ability to be a potent redox mediator. By turning the ''rib''-gene cluster from ''Shewanella oneidensis'' into a BioBrick and subsequently cloning it into the desired chassis ''Escherichia coli'', the iGEM Team Bielefeld was able to significantly raise the amount of riboflavin produced by ''E. coli''. <br> This means that the transformation of ''E. coli'' with <bbpart>BBa_K1172303</bbpart> and <bbpart>BBa_K1172306</bbpart>, respectively, represents a viable option when considering genetic optimization of microorganisms intended for usage in Microbial Fuel Cells (MFC).[[Team:Bielefeld-Germany/Project/Riboflavine#Results| Read more]].</p> | ||
==Phenazine== | ==Phenazine== | ||
| + | |||
| + | |||
*<p align="justify">There are no practical results in this section. The project was left aside after unsuccessful attempts to amplify the phenazine-coding fragment from ''Pseudomonas fluorescens sp.''</p> | *<p align="justify">There are no practical results in this section. The project was left aside after unsuccessful attempts to amplify the phenazine-coding fragment from ''Pseudomonas fluorescens sp.''</p> | ||
| Line 106: | Line 112: | ||
==Cytochromes== | ==Cytochromes== | ||
| + | |||
| + | |||
*<p align="justify">We isolated the ''mtrCAB'' gene cluster from ''Shewanella oneidensis'' MR-1 and cloned it into the backbone pSB1C3, generating the BioBrick <bbpart>K1172401</bbpart>. This gene cluster was combined with three promoters and ribosome binding sites of varying strength, thus engineering the devices <bbpart>K1172403</bbpart>, <bbpart>K1172404</bbpart>, <bbpart>K1172405</bbpart>. We transformed these devices into our host organism ''Escherichia coli'', but could not verify correct expression and localization. [[Team:Bielefeld-Germany/Project/Cytochromes#Results| Read more]].</p> | *<p align="justify">We isolated the ''mtrCAB'' gene cluster from ''Shewanella oneidensis'' MR-1 and cloned it into the backbone pSB1C3, generating the BioBrick <bbpart>K1172401</bbpart>. This gene cluster was combined with three promoters and ribosome binding sites of varying strength, thus engineering the devices <bbpart>K1172403</bbpart>, <bbpart>K1172404</bbpart>, <bbpart>K1172405</bbpart>. We transformed these devices into our host organism ''Escherichia coli'', but could not verify correct expression and localization. [[Team:Bielefeld-Germany/Project/Cytochromes#Results| Read more]].</p> | ||
==Nanowires== | ==Nanowires== | ||
| + | |||
| + | |||
*<p align="justify">After the successful amplification of the corresponding ''Geobacter sulfurreducens'' gene clusters, the transformation of these DNA sequences into the BioBrick form via Gibson Assembly led to substantial problems. Because of this and additional issues, the nanowire project was stopped until further notice to concentrate on the other projects. [[Team:Bielefeld-Germany/Project/Nanowires#Results| Read more]].</p> | *<p align="justify">After the successful amplification of the corresponding ''Geobacter sulfurreducens'' gene clusters, the transformation of these DNA sequences into the BioBrick form via Gibson Assembly led to substantial problems. Because of this and additional issues, the nanowire project was stopped until further notice to concentrate on the other projects. [[Team:Bielefeld-Germany/Project/Nanowires#Results| Read more]].</p> | ||
==Biosafety== | ==Biosafety== | ||
| + | |||
| + | |||
*<p align="justify">For further real world applications of the Microbial Fuel Cell outside the laboratory, we designed three novel Biosafety-Systems called araCtive, TetOR alive and Lac of Growth. These Biosafety-Systems are characterized by a higher plasmid stability and a double-kill switch mechanism, providing a higher resistance towards undesirable mutations. All three Biosafety-Systems were analyzed and compared and with each other, resulting in three functional Biosafety-Systems, that differ in leakiness of the toxic gene product and can therefore be used for any desirable application [[Team:Bielefeld-Germany/Biosafety/Biosafety_System| Read more]].</p> | *<p align="justify">For further real world applications of the Microbial Fuel Cell outside the laboratory, we designed three novel Biosafety-Systems called araCtive, TetOR alive and Lac of Growth. These Biosafety-Systems are characterized by a higher plasmid stability and a double-kill switch mechanism, providing a higher resistance towards undesirable mutations. All three Biosafety-Systems were analyzed and compared and with each other, resulting in three functional Biosafety-Systems, that differ in leakiness of the toxic gene product and can therefore be used for any desirable application [[Team:Bielefeld-Germany/Biosafety/Biosafety_System| Read more]].</p> | ||
<br><br><br> | <br><br><br> | ||
Revision as of 20:33, 27 October 2013
Results
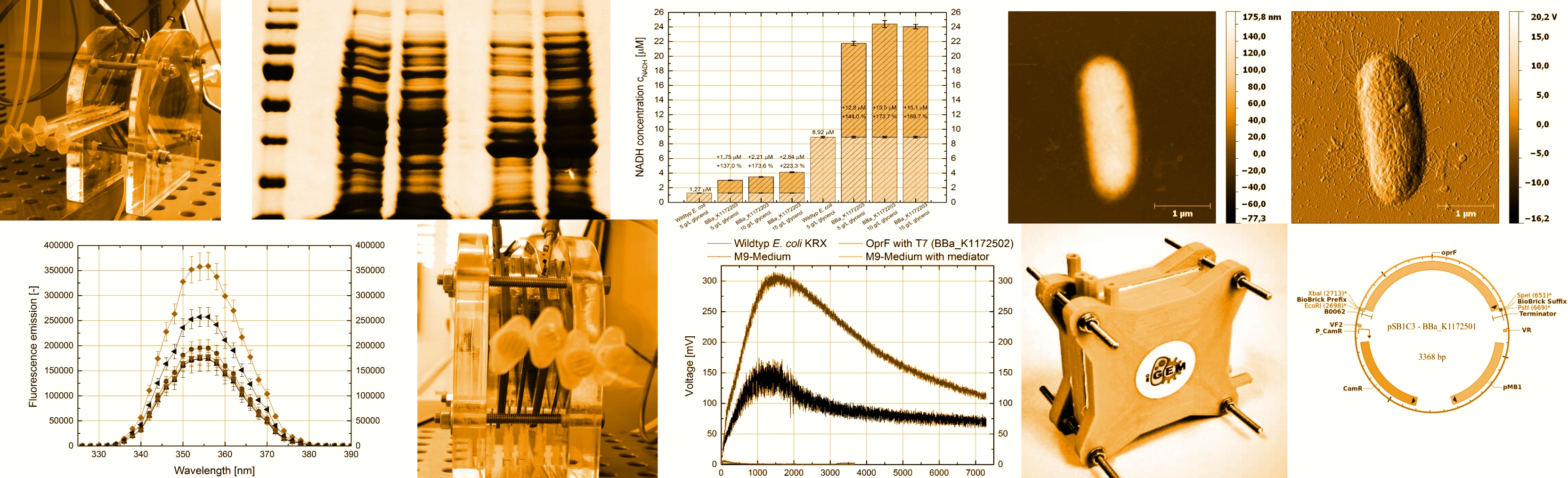
MFC
After extensive optimization, we were able to build a well-functioning Microbial Fuel Cell and establish a protocol for measuring the power output of our electricity generating cultures. We built a stack of five Fuel Cells which was successfully used to power different LEDs and the motor of a small fan. Furthermore, a 3D model was designed, which can be printed out using a 3D printer. This model was made available for download on our wiki, so anyone interested can build a fuel cell of their own. Read more.
Glycerol dehydrogenase
We demonstrate that engineering E. coli by introduction of the oxidoreductase glycerol dehydrogenase via gene manipulation can greatly improve the mediator production and power generation. We can show an extreme increase in the intracellular- and extracellular NADH concentration. This leads to 40 % enhanced average electric power in our Microbial Fuel Cell. The overexpression of the glycerol dehydrogenase from Escherichia coli is a great genetic optimization for electron shuttle-mediated extracellular electron transfer from bacteria to electrodes. Read more.
Riboflavin
Riboflavin possesses the ability to be a potent redox mediator. By turning the rib-gene cluster from Shewanella oneidensis into a BioBrick and subsequently cloning it into the desired chassis Escherichia coli, the iGEM Team Bielefeld was able to significantly raise the amount of riboflavin produced by E. coli.
This means that the transformation of E. coli with <bbpart>BBa_K1172303</bbpart> and <bbpart>BBa_K1172306</bbpart>, respectively, represents a viable option when considering genetic optimization of microorganisms intended for usage in Microbial Fuel Cells (MFC). Read more.
Phenazine
There are no practical results in this section. The project was left aside after unsuccessful attempts to amplify the phenazine-coding fragment from Pseudomonas fluorescens sp.
Porins
We heterologously expressed the porin protein OprF from Pseudomonas fluorescens in Escherichia coli. This leads to dramatically increased membrane permeability and a much higher current output in comparison to its parental strain (E. coli KRX) caused by improved electron shuttle-mediated extracellular electron transfer. This is a great genetic strategy to improve electricity generation by microorganisms. Read more.
Cytochromes
We isolated the mtrCAB gene cluster from Shewanella oneidensis MR-1 and cloned it into the backbone pSB1C3, generating the BioBrick <bbpart>K1172401</bbpart>. This gene cluster was combined with three promoters and ribosome binding sites of varying strength, thus engineering the devices <bbpart>K1172403</bbpart>, <bbpart>K1172404</bbpart>, <bbpart>K1172405</bbpart>. We transformed these devices into our host organism Escherichia coli, but could not verify correct expression and localization. Read more.
Nanowires
After the successful amplification of the corresponding Geobacter sulfurreducens gene clusters, the transformation of these DNA sequences into the BioBrick form via Gibson Assembly led to substantial problems. Because of this and additional issues, the nanowire project was stopped until further notice to concentrate on the other projects. Read more.
Biosafety
For further real world applications of the Microbial Fuel Cell outside the laboratory, we designed three novel Biosafety-Systems called araCtive, TetOR alive and Lac of Growth. These Biosafety-Systems are characterized by a higher plasmid stability and a double-kill switch mechanism, providing a higher resistance towards undesirable mutations. All three Biosafety-Systems were analyzed and compared and with each other, resulting in three functional Biosafety-Systems, that differ in leakiness of the toxic gene product and can therefore be used for any desirable application Read more.
 "
"