Team:Calgary/Project/OurSensor/Detector
From 2013.igem.org
| Line 168: | Line 168: | ||
<img src="https://static.igem.org/mediawiki/2013/9/95/Calary2013TALEABlotWithKinetics.png" width="70%" height="70%"> | <img src="https://static.igem.org/mediawiki/2013/9/95/Calary2013TALEABlotWithKinetics.png" width="70%" height="70%"> | ||
<figcaption> | <figcaption> | ||
| - | <p><b>Figure 24.</b> (A) Dot blot of ferriTALE A exposed to FAM labeled DNA containing the A target sequence. 1.5ug of TALEA+Kcoil and 1ug of ferritin with Ecoil were incubated for 1 | + | <p><b>Figure 24.</b> (A) Dot blot of ferriTALE A exposed to FAM labeled DNA containing the A target sequence. 1.5ug of TALEA+Kcoil and 1ug of ferritin with Ecoil were incubated for 1 hour to make the ferriTALE complex and the complex was blotted on a strip. The blots were then exposed to 1.66 mM FAM-labeled [A] (TALEA target site) for 1 to 90 minutes as indicated on the strips. "x" is a ferriTALE that was exposed to FAM labeled DNA prior to being blotted onto the nitrocellulose. The kinetics from the densitometry is shown in section B of the figure. The Kd from this plot was determined to be 293nM</p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| Line 175: | Line 175: | ||
<img src="https://static.igem.org/mediawiki/2013/d/db/Calgary2013TALEBBlotWithKinetics.png" width="70%" height="70%"> | <img src="https://static.igem.org/mediawiki/2013/d/db/Calgary2013TALEBBlotWithKinetics.png" width="70%" height="70%"> | ||
<figcaption> | <figcaption> | ||
| - | <p><b>Figure 25.</b> (A) Dot blot of ferriTALE B exposed to FAM labeled DNA containing the B target sequence. | + | <p><b>Figure 25.</b> (A) Dot blot of ferriTALE B exposed to FAM labeled DNA containing the B target sequence. 1ug of ferritin fused to Ecoil was incubated with 2ug of TALEB fused to kcoil for 1hour to make the FerriTALEB complex. Subsequently the complex was blotted on the nitrocellulose strip. The blots were then exposed to 1.66 mM FAM labeled DNA for 1 to 90 minutes as indicated on the strips. The controls are to the right, with "ftn" being ferritin only, "np" being no protein, and "D-" being no DNA exposure. The kinetics from the densitometry is shown in section B of the figure. The Kd from this plot was determined to be 66nM.</p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
Revision as of 00:21, 29 October 2013
Detector
Detector

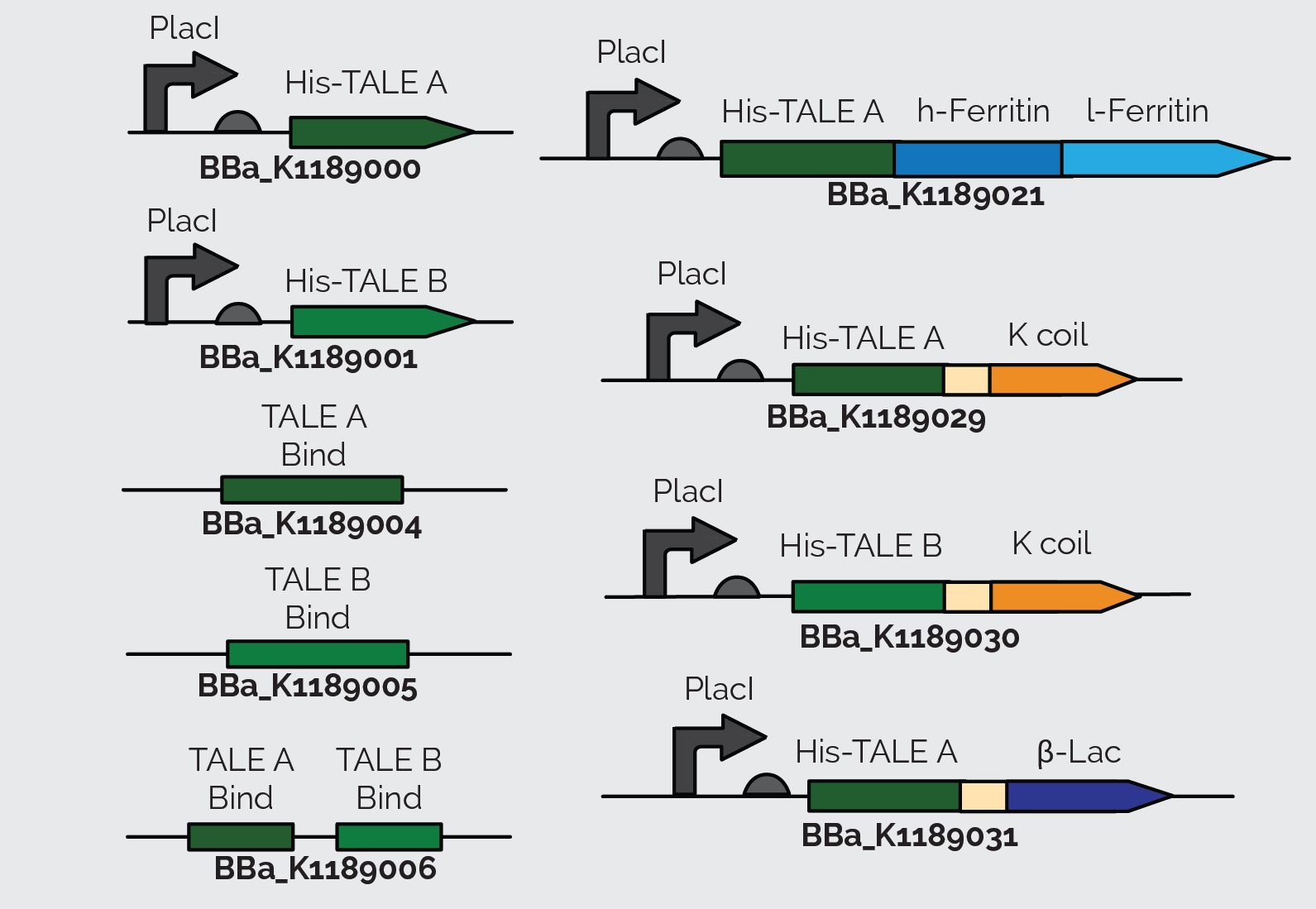
For the DNA detector in our system, we decided to use Transcription Activator-Like Effectors (TALEs). These are naturally occurring proteins that specifically bind to DNA. TALEs are an advantageous tool in synthetic biology because they can be engineered to recognize any chosen DNA sequence as long as it begins with a thymidine. For our system, we want them to bind to a sequence in enterohemorrhagic E. coli (EHEC). We have engineered TALEs that are specially designed to bind segments of the stx2 gene in the EHEC genome (BBa_K1189032 and BBa_K1189033). Click here to find out more about how we designed our TALEs. This summer we also worked with TALEs from the iGEM Parts Registry to build and test our constructs, for our proof of concept. To see what we accomplished with our TALEs, check out our results section. To learn more about TALE proteins, see our background section.
Background
Transcription Activator-Like Effectors (TALEs) are proteins produced by bacteria of the genus Xanthomonas and secreted into plant cells. These naturally occurring TALEs play a key role in bacterial infection, as they are responsible for upregulation of the host genes required for pathogenic growth and expansion (Mussolino & Cathomen, 2012).
All TALEs are made up of three main parts: N-terminus, DNA binding domain, and C-terminus. The N-terminus contains a type III secretion signal (T3SS), which allows the proteins to be translocated from the bacterium and into the plant cell. The C-terminus contains nuclear localization signals (NLS) and an acidic activation domain (AAD). The DNA binding domain, also termed repeat region, mediates DNA recognition through tandem repeats of 33 to 35 amino acids residues each (Bogdanove et al., 2010). The binding domain usually comprises 15.5 to 19.5 single repeats (Figure 1). The last repeat, close to the C-terminus, is called “half-repeat,” because it generally is 20 amino acids in length. Although the modules have conserved sequences, polymorphisms are found in residues 12 and 13, also known as “repeat-variable di-residues” (RVD). RVDs can be specific for one nucleotide or a number of nucleotides; therefore, 19.5 repeat units target a specific 20-nucleotide sequence in the DNA (Mussolino & Cathomen, 2012).

Figure 1.(A) Schematic representation of a TAL effector with the DNA binding domain in red. (B) 3D structure of TALEs obtained from our team’s work in Autodesk Maya. To learn more about our modeling, click here.
When in contact with the DNA, the TALE aligns the N-terminal to the 5' end of the DNA and the C-terminal to the 3’ end of the DNA. Each repeat is made of two alpha helices connected by a three-residue loop, two of which amino acids comprise an RVD. Although both amino acids 12 and 13 are responsible for base specificity, the TALE-DNA interaction happens through intermolecular bonds between residue 13 and the target base in the major groove of the DNA. Residue 12 plays a role in stabilizing the RVD loop (Meckler et al., 2013).
Over 20 different RVDs have been identified in TAL effectors. However, four of them appear in 75% of the repeats: HD, NG, NI and NN (Bogdanove et al., 2010). Quantitative analysis of DNA-TALE interactions by Meckler et al. (2013) revealed that the binding affinity is affected by the RVDs in the following order (The letters in brackets show the base that the RVD binds to): NG (T) > HD (C) ~ NN (G)>> NI (A) > NK (G). NG, specific to thymine, and HD, specific to cytosine, are strong RVDs. NN binds both guanine and adenine, but it prefers guanine. NK also interacts with guanine, but with 103-fold lower affinity. NI is specific for adenine, but it has low affinity when compared to strong RVDs such as NG and HD (Meckler et al., 2013). Although less common, another naturally occurring RVD, NH, was described to bind strongly to guanine (Cong et al., 2013). NS binds to any of the four bases and it is present in naturally occurring TALEs such as AvrBs3 from Xanthomonas campestris (Boch et al., 2009).
In addition to RVDs, the DNA binding affinity is also subject to polarity effects. Point mutations at the 5’ end of the target sequence affect TALE-DNA recognition more than the ones at the 3’ end (Meckler et al., 2013). Taking this in consideration, recommendations for TALE design include incorporation of strong RVDs close to the N-terminus (Streubel et al., 2012).
Due to the modular and easy to engineer nature of TALEs they can be designed to bind virtually any DNA sequence, making them a powerful tool in synthetic biology. They have been extensively used for genome editing in the literature (Mussolino et al, 2011) by fusing DNA cleavage domains of endonucleases to serve as custom restriction enzymes (Beurdeley et al., 2013). These engineered proteins are termed TALENs or Transcription Activator-Like Effector Nucleases. TALENs are also used in gene knockout because they promote gene disruption (Bogdanove & Voytas, 2011). Slovenia 2012 iGEM team designed and created repressor TAL effectors by adding repressor and activator domains to TALEs to control gene expression.
Our team, however, proposes an innovative application for TAL effectors: Using TALE as a nucleotide biosensor. We engineered TALEs to detect entero-haemorrhagic bacteria in feces of BBa_K782004 in cattle populations. As sensors, TALEs can bind to specific regions of the Shiga toxin II gene (stx2) and capture the DNA of interest from a feces sample, making it available for a second TALE, whose binding domain is specific for another region of stx2. This second TALE is connected to a reporter, which makes the TALE-DNA interaction visible within a short period of time.
Engineered TALEs

To design the TALEs that bind to EHEC we had to take a few things into consideration. Firstly, we had to carefully identify regions that would both be optimal for TALE binding and be specific to EHEC species. After thorough literature search we determined that Shiga Toxin, stx2 is a gene found in all EHEC organisms. Additionally, we insured that the TALE binding sequences were not found in organisms surrounding the cattle such as grass consumed by the cattle, the cow’s DNA, or another commensal microorganism in the cow gut. The stx2 gene produces shiga toxin that causes illness in humans. Shiga toxin is an AB5 toxin with 5 B subunits that bind to the globotriaosylceramide (Gb3) and then the A subunit is internalized by the host cell and cleaves ribosomal RNA. Cattle do not have the Gb3 as a surface marker and therefore carry EHEC asymptomatically. Since TALEs do not have 100% specificity we designed a two TALE system to increase the specificity and reduce the false negatives. We designed two TALEs that bind to two different regions on the stx2 gene.
In addition to maximizing specificity, we also tried to design both TALEs to have the highest possible binding affinity. Since our final system will be a strip that will be dipped into several solutions we require the TALEs to bind strongly to the DNA to keep it in place on our strip. Thus, if our TALEs do not bind strongly to the DNA, the system would produce false negatives. To increase the binding affinity of the TALEs, we picked the pyrimidine-rich regions of the stx2 gene. The relative binding affinity of different RVDs to their respective nucleotides is as follows: NG (1)> NN (0.18) ~ HD (0.16) >> NI (0.0016)> NK (0.00016) where they bind to T, G, C, A, and G, respectively (Meckler et al. 2013). Therefore, picking regions rich in thymines and cytosines would greatly enhance the binding affinity of the TALE to the DNA segment.
Another important factor in the selection of the two TALE target sequences was the distance between them. The distance had to be great enough so that the TALE and the FerriTALE could bind without steric hindrance but short enough so that the possibility of these two target sequences shearing and separating is reduced. In order for our system to work, the two regions of DNA that the TALEs bind to must remain contiguous, i.e., on the same stretch of DNA. Therefore, we picked target sequences as close to each other as possible to decrease the chances of a cut between the two target sites. We determined the ideal distance between the two binding sites were 200 bp.
The TALEs were designed to be extremely specific. Based on the considerations explained above, a number of possible target sites were selected. To determine the most specific pair, we conducted BLAST searches on each candidate TALE target sequence separately. As expected, we observed a huge number of alignments with EHEC strains. We also found some partial alignments in non-EHEC organisms, which were screened to determine if they can be found in the environment of the cattle. For example, consider the two selected target sites to be [1] and [2]. If [1] had a 90% alignment to a region in the human genome, we checked to see if [2] could also be found in the human genome. If it was found, then we checked to see whether both [1] and [2] were found on the same chromosome; if they were found on different chromosomes, meaning they were not physically attached, the system would not detect it as a false positive. If they were found on the same chromosome, we determined which end of the target sequences aligned with the similar DNA sequence. This is important as TALEs are polar and bind significantly stronger to the 5’ end of their target sequence compared to the 3’ end (Meckler et al. 2013). Therefore, if the alignment included the 0th to 10th nucleotide of the target sequence, that specific two target site combination could be ruled out. To get a better idea of how we designed the TALEs, please refer to Figure 3 below.

Figure 3.This flow chart shows the process to design TALEs specific to stx2 gene. First we picked two target sequences ([1] and [2]) based on the consideration mentioned in the above text. Then we used this flowchart to ensure the combination of the two target sites would not produce a false positive.

Figure 4. Alignment of both of the chosen EHEC target sequences. Notice how the target sequences start with a T and are pyrimidine rich. The two TALEs (BBa_K1189032 and BBa_K1189033) are designed to bind to the 19 bp target sequences.
Proof of Concept
The TALEs are a very fundamental part of our project. In order to have a functioning system for E. coli detection, we need to have proteins that will successfully recognize and bind the Stx2 gene. However, before we can actually create and use TALEs to bind to enterohemorrhagic E. coli DNA, wanted to build a proof of concept to optimize our assays and to ensure that TALEs work in our hands. On the iGEM parts registry we found TALEs that 2012 Slovenia iGEM team synthesized for a previous project. TALEs are very large proteins and are expensive to synthesize therefore, we decided to use these TALEs as a proof of concept. We also saw this as an opportunity to use and build upon parts made by former iGEM teams. Thus, we ordered their three TALEs: TALE A (BBa_K782004), TALE B (BBa_K782006), and TALE D (BBa_K782005).
We used TALE A(BBa_K782004) and TALE B (BBa_K782006) to build the proof of concept and resume testing. To test our TALEs, we synthesized the target sequences that bind to Slovenia's TALEs. We made similar variants of the target site by changing 1-2 bp at a time to characterize the binding affinity of the TALEs (BBa_K1189004, BBa_K1189005). Consequently, we can determine the specificity with which the proteins bind and how they might respond to DNA not belonging to enterohemorrhagic E. coli. This will help us define how specific we expect our final system to be.
When we sequenced TALE B (BBa_K782006), we discovered that it had a small mutated segment. We expected a sequence of AGCAATGGG in the repeat variable di-residue of the second repeat. However, the sequence was actually TCCCACGAC. This meant that the required target sequence at this position was a C, and not a T, as the parts registry web page indicates. The two TALEs also had a Kozak sequence at the front of the sequence which blocked expression in E. coli as shown in Figure 23. We removed the Kozak sequence before using TALE A and TALE B to construct BBa_K1189000 and BBa_K1189001 respectively.
Since, our system uses two TALEs we made a plasmid containing both TALE A and B target site annotated [A] and [B] respectively with some non specific nucleotides in between, such that the distance between [A] and [B] is similar to the same distance between the two target sites of the TALEs we engineered for EHEC detection (appx. 200bp).
We also incorporated TALE A (BBa_K782004) and TALE B (BBa_K782006) in our other constructs. We synthesized TALE A and TALE B with a K coil under a IPTG inducible promoter. The K coil binds to E coil, its binding partner in vitro which is fused to our Prussian blue reporter.
As a back up plan, we considered using β-lactamase as a reporter. Therefore, we synthesized TALE A linked to β-lactamase (BBa_K1189031). In all of TALE-A bound to the K-coil under a LacI promoter(BBa_K1189029), TALE-B bound to the K-coil under a LacI promoter (BBa_K1189030), and TALE-A bound to a β-lactamase under a LacI promoter (BBa_K1189031) we tried to further improve the TALEs, so that they are more useful to us and the general iGEM community, by codon optimizing the TALEs for E. coli. We also took out the nuclear localization sequence (NLS) on the C-terminus of the TALEs, since E. coli does not have a nucleus.
These parts were also designed in a way so that they could be used for a wide variety of applications. There is a KasI restriction site incorporated immediately following the TALE. This introduces the chemically inert amino acids, alanine and glycine, which are what the linker is made of and would not disrupt the functionality of either the TALE or the protein it is fused to. This allows other teams to easily switch out the TALE in the part with their own engineered TALE. This means that our system can potentially be used as a platform technology. By designing a TALE specific to any DNA target site, any one can use this technology to sense their DNA of interest.
Results
TALE Target Sequences
We produced specific DNA target sequences for both TALE A [A] and TALE B [B]. We designed primers that would produce these target sequences in addition to about 480bp of junk DNA (to facilitate cloning) in a PCR reaction. Our target sequences were inserted into a pSB1C3 backbone. Our verification digest of the target sequences is shown in Figure 15. The backbone is the highest band at around 2070bp and our target sequences are represented by the lowest bands at around 500bp. These target sequences were sent for sequencing and have been sequence verified.

Figure 16. Verification digest of minipreps of transformed mutated TALE B and non mutated TALE A and TALE B target sequences and BBa_J04500 cut with AflII and PstI. All of the TALE target sequences lanes except the twelfth lanes show bands at 500bp and 2100bp, the TALE target sequences and backbone are expected at 501bp and 2070bp. No contamination is observed in the negative control without DNA. No unexpected sized bands are seen. The gel was 1% agarose and was run at 100 V for an hour.
Once the individual target sequences were made, we designed new primers to create a target sequence construct with both the target sequences together. We did a KAPA PCR to produce our linear product. The PCR product consisted of the two target sequences, with about 200bp of junk DNA in between, and biobrick cut sites on either side. The results of the PCR are shown in Figure 16. We then inserted the linear PCR product into a pSB1C3 backbone. This construct was successfully transformed, mini prepped and sequence verified. Because TALE B was discovered to have a mutation, we have two versions of this construct. One has the original TALE B target sequence we created before we realized there was a mutation. The other contains the new target sequence we created for the mutated TALE B.

Figure 17. Kapa HiFi PCR using custom primers to extract construct of combined TALE target sequences [A] and [B]. The construct of the two target sequences contained about 200bp of junk DNA between the two target sites and was flanked by the biobrick cut sites on both side. Every lane shows bands at 500 bp and was expected at about 507bp. Some unexpected amplification is seen for this PCR. No contamination is observed in the negative control without template DNA. The gel was 1% agarose and was run at 100 V for an hour.
Protein Purification
After getting our DNA constructs ready, we moved on to express and purify them. We first did a mini expression of TALE A (BBa_K1189000) and TALE B (BBa_K1189001) to optimize expression of these TALEs. Figure 17 shows the western blot we performed on the crude lysate of TALE A and TALE B. Following optimization we purified our TALEs using affinity chromatography through Ni-NTA columns.
Figure 18. Western Blot of crude lysate of TALE A under a LacI promoter and BBa_J04500+TALE B.The blots were probed with anti-His antibody at a concentration of 1:1000 raised in mice and probed with a anti-mouse secondary antibody at a concentration of 1:20000. 
Following the same conditions for large scale protein expression and purification we were able to purify TALE A linked to Kcoil (BBa_K1189029) and TALE-A linked to β lactamase under a LacI promoter (BBa_K1189031). Figure 18 shows the western blot we performed on TALE A linked to K coil(BBa_K1189030) and Figure 19 shows the western blot for TALE-A linked to β lactamase under a LacI promtoer(BBa_K1189031). Figures 20 and 21 show the SDS-PAGE we ran on the elutions we collected after Ni-NTA purification of TALE B linked to Kcoil (BBa_K1189030) and TALE-A linked to βlactamase under a LacI promoter (BBa_K1189031), respectively.
Figure 19. Western Blot of TALE A with a Kcoil (BBa_K1189029).The blots were probed with anti-His antibody at a concentration of 1:1000 raised in mice and probed with a anti-mouse secondary antibody at a concentration of 1:20000. Figure 20.Western Blot of TALE-A linked to β-lactamase (BBa_K1189031).The blots were probed with anti-His antibody at a concentration of 1:1000 raised in mice and probed with a anti-mouse secondary antibody at a concentration of 1:20000. Figure 21.SDS Page gel of pSBIC3+BBa_J04500+His+TALE A+Link+Kcoil. Bands were expected and seen at 82kDa. The purified lysate from E. coli BL21 cells was run through the AKTA, an automatic His tag affinity chromatography machine. Figure 22.SDS Page gel of TALE A linked to β-lac under a LacI promoter.



We initially used the TALE A (BBa_K782004) and TALE B (BBa_K782006) from the parts registry and added a lacI promoter and RBS (BBa_J04500) upstream of it. However, since these TALEs were initially used in eukaryotic cells, they have a Kozak sequence at the start of the TALE A (BBa_K782004) and TALE B (BBa_K782006), which prevents the expression of these proteins. To improve the part and make it useful to everyone using E. coli as a chassis, we removed the Kozak sequence from the TALEs. All the parts that we submitted to the registry have the Kozak sequence removed and can be expressed in E. coli, as shown above. The western blot in Figure 22 below shows expression of TALE B with (BBa_J04500 + BBa_K782006) and TALE B without (BBa_K1189001) the Kozak sequence in E. coli.
Figure 23. Western blot of crude samples of TALE B with a his tag without and with the Kozak sequence. The blots were probed with anti-His antibody at a concentration of 1:1000 raised in mice and probed with a anti-mouse secondary antibody at a concentration of 1:20000. 
For our system, we designed two TALEs for two separate segments of the stx2 gene, which should dramatically increase the specificity for EHEC organisms. Once the individual EHEC target sequences were made, we designed new primers to create a target sequence construct with both the EHEC target sequences together. We did a KAPA PCR to produce our linear product. The PCR product consisted of the two target sequences, with about 200bp of junk DNA in between, and biobrick cut sites on either side. We then inserted the linear PCR product into a pSB1C3 backbone. This construct was successfully transformed, mini prepped and sequence verified. Figure 23 below shows the two TALEs (BBa_K1189032 and BBa_K1189033) attached together.
Figure 24. (A) Dot blot of ferriTALE A exposed to FAM labeled DNA containing the A target sequence. 1.5ug of TALEA+Kcoil and 1ug of ferritin with Ecoil were incubated for 1 hour to make the ferriTALE complex and the complex was blotted on a strip. The blots were then exposed to 1.66 mM FAM-labeled [A] (TALEA target site) for 1 to 90 minutes as indicated on the strips. "x" is a ferriTALE that was exposed to FAM labeled DNA prior to being blotted onto the nitrocellulose. The kinetics from the densitometry is shown in section B of the figure. The Kd from this plot was determined to be 293nM Figure 25. (A) Dot blot of ferriTALE B exposed to FAM labeled DNA containing the B target sequence. 1ug of ferritin fused to Ecoil was incubated with 2ug of TALEB fused to kcoil for 1hour to make the FerriTALEB complex. Subsequently the complex was blotted on the nitrocellulose strip. The blots were then exposed to 1.66 mM FAM labeled DNA for 1 to 90 minutes as indicated on the strips. The controls are to the right, with "ftn" being ferritin only, "np" being no protein, and "D-" being no DNA exposure. The kinetics from the densitometry is shown in section B of the figure. The Kd from this plot was determined to be 66nM.

 "
"
