Team:Colombia Uniandes/Parameters
From 2013.igem.org
(→Nickel removal system) |
(→Nickel removal system) |
||
| Line 77: | Line 77: | ||
'''Sensitivity Analysis''' | '''Sensitivity Analysis''' | ||
| - | + | A complete sensitivity analysis was performed in order to see which parameters have a significant importance in the model. You can see here the analysis for each of the 12 parameters of the system. [[File:SensNicke.pdf]] | |
Revision as of 16:36, 1 September 2013
Parameters
The second step in the deterministic model is to find the parameters that define the system. There are 3 possible ways to do this:
- Literature: Many of the genes that we use in iGEM have been characterised before, and the constants that model their behaviour have been found. Sometimes an exact value is not found, so we have to find at least the order of magnitude or a range where the parameter can be found
- Experiments: This is the most reliable way of knowing the parameters. But in our case we need the to perform de deterministic model first in order to see if the system works as expected in the lab. So the experimental way can only work to check if the parameters were correctly found.
- Screening: If the parameters we are looking of are not found in literature we proceed to perform a screening of parameters as described in Colombia 2012 Team Wiki
Below is shown how we found the parameters for both of the projects:
Glucocorticoid sensor
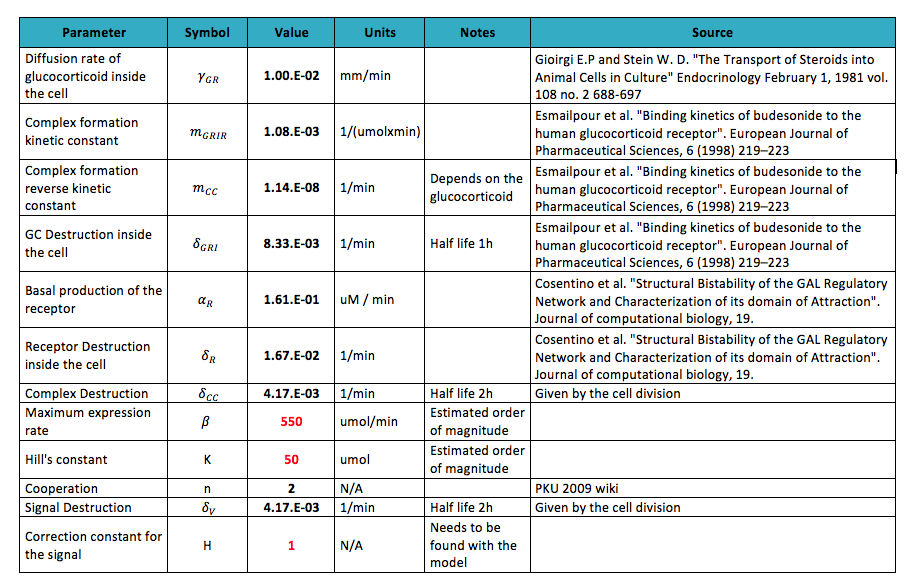
After performing the literature research for the parameters the following values were found:
- The values for these parameter varied a lot depending on the glucocorticoid detected; as in the lab is planned to test the circuit with Dexamethasone all the parameters were done for this glucocorticoid.
- The values in red are the parameters that could not be found. An estimated order of magnitude was assessed supported in the literature read.
- As the part of the chimera that is going to activate transcription is Gal4p the values for the activation of the gene are assumed to be the same as the pure protein.
- Some of the destruction rates are not found so it is assumed the concentration of the protein is significantly reduced only when the cell divides
Sensitivity Analysis
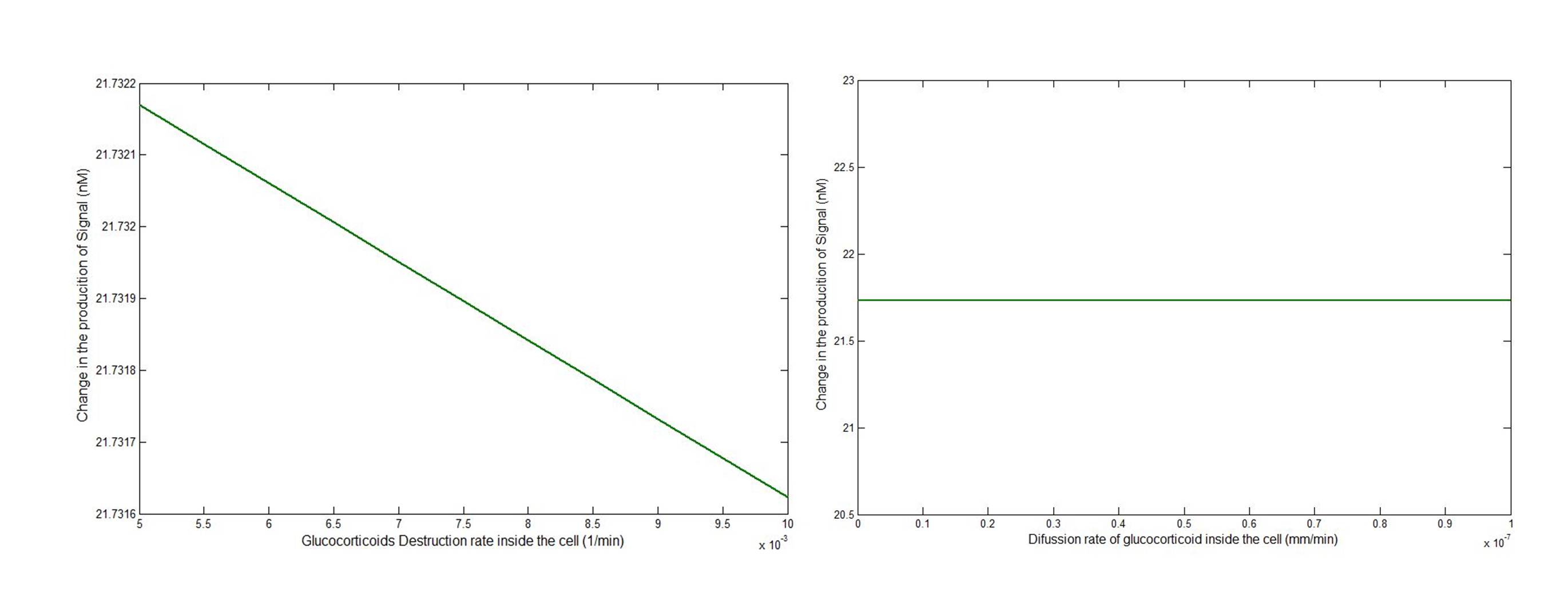
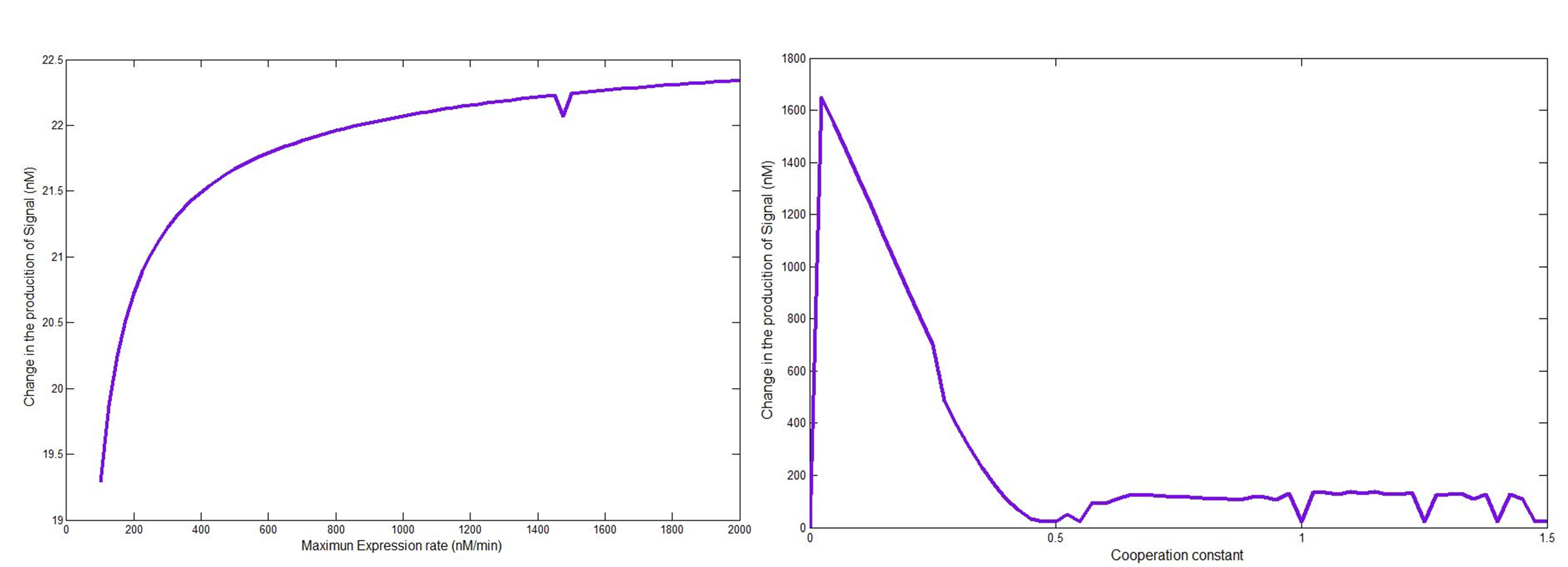
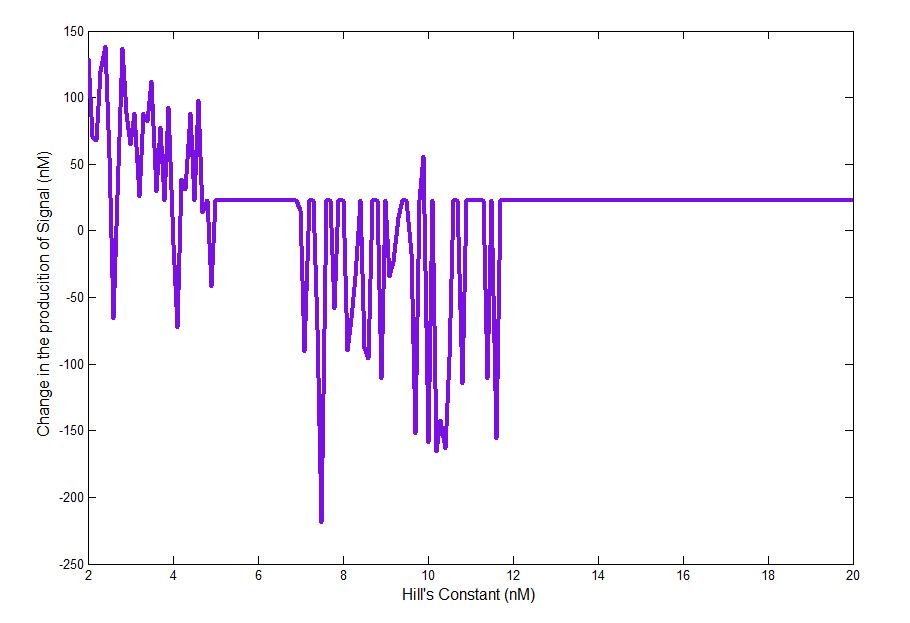
In order to see which if the parameters had a significant effect in the signal response, we performed a Sensitivity analysis. For each parameter, we took the value found in the literature (or guessed value) and evaluated the model in a range that varied from one order of magnitude below to one order of magnitude greater. Depending on how much the response varied to each parameter the screening was planned.
After perfoming the analysis we found some parameters that had little effect on the desired response of the system. For example the destruction rate of Glucocorticoid inside the cell and the reverse kinetic constant for the formation of the complex:
The parameters related to the hill expression had a significant effect on the desired response, for this reason and given the fact that two of the three parameters had an estimated order of magnitud the screening of parameters was focused on this part of the model.
You can see the complete Sensitivity Analysis here.
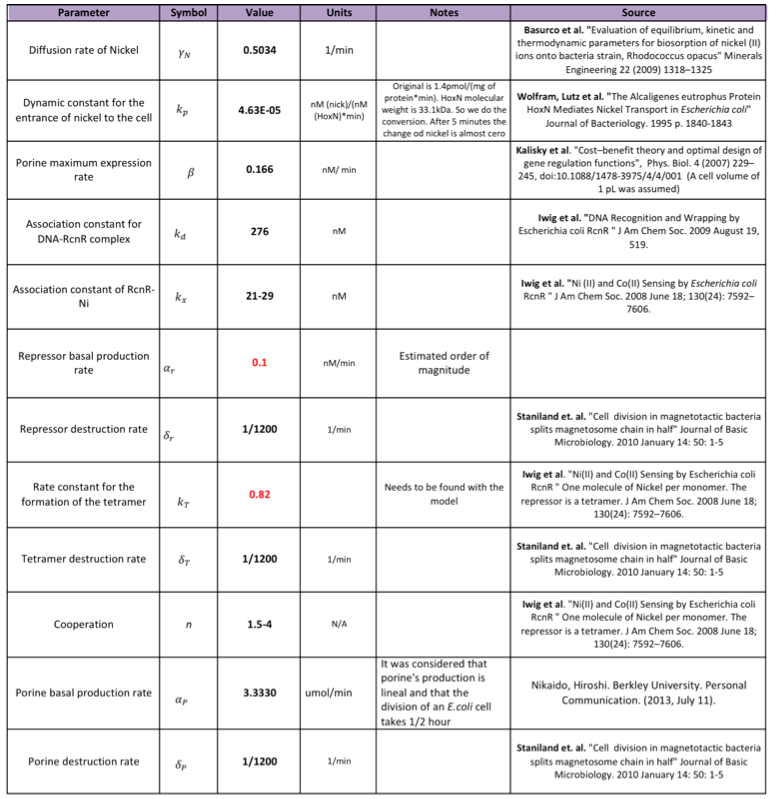
Nickel removal system
After a research for parameters in the literature research was done the following values were found:
- The values in red are the parameters that could not be found. An estimated order of magnitude was assessed supported in the literature read.
- Some of the destruction rates are not found so it is assumed the concentration of the protein is significantly reduced only when the cell divides
Sensitivity Analysis
A complete sensitivity analysis was performed in order to see which parameters have a significant importance in the model. You can see here the analysis for each of the 12 parameters of the system. File:SensNicke.pdf
 "
"