Team:Bielefeld-Germany/Project/Riboflavine
From 2013.igem.org
| Line 170: | Line 170: | ||
[[File:igem_bielefied2013_ribcluster.png|thumb|400px|left|'''Figure 5:''' Initial ''rib''-gene cluster from ''Shewanella oneidensis MR-1''. Three illegal restriction sites are marked with an asterisk]]<br> | [[File:igem_bielefied2013_ribcluster.png|thumb|400px|left|'''Figure 5:''' Initial ''rib''-gene cluster from ''Shewanella oneidensis MR-1''. Three illegal restriction sites are marked with an asterisk]]<br> | ||
[[File:igem_bielefied2013_ribcluster_cut3.png|thumb|400px|left|'''Figure 6:''' The strategy for base substitution: we designed three pairs of primers annealing on each illegal restriction site. As a result we amplified three fragments. For Primer Sequences look up [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/September Labjournal September]]]<br> | [[File:igem_bielefied2013_ribcluster_cut3.png|thumb|400px|left|'''Figure 6:''' The strategy for base substitution: we designed three pairs of primers annealing on each illegal restriction site. As a result we amplified three fragments. For Primer Sequences look up [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/September Labjournal September]]]<br> | ||
| - | [[File | + | [[File:Igem bielefied2013 ribcluster 2.png|thumb|400px|left|'''Figure 5:''' Ready ''rib''-gene cluster Biobrick from ''Shewanella oneidensis MR-1'' without illegal restriction sites.]] |
[[File:Igem bielefied2013 ribonorm.png|thumb|400px|left|'''Figure 7:'''Initial ''norM'' gene from ''Shewanella Oneidensis MR-1'' with one illegal restriction site]]<br> | [[File:Igem bielefied2013 ribonorm.png|thumb|400px|left|'''Figure 7:'''Initial ''norM'' gene from ''Shewanella Oneidensis MR-1'' with one illegal restriction site]]<br> | ||
[[File:igem_bielefied2013_ribonorm_ohne.png|thumb|400px|left|'''Figure 8:''' A strategy to remove an illegal restriction site: we created primers, that amplified two fragments X and Y. For Primer Sequences look up [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/September Labjournal September]]]<br> | [[File:igem_bielefied2013_ribonorm_ohne.png|thumb|400px|left|'''Figure 8:''' A strategy to remove an illegal restriction site: we created primers, that amplified two fragments X and Y. For Primer Sequences look up [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/September Labjournal September]]]<br> | ||
Revision as of 16:30, 4 October 2013
Riboflavin
Overview

Tubes from left to right:
- Tube 1: A 72-hour culture of E. Coli KRX [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1172306 BBa_K1172306]
- Tube 2: A 72-hour culture of E. Coli KRX [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1172306 BBa_K1172306], supernatant
- Tube 3: A 72-hour culture of E. Coli KRX [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1172306 BBa_K1172306], cell lysate
- Tube 4: A 72-hour culture of a wild type E. Coli KRX, supernatant
- Tube 5: A 72-hour culture of a wild type E. Coli KRX, cell lysate
[http://de.wikipedia.org/wiki/Riboflavin Riboflavin], or Vitamin B2 is a redox-active substance that plays an
essential role in living cells. Secreted into the medium, it can be
effectively used by some bacteria for electron transfer. Presence of riboflavin in anaerobic cultures leads to higher current flow in a
microbial fuel cell, which made riboflavin overproduction a suitable target for optimisation of our MFC.
We have shown that cloning of the riboflavin cluster from a metal-reducing bacterium Shewanella oneidnsis MR-1 in E. coli is sufficient to achieve
significant riboflavin overproduction detectable both in supernatant and in cells.
Theory
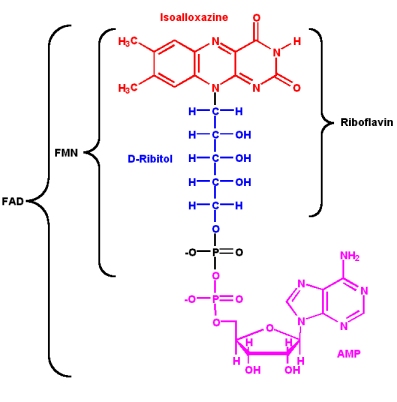
Since its discovery in 1879 and its first structural characterisation in the 1930’s, a lot of properties of riboflavin were elucidated. This substance is a precursor of FMN (flavin mononucleotide) and FAD (flavin adenine dinucleotide), which play an essential role as cofactors in many oxidative processes.
The modern name Riboflavin, also named Lactoflavin, is composed of two parts: «ribo» indicating the presence of the sugar alcohol ribitol, and «flavin» meaning «yellow»; to accentuate the yellow coloring of the oxidized molecule. Chemically this substance consists of two functional subunits, an already mentioned short-chain ribitol and a tricyclic heterosubstituted isoalloxazine ring.
The latter, also known as a riboflavin ring, exists in three redox states and is responsible for the diverse chemical activity of riboflavin. A fully oxidized quinone, a one-electron semiquinone and a fully reduced hydroquinone states are the three stages of riboflavin oxidation. In an aqueous solution, the quinone (fully oxidized) form of riboflavin has a typical yellow coloring. It becomes red in a semi-reduced anionic or blue in a neutral form and is colorless when fully reduced.
All these forms are present in different proportions in a living cell, making previous oxidation a necessary step if riboflavin analysis is to be conducted. Flavins have a typical yellow-green fluorescence in the UV light. The peaks of absorbance occur at 223, 266, 373 and 445 nm. The maximum fluorescence emission of the neutral solution is at 535 nm [Charles A. Abbas et al.,[http://mmbr.asm.org/content/75/2/321.full#ref-292 2011]]. These fluorimetric properties are widely used in the analysis of riboflavin.
Due to its structure, which allows a transfer of two electrons from hydrogen and hydrid ions, riboflavin can be imagined as a potential electron shuttle. It was previously known, that the electron transfer from the outer membrane-associated proteins to an inorganic electron acceptor is the main limiting growth factor for Fe(III)-reducing prokaryotes, so a few mechanisms were discovered, which showed how this process can be enhanced. One of them was a secretion of water-soluble redox mediators. It was proven, that secretion of riboflavin and FMN enhances the rate of insoluble mineral oxides reduction. Indeed, Shewanella Oneidensis, a facultative Fe-III respiring bacterium uses secreted riboflavin as its electron transmitter [Harald von Canstein et al.,[http://aem.asm.org/content/74/3/615.full 2008]]. Considering this acknowledgement, we decided to overproduce riboflavin in E.Coli to improve its efficiency in our MFC.
First of all, we have searched for a suitable microorganism, which has an active riboflavin cluster with known coding sequences. We have already used the remarkable versatility of Shewanella in order to clone the anaerobic respiratory chain (mtrCAB cluster), so now we were able to skip some initial steps, like the whole genome DNA isolation and advanced rapidly to more specific steps.
Before planning the cloning strategy, we checked [http://parts.igem.org/Main_Page the Parts registry] for any parts we could use or enhance. A [http://parts.igem.org/Part:BBa_K769203 ribC part] was listed, but it was neither submitted nor available.
We also had a close look on the [http://www.chem.qmul.ac.uk/iubmb/enzyme/reaction/misc/riboflavin2.html riboflavin biosynthesis pathway]. This process is well studied and a lot of appropriate literature is available. There are three types of riboflavin overproducers used in industry: yeast (Candida famate), fungi (Ashbya gossypii), and bacteria (Bacillus subtilis) [Overview, Seong Han Lim et al., [http://www.bbe.or.kr/storage/journal/BBE/6_2/6657/articlefile/article.pdf 2001]], but a prevailing majority of microorganisms also synthesise riboflavins in low concentrations. In E.Coli, for instance, genes are scattered through the whole genome and riboflavin is produced constitutively.
The biosynthesis of riboflavin starts with ribulose-5-phosphate and GTP, converted to formate and DHBP (L-3,4-dihydroxybutan-2-one 4-phosphate). The final stage involves forming of the third isoalloxazine ring by an exchange of a 4-carbon part, catalysed by riboflavin synthetase (EC 2.5.1.9). We assumed, that the operon in Shewanella could be similar to a well-studied [http://www.ncbi.nlm.nih.gov/pubmed/8159171 rib-operon] of Bacillus Subtilis. The operon is transcribed as a one polycistronic RNA, making a single promoter sufficient. Introduction of multiple copies of a ribA gene (coding for GTP cyclohydrolase II (EC 3.5.4.25)), comprised in the rib-operon, results in riboflavin overproduction in B.Subtilis [Hohmann H.P.,Stahmann K.P. 2010. Biotechnology of riboflavin production, p. 115–139.], so we predicted a notable riboflavin synthesis gain following a successful introduction of a multiple-copy plasmid harbouring the rib-operon under an active promoter.
Genetic Approach
Riboflavin Cluster
Below we shortly describe each functional member of this cluster.
- Gene: RibD http://www.ncbi.nlm.nih.gov/gene/1171145 Sequence, 1145 bp
- Protein: bifunctional diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino) uracil reductase RibD
- Enzyme: (EC: 3.5.4.26)
- Molecular weight: 41,257 Da
- Gene: SO_3468 http://www.ncbi.nlm.nih.gov/gene/1171144 Sequence, 656 bp
- Protein: Riboflavin synthase alpha subunit RibC-like protein
- Enzyme: EC 2.5.1.9
- Molecular weight: 23,483 Da
- Gene: RibBA → ribA & ribB http://www.ncbi.nlm.nih.gov/gene/1171143 Sequence, 1103 bp
- ribA
- Protein: GTP cyclohydrolase-2
- Enzyme: EC 3.5.4.25
- Molekular weight: 22,852 Da
- ribB
- Protein: 3,4-dihydroxy-2-butanone-4-phosphate synthase
- Enzyme: EC 3.5.4.25
- Molecular weight: 22,956 Da
- Gene: ribE http://www.ncbi.nlm.nih.gov/gene/1171142 Sequence, 476 bp
- Protein: 6,7-dimethyl-8-ribityllumazine synthase (aka: riboflavin synthase beta subunit RibE)
- Enzyme: EC 2.5.1.78
- Molecular Weight: 16,689 Da
Supplementary Genes
We have also separately amplified some other genes, that theoratically could be important for riboflavin synthesis or regulation.
- Gene: ribC http://www.ncbi.nlm.nih.gov/gene/1170021 Sequence, 621 bp
- Protein: 6,7-dimethyl-8-ribityllumazine synthase alpha subunit RibC, analogous to EC 2.5.1.9
- Molecular Weight: 22,110 Da
- Gene: nusB (full name: N utilization substance protein B homolog), http://www.ncbi.nlm.nih.gov/gene/1171141 Sequence, 403 bp
- Protein: transcription antitermination factor
- Molecular weight: 14,740 Da
- Gene: norM, http://www.ncbi.nlm.nih.gov/gene/1170020 Sequence, 1380 bp
- Protein: probable multidrug eflux protein NorM
- Molecular weight: 50,490 Da
Results


References
C. A. Abbas and A. S. Sibirny. (2011) Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. [http://mmbr.asm.org/content/75/2/321.full#ref-292| Microbiology and Molecular Biology Reviews 75(2): 321-360].
Hohmann H. P., Stahmann K. P. (2010). Biotechnology of riboflavin production, p. 115–139. In Mander L., Liu H. W. (ed.), Comprehensive natural products. II. Chemistry and biology, vol. 7. Cofactors. Elsevier, Philadelphia, PA.
von Canstein H., Ogawa J., Shimizu S., Lloyd J. R. (2008). Secretion of flavins by Shewanella species and their role in extracellular electron transfer. [http://aem.asm.org/content/74/3/615.full Appl. Environ. Microbiol. 74:615–623].
Bacher A., et al. (2001). Biosynthesis of riboflavin. [http://www.ncbi.nlm.nih.gov/pubmed/11153262 Vitam. Horm. 61:1–49.]
Tesliar G. E., Shavlovskii G. M. (1983). Localization of the genes coding for GTP cyclohydrolase II and riboflavin synthase on the chromosome of Escherichia coli K-12. Tsitol. Genet. 17:54–56. (In Russian.)
Seong Han Lim, Jong Soo Choi and Enoch Y. (2001). Park Microbial Production of Riboflavin Using Riboflavin Overproducers, Ashbya gossypii, Bacillus subtilis, and Candida famate: An Overview.[http://www.bbe.or.kr/storage/journal/BBE/6_2/6657/articlefile/article.pdf Biotechnol. Bioprocess Eng., 6: 75-88]
 "
"