Team:ETH Zurich/Infoproc
From 2013.igem.org
| Line 7: | Line 7: | ||
([https://2013.igem.org/Team:ETH_Zurich/Experiments_2 AHL]) by constitutive [https://2013.igem.org/Team:ETH_Zurich/pre_proc LuxI production]. <br><br> <b>Pre-Processing</b>: The agar minefield consists of mine and non-mine colonies in a <b>honey-comb grid</b>. The colonies are placed on the edges of each hexagon, except the center. This way, each colony is restricted to three neighboring colonies. The senders secrete the signalling molecule AHL that [https://2013.igem.org/Team:ETH_Zurich/Experiments_2#diffusion_experiment diffuses] through the agar. Depending on the amount of diffused AHL that is processed in the non-mines, different possible outcomes indicate the number of mines: <b>0 mines, 1 mine and 2 mines around</b> (to find out more on the grid pattern, please click [https://2013.igem.org/Team:ETH_Zurich/pre_proc here]). In the receiver cells, the signaling molecule forms a complex with the inactive LuxR to form an active complex AHL-luxR.<br><br><b>Processing</b>: by using [https://2013.igem.org/Team:ETH_Zurich/Processing_2 different sensitive P<sub>LuxR</sub> promoters] the AHL input concentration is translated into different outputs using specific reporters : GFP/RFP and later different | ([https://2013.igem.org/Team:ETH_Zurich/Experiments_2 AHL]) by constitutive [https://2013.igem.org/Team:ETH_Zurich/pre_proc LuxI production]. <br><br> <b>Pre-Processing</b>: The agar minefield consists of mine and non-mine colonies in a <b>honey-comb grid</b>. The colonies are placed on the edges of each hexagon, except the center. This way, each colony is restricted to three neighboring colonies. The senders secrete the signalling molecule AHL that [https://2013.igem.org/Team:ETH_Zurich/Experiments_2#diffusion_experiment diffuses] through the agar. Depending on the amount of diffused AHL that is processed in the non-mines, different possible outcomes indicate the number of mines: <b>0 mines, 1 mine and 2 mines around</b> (to find out more on the grid pattern, please click [https://2013.igem.org/Team:ETH_Zurich/pre_proc here]). In the receiver cells, the signaling molecule forms a complex with the inactive LuxR to form an active complex AHL-luxR.<br><br><b>Processing</b>: by using [https://2013.igem.org/Team:ETH_Zurich/Processing_2 different sensitive P<sub>LuxR</sub> promoters] the AHL input concentration is translated into different outputs using specific reporters : GFP/RFP and later different | ||
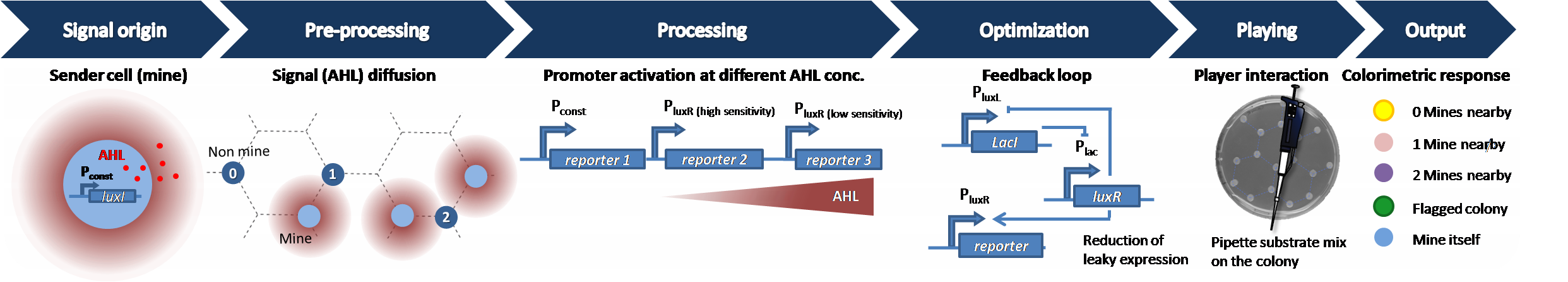
| - | [https://2013.igem.org/Team:ETH_Zurich/Experiments_7 hydrolases].<br><br><b>Optimization</b>: after the [https://2013.igem.org/Team:ETH_Zurich/Experiments_6 proof-of-principle with GFP] we carried out the first trials using the hydrolases as reporter system. We then concluded that our reporter system set-up is <b>leaky and start to review the circuit</b> to reduce the leakiness. (Please see the optimization part for more [https://2013.igem.org/Team:ETH_Zurich/Circuit details]).<br><br><b> Player interaction </b>: the player pipets a <b>substrate-mix</b> on a colony, which leads to a color change of the colony. This gives the player information to logically carry out the next move in the game. So the left click in the computer game is pipetting a substrate mixture on the colony in the bio-game. As a goodie we also included the right click in the computer game (<b>flagging</b>) by adding either a flagging solution converted by the chromosomal expressed lacZ or by adding the <i>Remazol blue dye</i> which is, in contrats to the lacZ flagging, removable. For more details please click [https://2013.igem.org/Team:ETH_Zurich/Play here]<br><br><b>Output</b>: within <b>minutes</b> after the addition of the substrate-mix a change in color due to the conversion of the specific substrates by the different expressed hydrolases according to the different amount of AHL dependent on the number of surrounding mines, indicates the identity of the played colony. The color output is based on an <b>overlay</b> of different expressed hydrolases in the different situations.</p> <br clear="all">[[File:Infoproc24.png| | + | [https://2013.igem.org/Team:ETH_Zurich/Experiments_7 hydrolases].<br><br><b>Optimization</b>: after the [https://2013.igem.org/Team:ETH_Zurich/Experiments_6 proof-of-principle with GFP] we carried out the first trials using the hydrolases as reporter system. We then concluded that our reporter system set-up is <b>leaky and start to review the circuit</b> to reduce the leakiness. (Please see the optimization part for more [https://2013.igem.org/Team:ETH_Zurich/Circuit details]).<br><br><b> Player interaction </b>: the player pipets a <b>substrate-mix</b> on a colony, which leads to a color change of the colony. This gives the player information to logically carry out the next move in the game. So the left click in the computer game is pipetting a substrate mixture on the colony in the bio-game. As a goodie we also included the right click in the computer game (<b>flagging</b>) by adding either a flagging solution converted by the chromosomal expressed lacZ or by adding the <i>Remazol blue dye</i> which is, in contrats to the lacZ flagging, removable. For more details please click [https://2013.igem.org/Team:ETH_Zurich/Play here]<br><br><b>Output</b>: within <b>minutes</b> after the addition of the substrate-mix a change in color due to the conversion of the specific substrates by the different expressed hydrolases according to the different amount of AHL dependent on the number of surrounding mines, indicates the identity of the played colony. The color output is based on an <b>overlay</b> of different expressed hydrolases in the different situations.</p> <br clear="all">[[File:Infoproc24.png|1270px|center|thumb|<b>Figure 1:Information processing from secreted signaling molecule to colorimetric response.</b> The signal diffuses through the agar from sender cells (light blue) to receiver cells (dark blue).The non-mine colonies are designed to distinguish between different concentrations of AHL and translate this information into expression of different hydrolases. The expression is driven by different P<sub>LuxR</sub> promoters that show different AHL sensitivities and serve as high pass filters. After an incubation time of 12 hours the player pipets a substrate on the colony. The hydrolase converts the substrate into a colored product which is visible by eye ]] |
<br clear="all"/> | <br clear="all"/> | ||
Revision as of 22:32, 26 October 2013
Information processing and project overview
Our game Colisweeper is played on an agar mine-field, which is a petridish with E.coli colonies. Some of these colonies are mines and others non-mines. The mines serve as the sender cells and the non-mines serve as the receiver cells.
Signal origin: The sender cells secrete the signaling molecule 3-oxo-N-hexanoyl-L-homoserine lactone
(AHL) by constitutive LuxI production.
Pre-Processing: The agar minefield consists of mine and non-mine colonies in a honey-comb grid. The colonies are placed on the edges of each hexagon, except the center. This way, each colony is restricted to three neighboring colonies. The senders secrete the signalling molecule AHL that diffuses through the agar. Depending on the amount of diffused AHL that is processed in the non-mines, different possible outcomes indicate the number of mines: 0 mines, 1 mine and 2 mines around (to find out more on the grid pattern, please click here). In the receiver cells, the signaling molecule forms a complex with the inactive LuxR to form an active complex AHL-luxR.
Processing: by using different sensitive PLuxR promoters the AHL input concentration is translated into different outputs using specific reporters : GFP/RFP and later different
hydrolases.
Optimization: after the proof-of-principle with GFP we carried out the first trials using the hydrolases as reporter system. We then concluded that our reporter system set-up is leaky and start to review the circuit to reduce the leakiness. (Please see the optimization part for more details).
Player interaction : the player pipets a substrate-mix on a colony, which leads to a color change of the colony. This gives the player information to logically carry out the next move in the game. So the left click in the computer game is pipetting a substrate mixture on the colony in the bio-game. As a goodie we also included the right click in the computer game (flagging) by adding either a flagging solution converted by the chromosomal expressed lacZ or by adding the Remazol blue dye which is, in contrats to the lacZ flagging, removable. For more details please click here
Output: within minutes after the addition of the substrate-mix a change in color due to the conversion of the specific substrates by the different expressed hydrolases according to the different amount of AHL dependent on the number of surrounding mines, indicates the identity of the played colony. The color output is based on an overlay of different expressed hydrolases in the different situations.
 "
"





