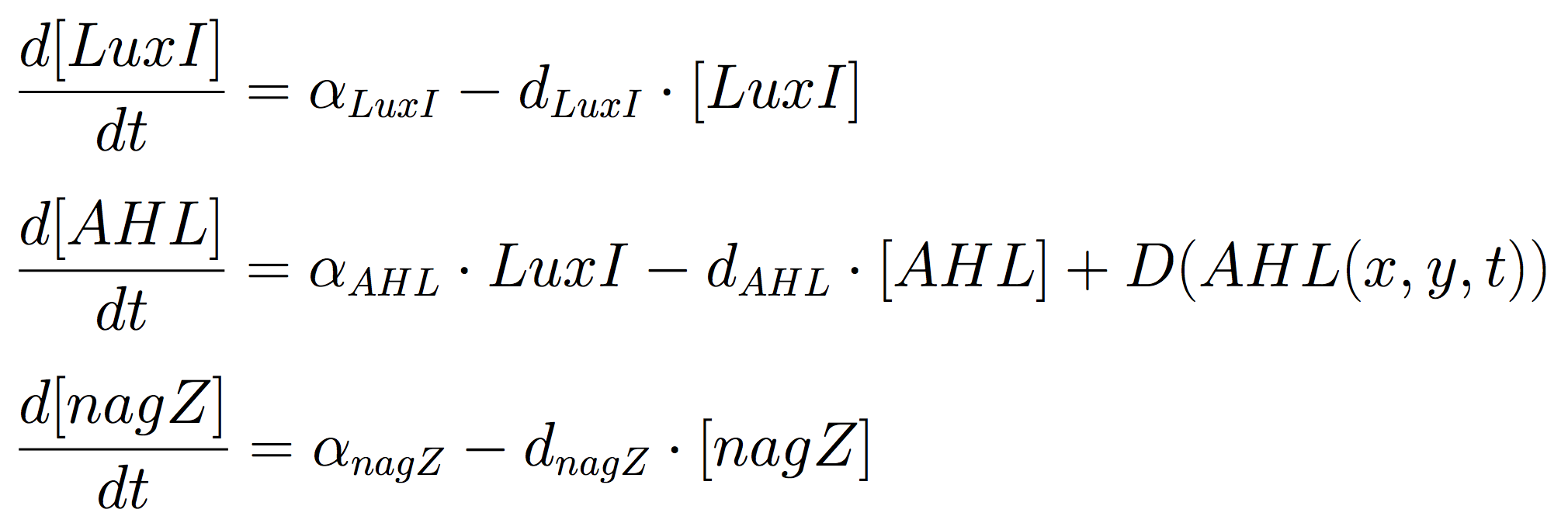Team:ETH Zurich/Modeling
From 2013.igem.org
Contents |
Circuit containing hydrolases
A simple seven-species model was used to model the spatiotemporal behaviour of a multicellular sender–receiver system. The model was based on differential equations with Hill functions that captured the activation of protein synthesis as a result of the presence of the signalling molecule.
Mine Cells
Mine Cells conduct the synthesis of the signalling molecule, by constitutive expression of luxI gene. Additionally, to reveal the nature of the cells, a coloured-substrate reaction is triggered upon addition of 5-Bromo-4-chloro-3-indoxyl-N-acetyl-beta-D-glucosaminide; given that the glycoside hydrolase NagZ is expressed constitutively.
The PDEs for the states involved in the sender module are given below:
Receiver Cells
Receiver cells are engineered to respond differently to two concentration levels of OHHL. Basically, cells should be capable of produce a visible response for the player, in order to discriminate between the presence of 0, 1 or 2 mine cells around them in the immediate vicinity in a three neighbours setup. This goal was achieved by regulated expression of two hydrolases, under control of Plux promoters, GusA and AES. Such enzymes can catalyze the hydrolysis of various chromogenic compounds giving rise to a colored response.
The intracellular species of interest in the receiver cells included LuxR, OHHL, LuxR/OHHL complex (denoted as R) and the hydrolases (GusA and AES).
To distinguish between OHHL-levels, the expression of the hydrolases is controlled by PLuxR promoters mutants, which are sensitive to different concentration of the dimer LuxR-OHHL (denoted as R) given by the number of surrounding mines.
 "
"