Team:Colombia Uniandes/Stochastic
From 2013.igem.org
Stochastic
To develop the stochastic model for both projects the following assumptions were made:
- Only one event happens at each time
- The union of the ribosome and the RNA is considered a permanent bond, not intermittent.
- Transcription and Translation are modelled as a single event
The scripting is based in the steps explained in Colombia Team 2012 wiki.
Nickel removal system
The biochemical reactions carried out in living systems, specifically in cells, follow in general a stochastic process. Transcription, signaling networks and systems within a cell can be modeled and simulated in time as Markovian processes. Gillespie algorithm (see references) gives a stochastic simulation method of these kinds of phenomena and offers a good resolution with an efficient use of computational resources.
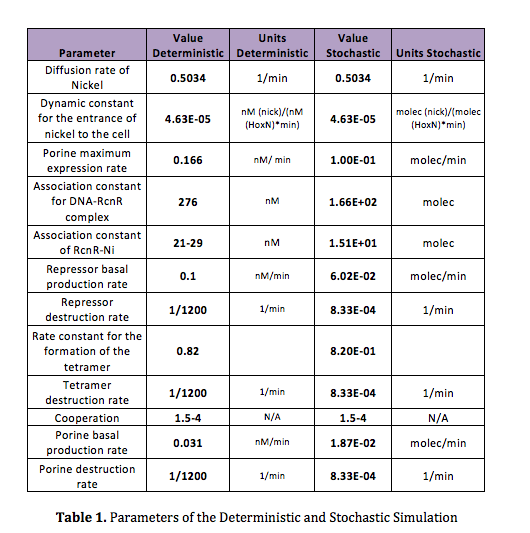
The advantage of this kind of modeling is that the differential equations that we use in the deterministic model can also be used in the stochastic model. However, in order to do the simulation for our nickel uptake system, we must start defining the parameters of the model in the correct units. For our purposes and the algorithm’s requirements, we need units of molecules per unite of time (in our case, minutes). In the following table we show the parameters used for deterministic simulation and the stochastic parameters. The new parameters where obtained by using Avogadro's number (6.02e23 molecules per mole) and an average cell volume of 10e-15 liters.
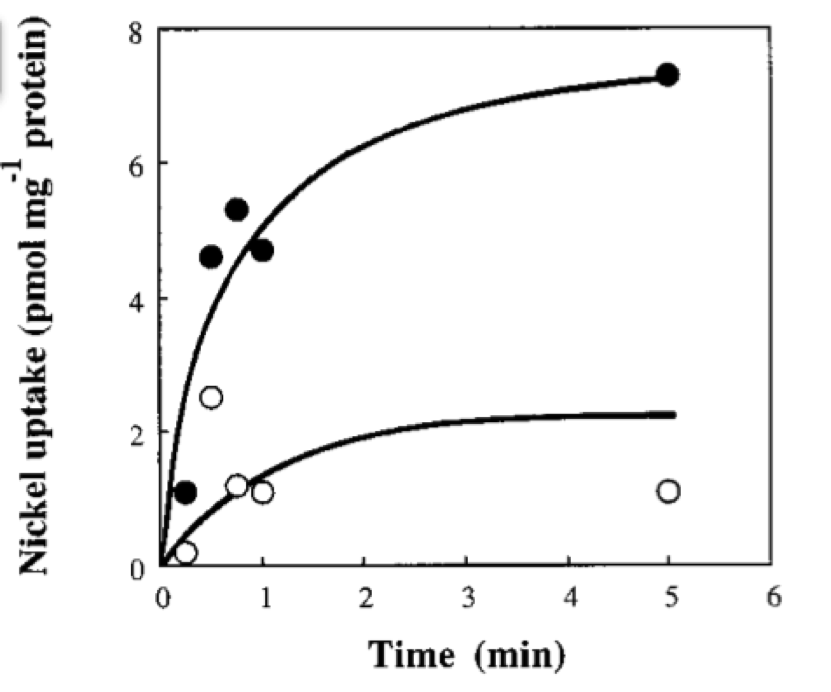
If we neglect the union of the nickel ion with the repressor, we can assume that nickel concentration in each cell is a function of the quantity of porine in the cell membrane and time. This function, known to be exponential, can be obtained by using an interpolation of data found in publications (Wolfram, Friedrich, & Eitinger, 1995). We could use a fitting function to describe the nickel uptake in the cell through time. However, we noticed that after 5 minutes, the curve reaches a steady state value. Given that we are not modeling the creation of mRNA and then the transcription (we are making an approximation for our time interests and computationally efficiency) we have a ~5min resolution. With this time resolution, it won’t matter if we fit the curve of figure 1 with higher detail. So we do an approximation based on the steady state of about 7pmolNickel/mgPorine
Nickel uptake of recombinant E. coli CC118. Solid circles, CC118(pCH231-Sm) containing functional hoxN; open circles, CC118(pCH231- P47) harboring an inactivated hoxN. The assay mix consisted of 100 nM 63NiCl2 and 10 mM MgCl2 in a 50 mM Tris-hydrochloride buffer (pH 7.5). Taken from: (Wolfram et al., 1995) </div>
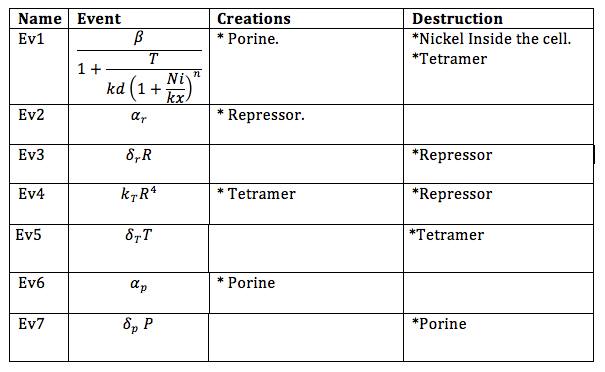
Now, the different events that may produce creation/generation and destruction/elimination are specified as follows based on the differential equations:
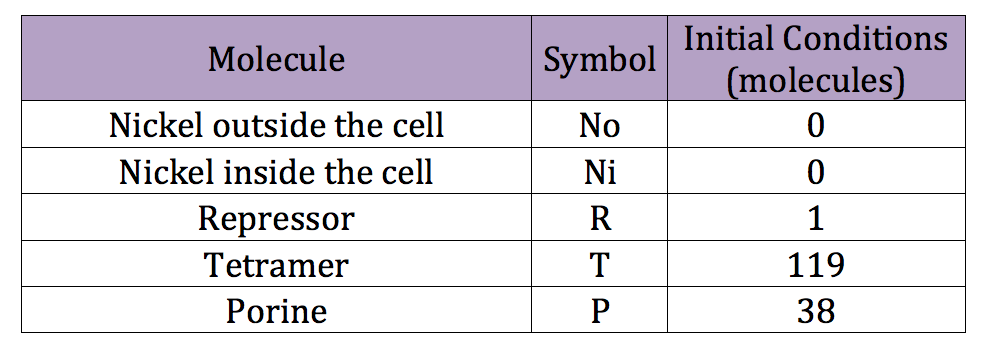
We can run a similar code used in the deterministic simulation to obtain reasonable initial conditions for the stochastic simulation. However this time we introduce the new, adequate parameters. The initial conditions obtained are listed in the next table
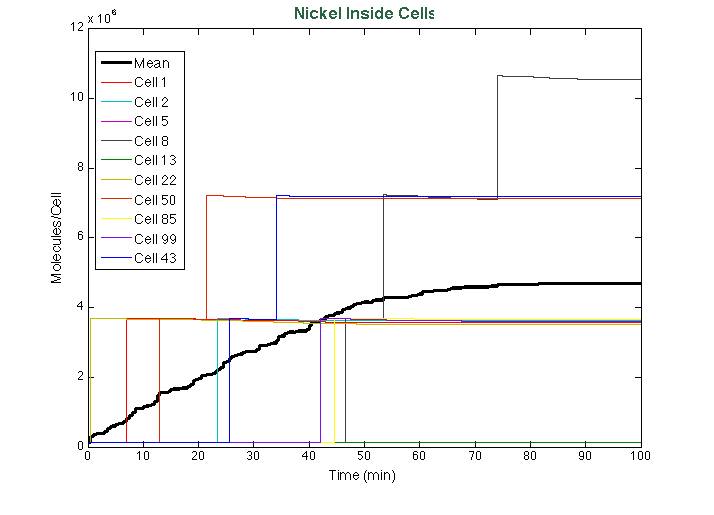
After running the scripts, these were the results of the simulation:
References=
Wolfram, L., Friedrich, B., & Eitinger, T. (1995). The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. Journal of bacteriology, 177(7), 1840–3. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=176814&tool=pmcentrez&rendertype=abstract
 "
"