Team:ETH Zurich/Experiments 5
From 2013.igem.org
| Line 10: | Line 10: | ||
<h1>PluxR random mutagenesis PCR</h1> | <h1>PluxR random mutagenesis PCR</h1> | ||
| - | <h1>PluxR promoter | + | <h1>PluxR promoter with different sensitivity analyzed using the Fortessa FACS analyzer</h1> |
| + | [[File:C6.png|left|600px|thumb|<b>Figure 2. Wild type sensitivy curve using the Fortessa FACS Analyzer</b>]] | ||
| + | <br clear="all"/> | ||
| + | [[File:G1.png|left|600px|thumb|<b>Figure 3. PluxR1 (G1) mutated promoter sensitivy curve using the Fortessa FACS Analyzer</b>]] | ||
| + | <br clear="all"/> | ||
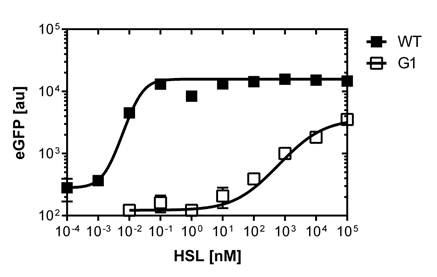
| + | [[File:Comparison.png|left|400px|thumb|<b>Figure 4. Comparison of the Wild Type and PluxR1 mutant sensitivy curve using the Fortessa FACS Analyzer</b>]] | ||
| + | <br clear="all"/> | ||
| + | |||
| + | <br clear="all"/> | ||
{{:Team:ETH_Zurich/templates/footer}} | {{:Team:ETH_Zurich/templates/footer}} | ||
Revision as of 18:04, 22 September 2013
Contents |
Native N-3-Oxo-Hexanoyl-l-Homoserine Lactone tests in liquid culture using the Tecan Infinite M200 PRO
In order to select mutated pLuxR promoters we need to know about the sensivity of the Wild-Type type. The test range was inspired form the paper about Evaluation of a focused libraryu of N-Acryl L-Homoserine lactone reveals a new set of potent quorum sensing modulators.The paper shows sensivities of the pLuxR promoter to different sets of AHL molecules in Vibrio fisheri. We need to readjuste the ranges for our E.coli DH5alpha strain in a second run and finally get our sensivity curve over the linear range of [0 nM],[0.25 nM],[0.5 nM],[1 nM],[2 nM],[3 nM],[4 nM],[5 nM],[10nM],[20 nM],[30 nM],[40 nM]and[50 nM],.
The experimental set-up included a 96-well plate filled with 180 uL LB media , 10uL of receiver cells and 10 uL of different AHL concentrations, everything in triplicates. (+ blank and negative control). The experiment runs for 16 hours in order to monitor the evolution of fluorescence and later choose the steady state to create the sensivity curve.The datas are analyzed with GraphPad Prism 6.0.
PluxR random mutagenesis PCR
PluxR promoter with different sensitivity analyzed using the Fortessa FACS analyzer
 "
"