Team:ETH Zurich/Experiments 7
From 2013.igem.org
Contents |
What about those hydrolases ?
Colisweeper depends largely on processing of molecular signals and generation of visible and distinguishable outputs accordingly. Next to our PLuxR promoter mutations, which detect ranges of OHHL concentrations, we made use of a reporter system which gives colorimetric responses and additionally provides that the visibility of output is only to be triggered by the player of Colisweeper: A set of orthogonal hydrolases which specifically cleave the chromogenic hydrolase substrates added by the player.
These hydrolases include the Citrobacter alkaline phospohatase, the Bacillus subtilis β-glucuronidase and the Escherichia coli acetylesterase, N-acetyl-β-glucosaminidase and β-galactosidase. To ensure specific enzyme-substrate reactions, we used a triple deletion Escherichia coli strain, lacking aes (acetylesterase), gusA (β-glucuronidase) and nagZ (N-acetyl-β-glucosaminidase).
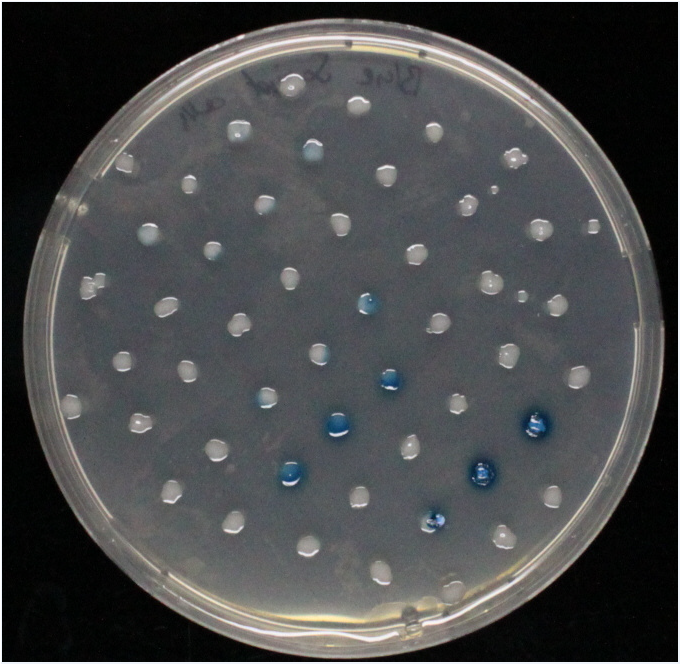
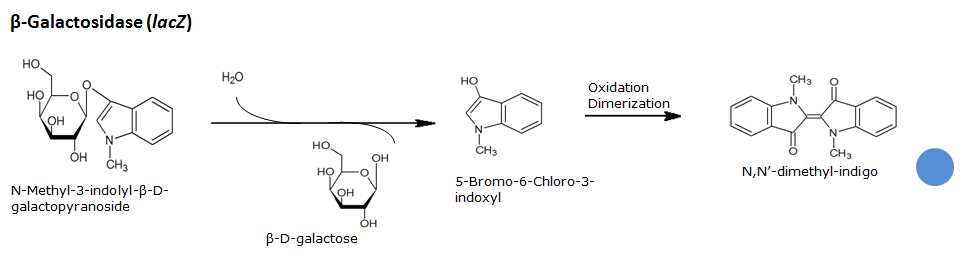
Beta-galactosidase(LacZ)
Chemical conversion
Colorimetric response in liquid culture
Colorimetric response on LB-Agar
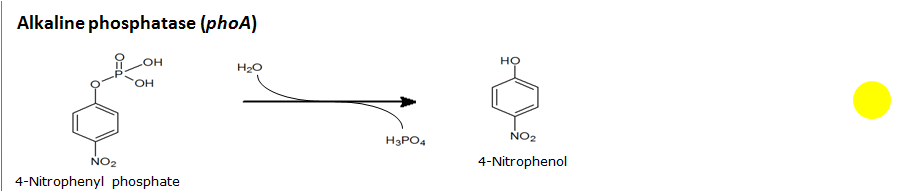
Alkaline phosphatase(PhoA)
Chemical conversion
Colorimetric response in liquid culture
Colorimetric response on LB-Agar
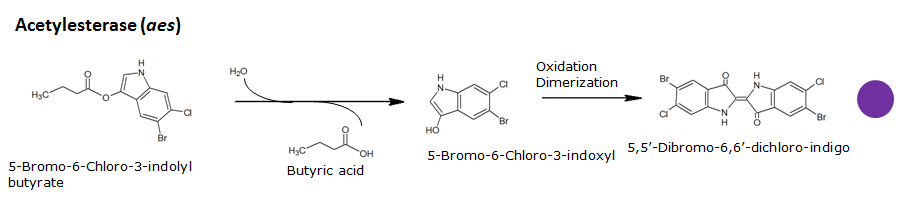
Acetyl esterase(Aes)
Chemical conversion
Colorimetric response in liquid culture
Colorimetric response on LB-Agar
Beta-glucuronidase(GusA)
Chemical conversion
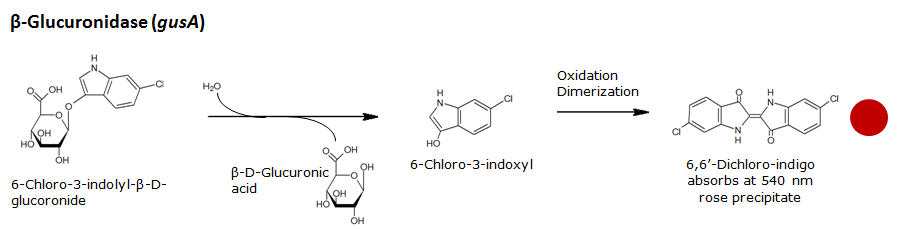
Colorimetric response in liquid culture
Colorimetric response on LB-Agar
β-N-Acetylglucosaminidase(NagZ)
Chemical conversion
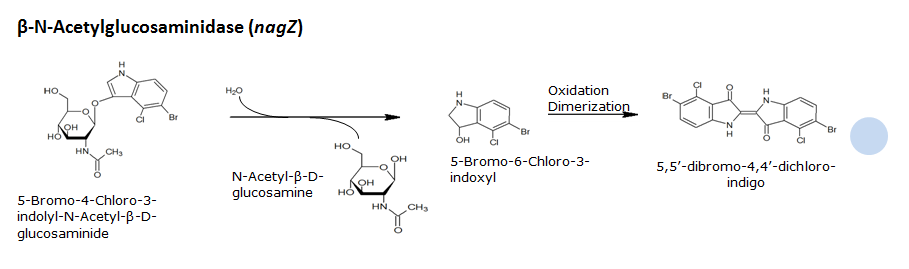
Colorimetric response in liquid culture
Colorimetric response on LB-Agar
 "
"