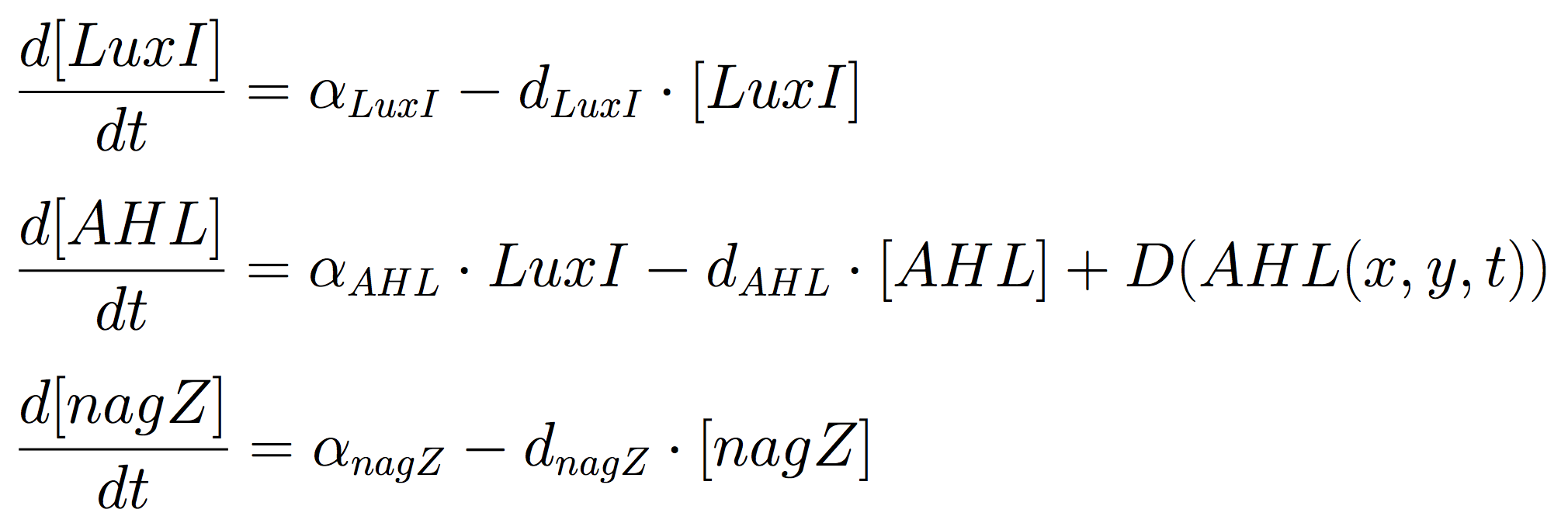Team:ETH Zurich/Modeling
From 2013.igem.org
| Line 6: | Line 6: | ||
color:white; | color:white; | ||
font-family: Verdana; | font-family: Verdana; | ||
| - | font-size: | + | font-size:18px; |
} | } | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
b, #eth_content { | b, #eth_content { | ||
color:black; | color:black; | ||
| Line 24: | Line 17: | ||
| - | < | + | <b>The digital bacterial-based minesweeper</b> |
| + | <br><br> | ||
| + | The change of the species concentrations in time is given by non-linear ordinary differential equations (ODEs), most of which follow Hill kinetics. The parameters we used in the model are derived from literature. | ||
<br> | <br> | ||
| - | + | <br clear="all"/> | |
| + | <hr> | ||
| - | + | <b>Mine Cells</b> | |
[[File:eqnSender.png|500px|thumb|Equation system 1: ODE for Mine Cells.]] | [[File:eqnSender.png|500px|thumb|Equation system 1: ODE for Mine Cells.]] | ||
The Mine Cells lead to the synthesis the signalling molecule, by constitutive expression of ''luxI'' gene. To reveal the nature of the cells, a coloured-substrate reaction is triggered upon addition of ''5-Bromo-4-chloro-3-indoxyl-N-acetyl-beta-D-glucosaminide''; given that the glycoside hydrolase ''NagZ'' is expressed constitutively. <br><br> The ODEs for the states involved in the sender module are given below: | The Mine Cells lead to the synthesis the signalling molecule, by constitutive expression of ''luxI'' gene. To reveal the nature of the cells, a coloured-substrate reaction is triggered upon addition of ''5-Bromo-4-chloro-3-indoxyl-N-acetyl-beta-D-glucosaminide''; given that the glycoside hydrolase ''NagZ'' is expressed constitutively. <br><br> The ODEs for the states involved in the sender module are given below: | ||
<br clear="all"/> | <br clear="all"/> | ||
| + | <hr> | ||
| - | + | <b>Biosensors</b> | |
| - | + | ||
[[File:eqnBiosensor.png|500px|thumb|Equation system 1: ODE for Mine Cells.]] | [[File:eqnBiosensor.png|500px|thumb|Equation system 1: ODE for Mine Cells.]] | ||
Our Biosensor cells are engineered to respond differently to high and medium concentrations of AHL. This cells are capable of discriminate between the presence of 1 or 2 mine cells around them in the immediate vicinity. To accomplish this task, the enzymes involved in the coloured-substrate reaction are sensitive to different concentrations of the dimer LuxR-AHL (denoted as R). | Our Biosensor cells are engineered to respond differently to high and medium concentrations of AHL. This cells are capable of discriminate between the presence of 1 or 2 mine cells around them in the immediate vicinity. To accomplish this task, the enzymes involved in the coloured-substrate reaction are sensitive to different concentrations of the dimer LuxR-AHL (denoted as R). | ||
<br clear="all"/> | <br clear="all"/> | ||
| + | <hr> | ||
Revision as of 10:16, 19 August 2013
The digital bacterial-based minesweeper
The change of the species concentrations in time is given by non-linear ordinary differential equations (ODEs), most of which follow Hill kinetics. The parameters we used in the model are derived from literature.
Mine Cells
The Mine Cells lead to the synthesis the signalling molecule, by constitutive expression of luxI gene. To reveal the nature of the cells, a coloured-substrate reaction is triggered upon addition of 5-Bromo-4-chloro-3-indoxyl-N-acetyl-beta-D-glucosaminide; given that the glycoside hydrolase NagZ is expressed constitutively.
The ODEs for the states involved in the sender module are given below:
Biosensors
Our Biosensor cells are engineered to respond differently to high and medium concentrations of AHL. This cells are capable of discriminate between the presence of 1 or 2 mine cells around them in the immediate vicinity. To accomplish this task, the enzymes involved in the coloured-substrate reaction are sensitive to different concentrations of the dimer LuxR-AHL (denoted as R).
 "
"