Team:ETH Zurich/Processing 2
From 2013.igem.org
Creation of a promoter library
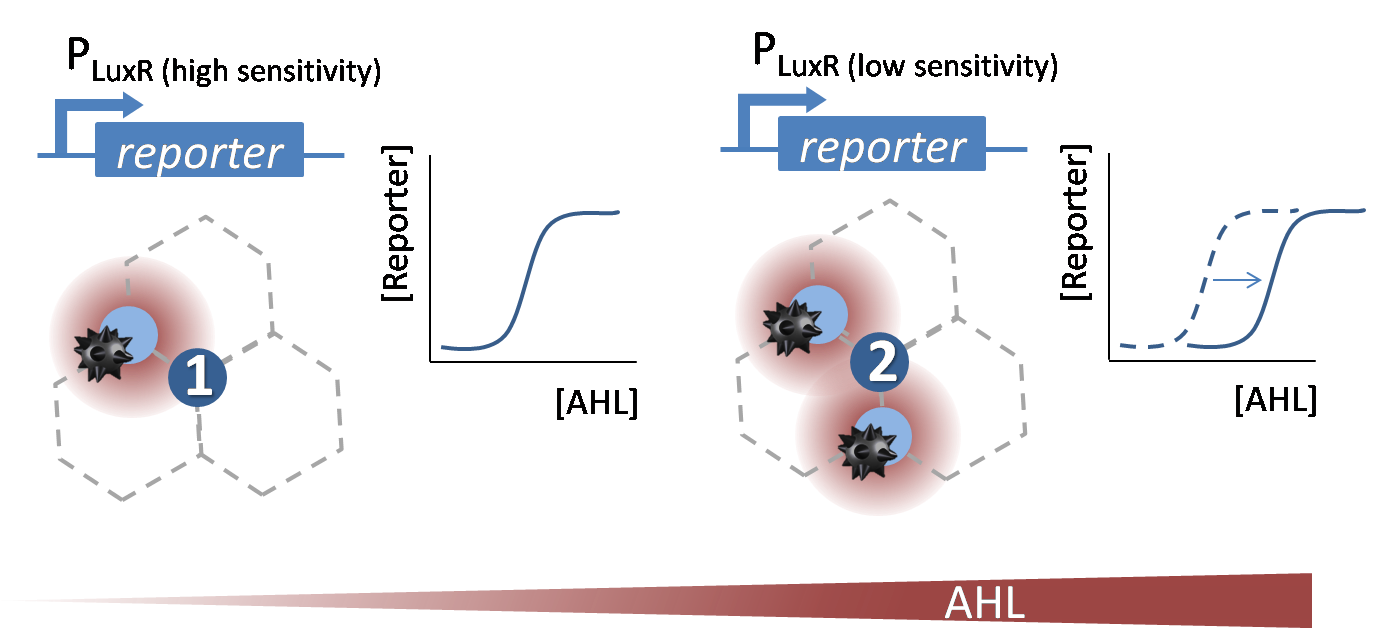
The wild-type PLuxR promoter (BBa_R0062)gets activated in presence of AHL. IN our project we need our promoter to distinguish between different levels of the AHL concentrations according to the different number of surrounding mines. Therefor we need to shift the dose-response curve of the initial BBa_R0062 promoter. We decided to do site directed saturation mutagenesis (see below).
Site directed saturation mutagenesis PCR of the BBa_R0062 PluxR

We did site directed saturation mutagenesis of specific sites of the lux box according to the results taken from literature Antunes et. al., 2007. (see Figure 2).
We mutated the promoter directly in the BBa_J09855 construct and cloned it in front an eGFP gene as a reporter to be able to screen for differences in the dose response curves. At first we incubated the transformed cells on an AHL containing plate to rule out all AHL-insensitive mutations. All colonies expressing GFP were then restreaked on a plate which contained no AHL, thus allowing us to rule out constitutive promoters. The remaining clones were then grown in luquid cultures on 96-well plates and tested for sensitivity over a broad range of AHL concentrations. Promising candidates were then measured in a flow cytometer to check for bistable promoters. The results of all these processes can be seen here.
 "
"