Team:SJTU-BioX-Shanghai/Project/Regulator/Integrating
From 2013.igem.org
(→Testing the Interface of CRISPRi and Light Sensors) |
|||
| Line 27: | Line 27: | ||
__NOTOC__ | __NOTOC__ | ||
<!----------------------------------------------------从这里开始写wiki---------------------------------> | <!----------------------------------------------------从这里开始写wiki---------------------------------> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | = | + | =From CRISPR to CRISPRi= |
<br> | <br> | ||
| - | + | <font size=4>CRISPR</font size=4> | |
| - | + | [[File:CRISPR_genome_editing.png|thumb|300px|right|CRISPR Genome Editing (Jinek et al., 2012)]] | |
| - | + | ||
| - | + | ||
| - | [[File: | + | |
<br> | <br> | ||
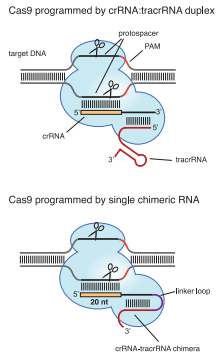
| - | + | CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and <b>CRISPR-associated system (Cas)</b> widely exist in Bacteria and Archea, endowing the cell with <b>adaptive immunity</b> to foreign DNAs. The immune process could be divided into three stages:<br> | |
| - | < | + | * <b>Adaptive Stage</b>. Fragments of foreign DNA (named protospacers) is incorporated into the proximal end of CRISPR.<br> |
| - | + | * <b>Expression Stage</b>. Precursor CRISPR RNAs (pre-crRNAs) are transcribed from CRISPR. Pre-crRNAs are then cleaved to produce <b>crRNAs</b>.<br> | |
| - | + | * <b>Interference Stage</b>. crRNA complements with target DNA (protospacers), forming a complex that is recognized and bound by Cas nuclease (Csn). Csn excises the target, leaving the target vulnerable to successive degradation. | |
| - | + | ||
<br> | <br> | ||
| - | + | Three types of CRISPR system have already been revealed, of which Type II CRISPR system is the <b>simplest</b> – one sole protein, <b>Cas9</b> (formerly named Csn1) is capable of all tasks once crRNA is complemented by <b>trans-activating crRNA (tracrRNA)</b>. Due to this simplicity, Type II CRISPR/Cas can be ectopically expressed as a tool for sequence-specific genome editing (Jinek et al., 2012). And to further simplify the tool, Jinek et al. linked crRNA and tracr RNA together, successfully creating <b>small guide RNA (sgRNA)</b> as an equivalent (Jinek et al., 2012). | |
| - | <br><br><br><br> | + | <br><br><br> |
| + | <font size=4>CRISPRi</font size=4><br> | ||
| + | [[File:CRISPRi.png|250px|right|thumb|CRISPRi Mechanism]] | ||
| + | <br> | ||
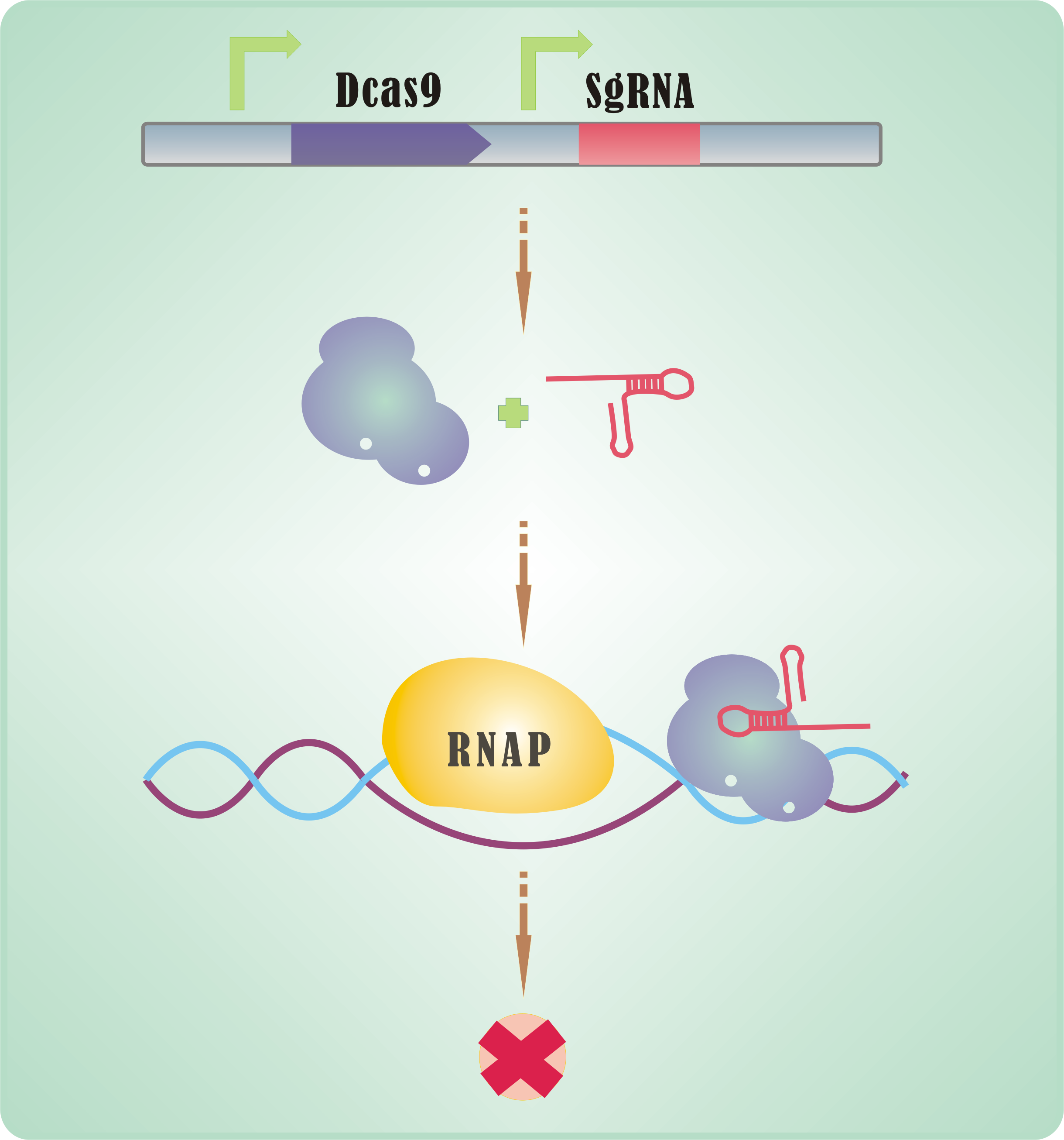
| + | With its <b>endonuclease activity crippled</b>, Cas9 can be used to hinder RNA polymerase and transcript elongation. Together with a proper sgRNA, Cas9 possessing such defect (named dCas9) can be used as an effective tool in expression interrogation – <b>CRISPR interference</b>, abbreviated as CRISPRi (Qi et al., 2013). | ||
| + | <br> | ||
| + | CRISPRi is promising tool for expression regulation, <b>versatile and easy-to-use</b>. Generally, a small shift in sgRNA would be enough for a new target. | ||
| + | <br><br><br> | ||
| - | + | =Key Parts of CRISPRi= | |
| - | + | <br> | |
| - | + | There are only 2 parts to be expressed -- dCas9 and sgRNA. Only <font size=4>2</font size=4>! | |
| - | + | ||
| - | + | ||
<br><br> | <br><br> | ||
| + | <font size=4>dCas9</font size=4> | ||
| + | <br><br> | ||
| + | As the signature protein of the type II CRISPR/Cas, Cas9, does not show any detectable similarity to any proteins in Type I and Type III systems, in that it is sufficient both to generate crRNA and to cleave the target DNA. This large protein (about 1000 amino acids) contains two predicted nuclease domains -- the N-terminal '''RuvC-like nuclease''' (RNAse H fold) and the '''HNH (McrA-like) nuclease''' domain that is located in the middle of the protein (Makarova et al., 2011), each of which would cleave one DNA strand. | ||
| + | To acquire dCas9, one point mutation is conducted on each nuclease, respectively, namely, '''D10A and H841A''' (Qi et al., 2013). | ||
| + | <br><br> | ||
| + | <font size=4>sgRNA</font size=4> | ||
| + | [[File:SgRNA.png|thumb|250px|right|Structure of sgRNA]] | ||
| + | <br> | ||
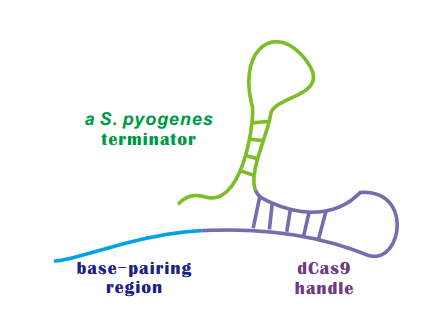
| + | If dCas9 acts as an executive, than sgRNA is the director of CRISPRi. | ||
| + | An sgRNA constitutes three parts: | ||
| + | * '''Base-Pairing Region''' | ||
| + | A 20 nt complementary region for specific DNA binding. | ||
| + | * dCas9 '''Handle''' | ||
| + | a 42 nt hairpin for Cas9 binding | ||
| + | * A '''terminator''' derived from S. pyogenes, 42nt | ||
| + | <br> | ||
| + | <b><font size=2.5>Design Criteria</font></b> (Qi et al., 2013) | ||
| + | * Binding specificity is determined by both sgRNA-DNA base pairing and a <b>protospacer adjacent motif (PAM), NGG</b>, upstream of target. | ||
| + | * Generally, the optimal length of the complementary region is <b>20nt</b>. | ||
| + | * If a sgRNA targets coding sequence, it has to be <b>complementary to the non-template (NT) strand</b>. But if a sgRNA targets the promoter, either strand would be acceptable. | ||
| + | * If a sgRNA targets coding sequence, generally CRISPRi would work better if the complementary region is <b>closer to the promoter</b>. | ||
| + | * The first 7 nt might be a "seed region" for binding, since any single mutation of dramatically decreased repression. | ||
<br><br> | <br><br> | ||
| - | = | + | <html> |
| + | <h1 style="color:grey;">References</h1> | ||
| + | <p style="color:grey;"> | ||
<br> | <br> | ||
| - | + | LEVSKAYA, A., CHEVALIER, A. A., TABOR, J. J., SIMPSON, Z. B., LAVERY, L. A., LEVY, M., DAVIDSON, E. A., SCOURAS, A., ELLINGTON, A. D., MARCOTTE, E. M. & VOIGT, C. A. 2005. Synthetic biology: engineering Escherichia coli to see light. Nature, 438, 441-2. | |
| + | <br/><br/> | ||
| + | TABOR, J. J., LEVSKAYA, A. & VOIGT, C. A. 2011. Multichromatic control of gene expression in escherichia coli. Journal of Molecular Biology, 405, 315-324. | ||
| + | </p></html> | ||
| + | |||
| + | |||
| - | |||
| - | |||
| - | |||
<!----------------------------------------------------到这里结束---------------------------------------> | <!----------------------------------------------------到这里结束---------------------------------------> | ||
</td> | </td> | ||
Revision as of 00:29, 28 September 2013
|
| ||
|
 "
"