Team:NTU-Taida/Notebook/Journal/July
From 2013.igem.org
Contents |
July
7/1~7/5
Finish RhlR and LasR circuit.
7/1
Transform:
- Positive feedback circuit:
Pc-A-C-B, Pc-A-C-E, Pc-A-C-B, Pc-D-F-B, Pc-D-F-E, Pc-D-F-G; - Control:
PLas-B, PLas-E, PLas-G, PRhl-B, PRhl-E, PRhl-G;
(A: RBS-RhlR-tt C: PRhl-RBS-RhlR D: RBS-LasR-tt F: PLas-RBS-LasR B: RBS-mCherry-tt E: RBS-GFPmut-tt G: RBS-mRFP-tt )
- New:
Pcin, CinR (resistant=C)
7/2
Result:
There’s no colonies at plates of Pcin and CinR.
Transform:
Pcin(resistant=A), CinR (resistant=K)
Inoculation and incubation at LB broth
- Positive feedback circuit: Pc-A-C-B, Pc-A-C-E, Pc-A-C-B, Pc-D-F-B, Pc-D-F-E, Pc-D-F-G;
- Control: PLas-B, PLas-E, PLas-G, PRhl-B, PRhl-E, PRhl-G; (A: RBS-RhlR-tt C: PRhl-RBS-RhlR D: RBS-LasR-tt F: PLas-RBS-LasR B: RBS-mCherry-tt E: RBS-GFPmut-tt G: RBS-mRFP-tt )
7/3
Inoculation and incubation at LB broth
Pcin(resistant=A), CinR (resistant=K)
Check
- Positive feedback circuit: Pc-A-C-B, Pc-A-C-E, Pc-A-C-B, Pc-D-F-B, Pc-D-F-E, Pc-D-F-G;
- Control: PLas-B, PLas-E, PLas-G, PRhl-B, PRhl-E, PRhl-G; (A: RBS-RhlR-tt C: PRhl-RBS-RhlR D: RBS-LasR-tt F: PLas-RBS-LasR B: RBS-mCherry-tt E: RBS-GFPmut-tt G: RBS-mRFP-tt )
7/4
Check and Plasmid DNA extraction (mini-prep):
- Positive feedback circuit: Pc-A-C-B, Pc-A-C-E, Pc-A-C-B, Pc-D-F-B, Pc-D-F-E, Pc-D-F-G;
- Control: PLas-B, PLas-E, PLas-G, PRhl-B, PRhl-E, PRhl-G; (A: RBS-RhlR-tt C: PRhl-RBS-RhlR D: RBS-LasR-tt F: PLas-RBS-LasR B: RBS-mCherry-tt E: RBS-GFPmut-tt G: RBS-mRFP-tt )
- Pcin(resistant=A), CinR (resistant=K)
7/5
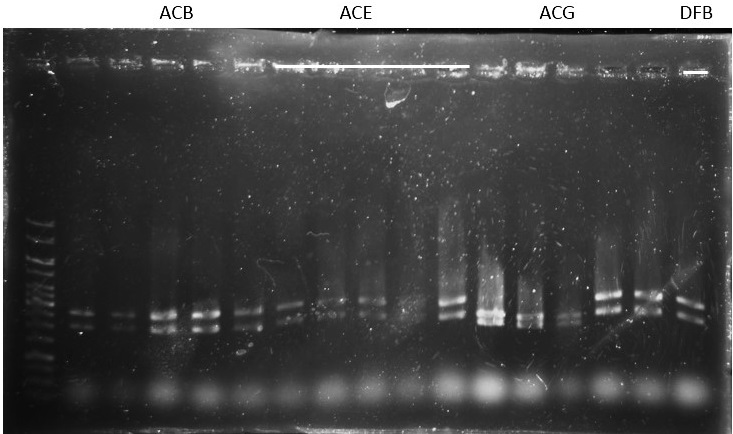
Check: (for plasmid ready to be sequenced)
ACB-4, ACE-5, ACG-1, DFB-1, DFE-2, DFE-3, DFE-4, DFE-5, DFG-1, RhlB-1, RhlE-2, RhlG-1, RhlG-2, LasB3, LasE1, LasG4 (8,9,11 well failed :DFE-4/DFE-5/ RhlB-1)
7/8~7/12
Construct the circuit for new receptor CinR
7/8
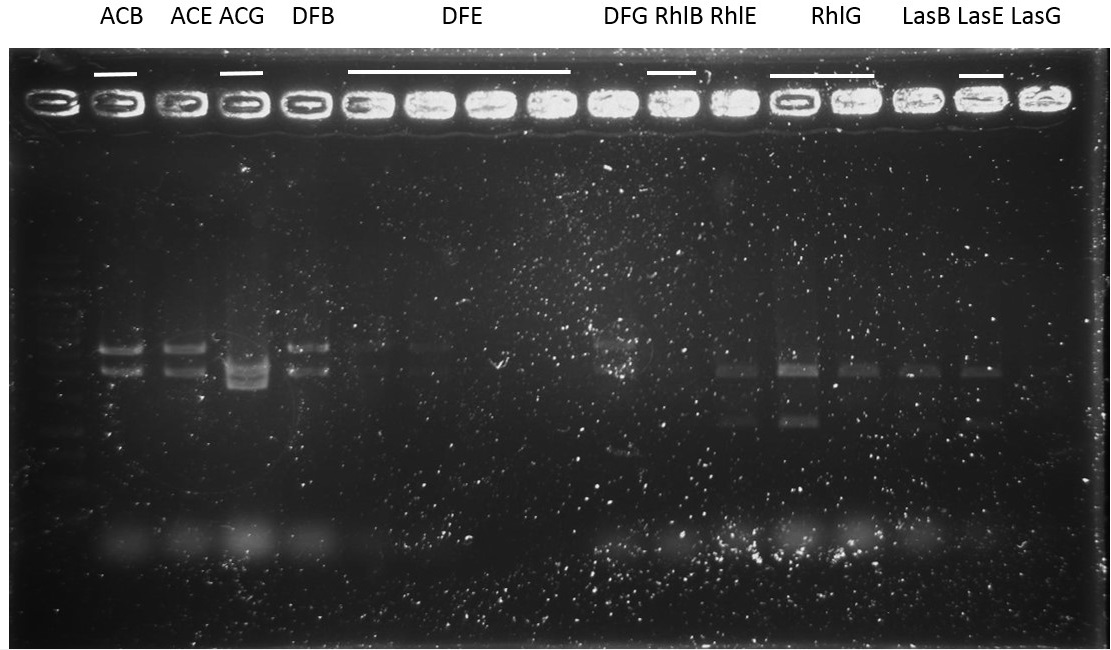
Sequence:
ACB-4, ACE-5, ACG-1, DFB-1, DFE-2, DFE-3, DFG-1, RhlE-2, RhlG-1, RhlG-2, LasB3, LasE1, LasG4
Primer list:
| Primer order | Name | sequence | |
|---|---|---|---|
| BMRC-120 | iGEM2013-pSB1A2 | attaccgcctttgagtgagc | R |
| BMRC-153 | iGEM2013-pSB1A2 | gtgccacctgacgtctaagaa | F |
| BMRC-122 | iGEM2013-mCherry | gccgtcctcgaagttcatcac | mcherry |
| BMRC-124 | iGEM2013-mRFP | aacggtaacaccaccgtc | RFP |
| BMRC-126 | iGEM2013-GFP | cttgtagttcccgtcatcttt | GFP |
Digestion, ligation, transform: 3A assembly and standard assembly both
B0030+CinR
7/9
Result:
The plate of B0030+CinR following 3A assembly was OK. The other following standard assembly failed unexpectedly.
Inoculation and incubation:
B0030+cinR
7/10
Plasmid DNA extraction (mini-prep):
B0030-cinR
Result:
A260/A280=1.3
Re-inoculation and incubation of plate of B0030-CinR
7/11
Plasmid DNA extraction (mini-prep):
B0030-cinR
Result:
7/12
Digestion: standard assembly only:
B0030 CinR
Result:
There’s no band for B0030
7/15~7/19
7/15
Ligation:
B0030+CinR (The certain content in the ependorf labeled B0030 undergone digestion was actually previously digested; thus, concentration was assumed to be low)
Transform:
7/16
Result:
Only one colony of plate(B0030-CinR) grew. CI are successfully incubated. pCI failed to grow.
Conclusion:
7/17
Digestion and ligation:
B0030 (eppendorf from early stage)
Transform:
B0030-CinR
RBS(B0030)
pCI(R0051)
LuxR(C0062)
pLux(Lux pR, R0062)
pConst(J23119)
mTagBFP(K592100)
Luciferase(J52008)
simple Las detecting system(K575024)
simple Rhl detecting system(K575033)
Sequence:
PcDFE, PcDFG, Pc ACB, Rhl B, Rhl E Total 16 tube
Sequence results:
No single plasmid was correct at all.
7/18
Inoculation and Incubation at LB broth:
B0030-CinR, B0030, LuxR, pLux, pConst(J23119), mTagBFP, Luciferase, simple Las detecting system(K575024), simple Rhl detecting system(K575033)
7/19
Check and Plasmid DNA extraction (mini-prep):
B0030-CinR, B0030, LuxR, pLux, pConst(J23119), mTagBFP, Luciferase, simple Las detecting system(K575024), simple Rhl detecting system(K575033)
7/22~7/31
7/22
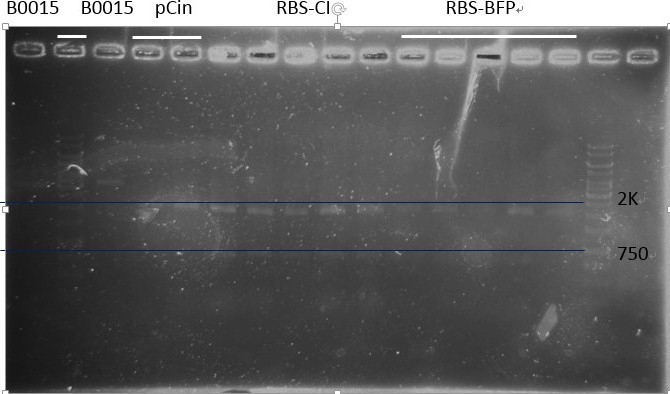
Digestion:
B0030, CI, mTagBFP, Luciferase, B0015(tt), pCin, ACin (RBS-CinR), BCin (RBS-CinR), B0030 (RBS), LuxR
Note:Because the gel for electrophoresis healed since we took away the comb too early, most of the digestion products overflew from the wells. And we almost couldn’t see the band after dyeing, so we redid the digestion step. The second time we finally got our material for ligation! But LuxR had very strange bands (too large), so we decided not use it. And retransformed it from official plasmid.
Ligation overnight at 16 degree incubator:
(1) B0030+CI
(2) B0030+mTagBFP
(3) B0030+Luciferase
(4) B0030-CinR+tt
(5) pCin+B0030-CinR
Transform:
(1) pCI (Amp, Chl): to check the drug resistance was correct.
(2) LuR (Chl)
(3) pCin (Amp)
Primer design and Sequence:
Design primers for AbaR, pqsR.
Start checking sequencing results.
7/23
Results:
Transformation: All 4 plates didn’t grow
Transform:
(1) B0030-CI (Amp)
(2) B0030-mTagBFP (Amp)
(3) B0030-Luciferase (Amp)
(4) [B0030-CinR]-tt (Amp)
(5) pCin-[B0030-CinR] (Amp)
(6)(7) pCI (Amp, Chl)
(8) pCin (Amp)
(9) LuxR (Chl) x2
Original biobrick:
(1) Got P.aeruginosa & A.baumannii (on blood agar plate)
(2) Each liquid culture x3 tubes
Human practice:
Visited the department of laboratory medicine in NTUH
7/24
Result:
(1) Transformation: all 10 plates didn’t growth…
(2) Debug: We took the wrong competent cell(HB101)…
(3) Sequencing: for functional assay, only the positive feedback of RhlR-GFP (ACE) was all correct. Others need to be redone.
Ligation at room temperature for 3 hours:
(1) B0030-CI
(2) B0030-mTagBFP
(3) B0030-Luciferase
(4) [B0030-CinR]-tt
(5) pCin-[B0030-CinR]
Transformation (correct competent cell – DH5α)
(1) B0030-CI (Amp)
(2) B0030-mTagBFP (Amp)
(3) B0030-Luciferase (Amp)
(4) [B0030-CinR]-tt (Amp)
(5) pCin-[B0030-CinR] (Amp)
(6)(7) pCI (Amp, Chl)
(8) pCin (Amp)
(9) LuxR (Amp): we took the wrong LuxR previously, but this time it was correct.
(10) B0015 (tt) (Amp)
Original biobrick:
(1) Extracted the whole genome of P. aeruginosa
7/25
Results
(1) Transformation: 7 growth except pCI and LuxR
Transformation
(1) PcA (pConst18C-B0030-RhlR-tt)
(2) PcD (pConst18C-B0030-LasR-tt)
(3) G5 (B0030-mRFP-tt)
(4) E1 (B0030-GFP-tt)
(5) B5 (B0030-mCherry-tt)
(6) C1 (pRhl-B0030-RhlR)
(7) F2 (pLas-B0030-LasR)
(8) pCI
(9) LuxR
No.1~7 are going to make stocks.
Inoculation and Incubation:
(No.1~5 cultured 5 tubes, 6 & 7 cultured 2 tubes, total 29 tubes)
(1) B0030-CI (Amp)
(2) B0030-mTagBFP (Amp)
(3) B0030-Luciferase (Amp)
(4) B0030-CinR-tt (Amp)
(5) pCin-B0030-CinR (Amp)
(6) pCin (Amp)
(7) B0015 (Amp)
Original biobrick:
(1) Extracted the whole genome of A. baumannii
(2) The concentration wasn’t very high
7/26
Results:
(1) Inoculation: forgot to put in 37 degree incubator…(put in 4 degree refrigerator)
(2) Transformation: 7 growth except pCI and LuxR…
(3) Debug: purified LuxR official plasmid, but the concentration was very~low
Official biobricks
(1) Take pCI and LuxR from 2012 official kit (at Life Science college)
Transformation
(1) pCI (Amp)
(2) LuxR (Amp)
(3) ACE (positive feedback of RhlR-GFP) (Amp): for functional assay
Transform at room temperature over weekend.
Inoculation and Incubation
(1) Keep inoculating at 37 degree incubator (since they didn’t growth very well)
(2) But the quality is uncertain
Make new Amp plates
7/29
Check:
(1)B0030-CI (Amp)
(2)B0030-mTagBFP (Amp)
(3)B0030-Luciferase (Amp)
(4)B0030-CinR-tt (Amp)
(5)pCin-B0030-CinR (Amp)
(6)pCin (Amp)
(7)B0015 (Amp)
B0015 contained a backbone of 3K. Check again!?
Inoculation and Incubation at LB broth:
(1)PcA (pConst18C-B0030-RhlR-tt)
(2)PcD (pConst18C-B0030-LasR-tt)
(3)G5 (B0030-mRFP-tt)
(4)E1 (B0030-GFP-tt)
(5)B5 (B0030-mCherry-tt)
(6)C1 (pRhl-B0030-RhlR)
(7)F2 (pLas-B0030-LasR)
(8)pCI
(9)LuxR
7/30
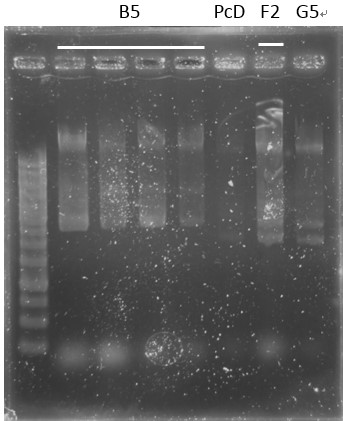
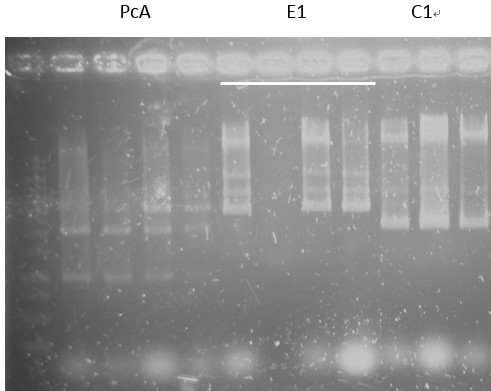
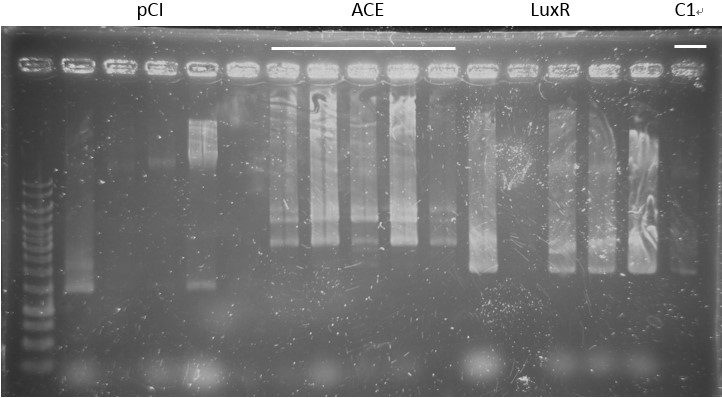
Check:
pCI-1, ACE, LuxR, C1, PcA, E1-1, C1, B5, PcD, F2, G5
Digestion, Ligation and Transformation:
Pc+AcinR
BcinR+C GFPmut(E1)
BcinR +C mCherry(B5)
B0030-mTagBFP+B0015
B0030-Luci+B0015
7/31
Digestion, Ligation and Transformation:
Pc+AcinR
BcinR+CGFPmut(E1)
B0030-mTagBFP+B0015
B0030-Luci+B0015
B0030-CI+B0015
pLas + RBS-LasR failed two times (without bands at gel extracting)
11 plate
Pc+AcinR 30 / 31
BcinR+CGFPmut(E1) 30 / 31
BcinR +CmCherry(B5) 30
B0030-mTagBFP+B0015 30/31
B0030-Luci+B0015 30/31
B0030-CI+B0015 30/31
Plasmid DNA Extraction (mini-prep):
| gene | name | vector | size(bp) | conc(ng/ul) | site |
|---|---|---|---|---|---|
| pCI-1 | Bba_R0051 | pSB1A3 | 49 | 136 | yellow box |
| pCI-2 | Bba_R0051 | pSB1A4 | 49 | 66.6 | yellow box |
| LuxR-1 | Bba_C0062 | pSB1A2 | 781 | 119 | yellow box |
| LuxR-3 | Bba_C0062 | pSB1A2 | 781 | 119.8 | yellow box |
| RhlR-GFPmut3-positive | ACE-2 | pSB1A2 | 2572 | 308.8 | yellow box |
| ACE-4 | pSB1A2 | 2572 | 287.9 | yellow box | |
| Pc-RBS-Rhl-tt | PcA-1 | pSB1A2 | 908 | 82.3 | yellow box |
| PcA-3 | pSB1A2 | 908 | 79.8 | yellow box | |
| Pc-RBS-Las-tt | PcD-2 | pSB1A2 | 902 | 72.4 | yellow box |
| PcD-3 | pSB1A2 | 902 | 81.5 | yellow box | |
| pLas-RBS-LasR | seguence is not correct | ||||
| pRhl-RBS-RhlR | C1-2 | pSB1A2 | 797 | 104.2 | yellow box |
| C1-3 | pSB1A2 | 797 | 119.2 | yellow box | |
| RBS-GFPmut3-tt | E1-1 | pSB1AK3 | 867 | 129.2 | yellow box |
| E1-3 | pSB1AK3 | 867 | 91.1 | yellow box | |
| RBS-mCherry-tt | B5-1 | pSB1AK3 | 858 | 105.1 | yellow box |
| B5-3 | pSB1AK3 | 858 | 73.2 | yellow box | |
| RBS-mRFP-tt | seguence is not correct |
 "
"