Team:NTU-Taida/Project/Overview
From 2013.igem.org
(→Overview) |
|||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NTU-Taida/Templates/Header}}{{:Team:NTU-Taida/Templates/Navbar}}{{:Team:NTU-Taida/Templates/Sidebar|Title=Overview}}{{:Team:NTU-Taida/Templates/ContentStart}} | {{:Team:NTU-Taida/Templates/Header}}{{:Team:NTU-Taida/Templates/Navbar}}{{:Team:NTU-Taida/Templates/Sidebar|Title=Overview}}{{:Team:NTU-Taida/Templates/ContentStart}} | ||
| + | =Overview= | ||
Our IGEM project aims to tailor an '''instant bacteria identification array'''. We chose '''Quorum sensing molecules (QS molecules)''' instead of traditional biochemical reactions as our main targets to achieve our goals. As a cell-cell communication pathway, Quorum sensing phenomenon regulates '''virulence gene expression''' and has recently been considered as a new target for therapeutic application. We proposed a novel method to identify bacteria by '''the expression pattern''' of many QS receptors from the intensity of fluorescent signals. | Our IGEM project aims to tailor an '''instant bacteria identification array'''. We chose '''Quorum sensing molecules (QS molecules)''' instead of traditional biochemical reactions as our main targets to achieve our goals. As a cell-cell communication pathway, Quorum sensing phenomenon regulates '''virulence gene expression''' and has recently been considered as a new target for therapeutic application. We proposed a novel method to identify bacteria by '''the expression pattern''' of many QS receptors from the intensity of fluorescent signals. | ||
| - | + | For the sakes of sensitivity and the consideration of time, we underwent process of cloning and designed both '''positive feedback''' and '''negative regulation circuits''' insides plasmids containing quorum sensing receptors (or transcriptional regulator) conjugated with florescent proteins. These plasmids were transformed into ''E. Coli'' DH5α. They would act as '''biosensors''' when meeting quorum sensing molecules. | |
| - | + | We would like to identify bacterial strains both '''between species''' and '''within species'''. For Gram negative species, AHL is a common quorum sensing molecule. Different receptors, however, target different kinds of '''AHL (Acylhomoserine Lactone)''' depending on its carbon number of the acyl group. We targeted to construct an array of QS sensors, consisting of sensor for 4-carbon, 8-carbon, 12-carbon and 14-carbon AHL. Because QS sensors are also able to sense AHL molecules with '''a closer number of carbons''', this biosensor array can detects 2~14 carbon AHLs. In addition to AHL, we also developed '''novel biosensor—PQS''' for new type of quorum sensing molecule in iGem—'''Quinolones'''. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | We would like to identify bacterial strains both '''between species | + | |
To achieve a complete array, a lot of '''measurements''' are indispensable. We manipulated simple qualitative experiments for its property and the fluorescent protein expression via polyacrylamide gel electrophoresis stained by coomassie blue. Next, we bought molecules including AHL and Quinolones and utilized '''ELISA plate reader''' and '''flow cytometer''' to calibrate the '''standard diagram''' (intensity of florescent protein—time) of different biosensors added with series dilution of AHL in 96-well plate. At the third stage, the '''supernatant of bacteria''' (between species and within species) were taken into experiments. By replacement of AHL or Quinolones with the supernatant of bacteria, we depicted new curves, and compared it with each other and also standard diagram. With the efforts of the teams, we proved our idea. The responses of different strains of bacteria to our array varied greatly. | To achieve a complete array, a lot of '''measurements''' are indispensable. We manipulated simple qualitative experiments for its property and the fluorescent protein expression via polyacrylamide gel electrophoresis stained by coomassie blue. Next, we bought molecules including AHL and Quinolones and utilized '''ELISA plate reader''' and '''flow cytometer''' to calibrate the '''standard diagram''' (intensity of florescent protein—time) of different biosensors added with series dilution of AHL in 96-well plate. At the third stage, the '''supernatant of bacteria''' (between species and within species) were taken into experiments. By replacement of AHL or Quinolones with the supernatant of bacteria, we depicted new curves, and compared it with each other and also standard diagram. With the efforts of the teams, we proved our idea. The responses of different strains of bacteria to our array varied greatly. | ||
| - | To put it into practice, we aligned different | + | To put it into practice, we aligned circuits of different quorum sensing receptors to form an array to test '''clinical samples''' like septum, blood, urine, etc. After collecting enough data of different clinical bacterial strains and build up a database, we hope to build up a new system for bacteria identification. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[File:NTU-Taida-overview-1.jpg|700px|thumb|center|Figure.3]] | ||
{{:Team:NTU-Taida/Templates/ContentEnd}}{{:Team:NTU-Taida/Templates/Footer|ActiveNavbar=Project, #nav-Modeling-Overview}} | {{:Team:NTU-Taida/Templates/ContentEnd}}{{:Team:NTU-Taida/Templates/Footer|ActiveNavbar=Project, #nav-Modeling-Overview}} | ||
Latest revision as of 03:24, 28 September 2013
Overview
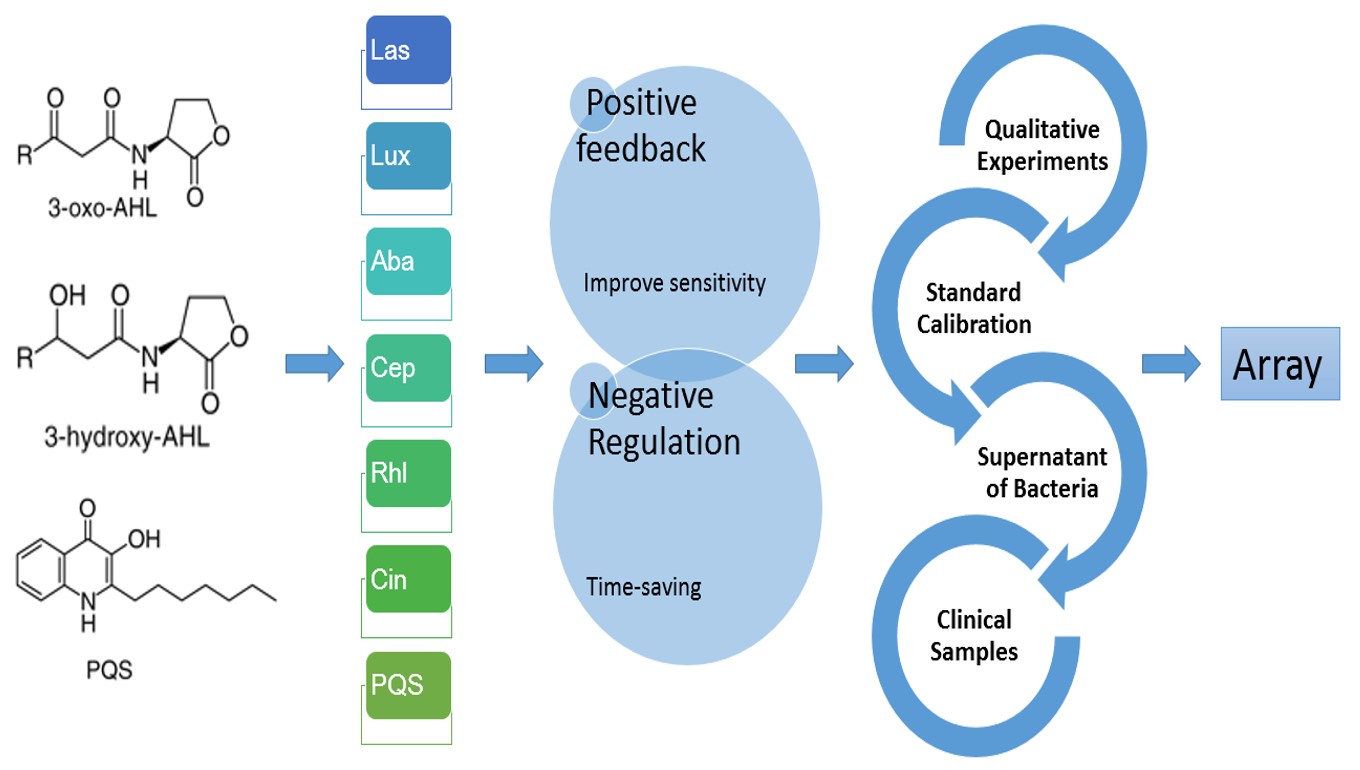
Our IGEM project aims to tailor an instant bacteria identification array. We chose Quorum sensing molecules (QS molecules) instead of traditional biochemical reactions as our main targets to achieve our goals. As a cell-cell communication pathway, Quorum sensing phenomenon regulates virulence gene expression and has recently been considered as a new target for therapeutic application. We proposed a novel method to identify bacteria by the expression pattern of many QS receptors from the intensity of fluorescent signals.
For the sakes of sensitivity and the consideration of time, we underwent process of cloning and designed both positive feedback and negative regulation circuits insides plasmids containing quorum sensing receptors (or transcriptional regulator) conjugated with florescent proteins. These plasmids were transformed into E. Coli DH5α. They would act as biosensors when meeting quorum sensing molecules.
We would like to identify bacterial strains both between species and within species. For Gram negative species, AHL is a common quorum sensing molecule. Different receptors, however, target different kinds of AHL (Acylhomoserine Lactone) depending on its carbon number of the acyl group. We targeted to construct an array of QS sensors, consisting of sensor for 4-carbon, 8-carbon, 12-carbon and 14-carbon AHL. Because QS sensors are also able to sense AHL molecules with a closer number of carbons, this biosensor array can detects 2~14 carbon AHLs. In addition to AHL, we also developed novel biosensor—PQS for new type of quorum sensing molecule in iGem—Quinolones.
To achieve a complete array, a lot of measurements are indispensable. We manipulated simple qualitative experiments for its property and the fluorescent protein expression via polyacrylamide gel electrophoresis stained by coomassie blue. Next, we bought molecules including AHL and Quinolones and utilized ELISA plate reader and flow cytometer to calibrate the standard diagram (intensity of florescent protein—time) of different biosensors added with series dilution of AHL in 96-well plate. At the third stage, the supernatant of bacteria (between species and within species) were taken into experiments. By replacement of AHL or Quinolones with the supernatant of bacteria, we depicted new curves, and compared it with each other and also standard diagram. With the efforts of the teams, we proved our idea. The responses of different strains of bacteria to our array varied greatly.
To put it into practice, we aligned circuits of different quorum sensing receptors to form an array to test clinical samples like septum, blood, urine, etc. After collecting enough data of different clinical bacterial strains and build up a database, we hope to build up a new system for bacteria identification.
 "
"