Team:Heidelberg/Delftibactin/MMCoA
From 2013.igem.org
(Difference between revisions)
Nils.kurzawa (Talk | contribs) m |
Nils.kurzawa (Talk | contribs) m |
||
| (10 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
<div class="container"> | <div class="container"> | ||
<!--Project Description--> | <!--Project Description--> | ||
| - | <div | + | <div> |
<h1><span style="font-size:140%;color:#FFCC00;">Methylmalonyl-CoA Pathway.</span><span class="text-muted" style="font-family:Arial, sans-serif; font-size:90%"> Making the whole thing work.</span></h1> | <h1><span style="font-size:140%;color:#FFCC00;">Methylmalonyl-CoA Pathway.</span><span class="text-muted" style="font-family:Arial, sans-serif; font-size:90%"> Making the whole thing work.</span></h1> | ||
| - | + | ||
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col-sm- | + | <div class="col-sm-12 col-md-6"> |
<!--Months--> | <!--Months--> | ||
| - | <ul class="pagination" style="margin-bottom:2%; margin-left: | + | <ul class="pagination" style="margin-bottom:2%; margin-left:17%;"> |
<!--<button type="button" class="btn btn-default">May</button> | <!--<button type="button" class="btn btn-default">May</button> | ||
| Line 55: | Line 55: | ||
</ol>--> | </ol>--> | ||
| - | <div class="carousel-inner" style="margin-top: | + | <div class="carousel-inner" style="margin-top:15%"> |
<div class="item active june first"> | <div class="item active june first"> | ||
| Line 62: | Line 62: | ||
<h1>Week 6</h1> | <h1>Week 6</h1> | ||
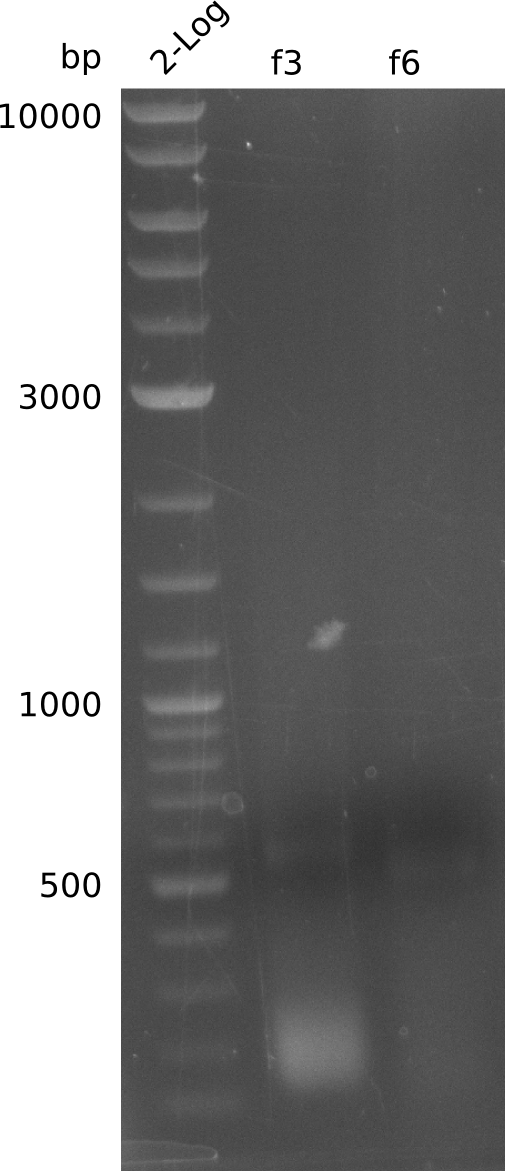
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In this week the first experiments for the genomic integration of the methyl-malonyl-CoA synthetase were set up. First of all the plasmids were transformed into TOP10 grown and purified. The first attempts on the amplification of the fragment didn't work at all and we ended up with a specific product of the wrong size.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 76: | Line 71: | ||
<h1>Week 7</h1> | <h1>Week 7</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After a validation of the template plasmid via restriction digest the PCR reaction was optimised. Unfortunately none of the cells transformed with the PCR product using electroporation could be positively screened, no matter how the PCR fragment was amplified.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 90: | Line 80: | ||
<h1>Week 8</h1> | <h1>Week 8</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">As the repetition of the colony PCR under different conditions was still negative, new PCR products were prepared and pooled. To eliminate possible errors fresh competent cells were prepared.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 104: | Line 89: | ||
<h1>Week 9</h1> | <h1>Week 9</h1> | ||
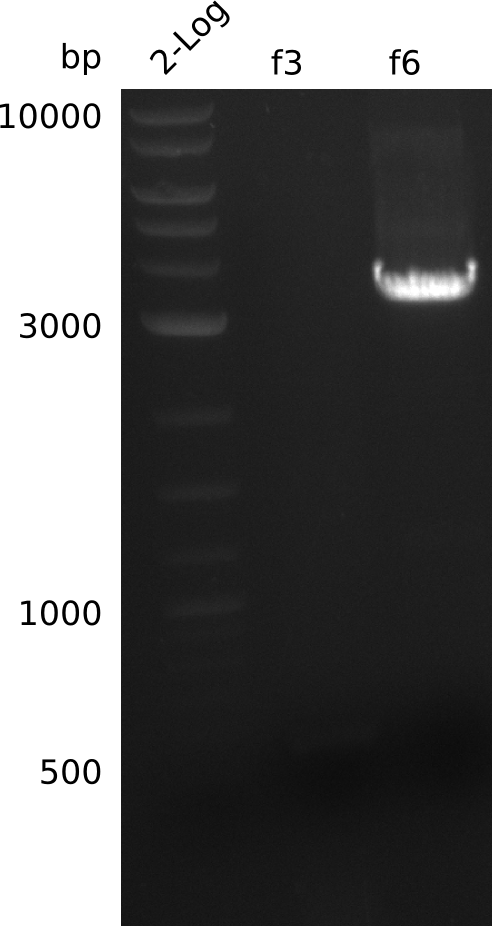
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After doing colony PCR again and getting inconclusive results, it was decided that the genomic integration failed using this experimental setup. Thus we started from scratch using a new primer pair and finally it turned out that the polymerase was causing the problems. By the end of the week BAP1-pKD46 could be transformed.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 118: | Line 98: | ||
<h1>Week 10</h1> | <h1>Week 10</h1> | ||
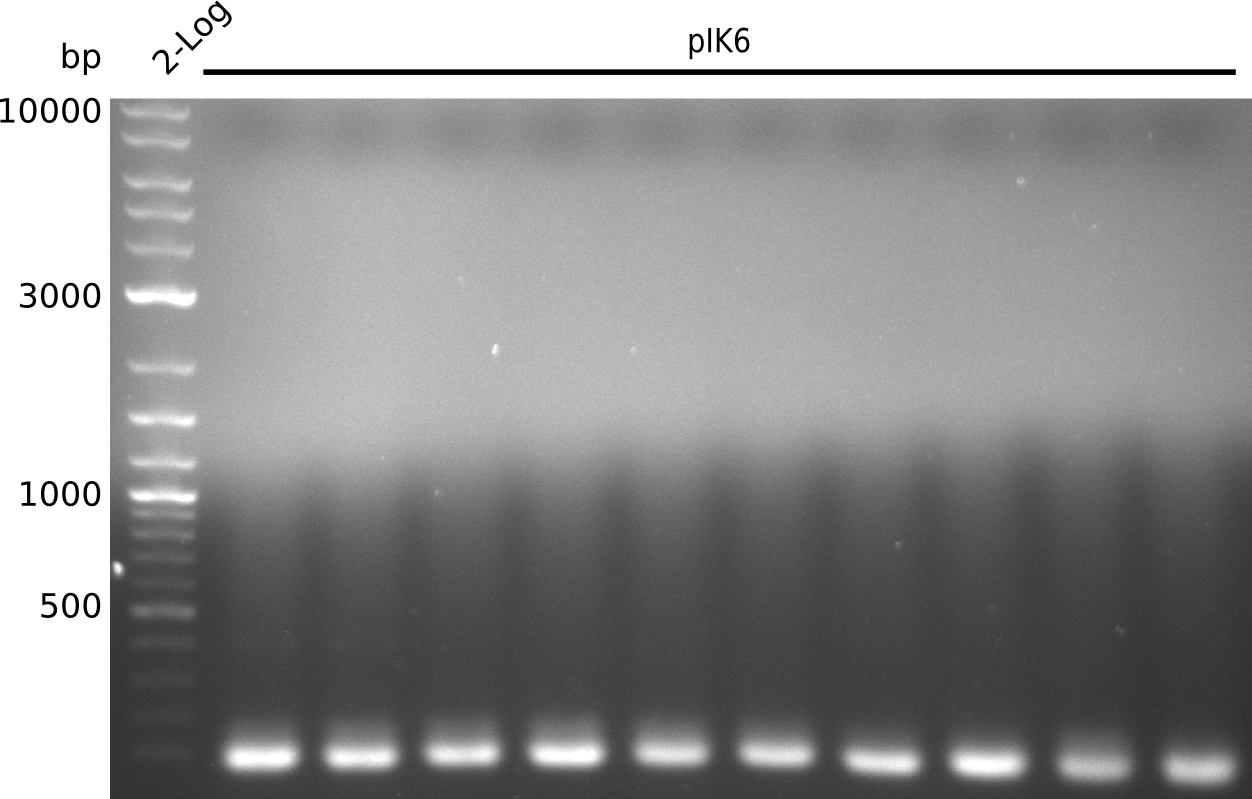
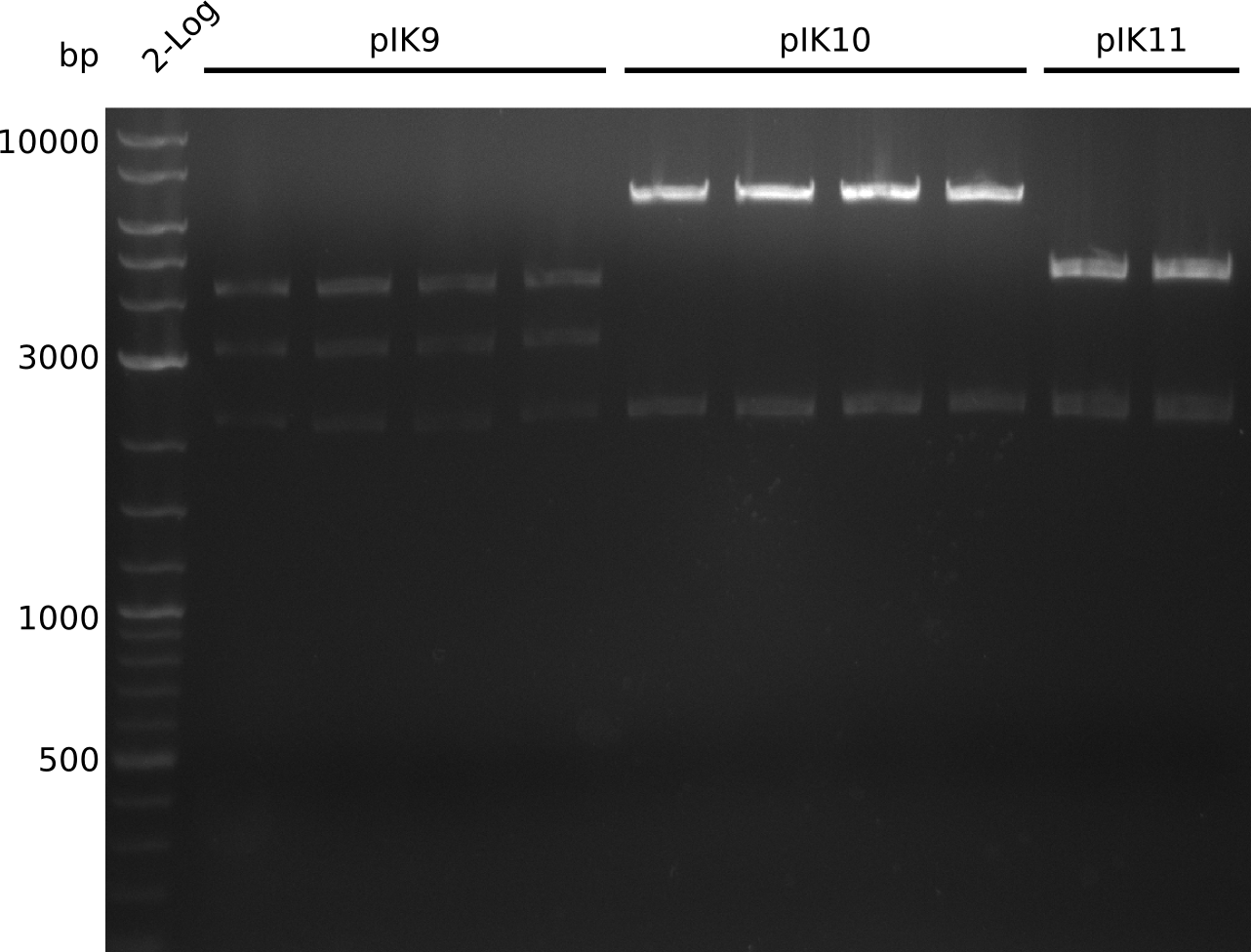
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Even after running several colony PCRs there were no positive clones to be found. Thus the pKD46 was digested for validation. On the gel a miniprep from the PCR-transformed cells was included. The identity of the pKD46 was fine and the miniprep didn't contain any DNA, as expected.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 131: | Line 106: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 11</h1> | <h1>Week 11</h1> | ||
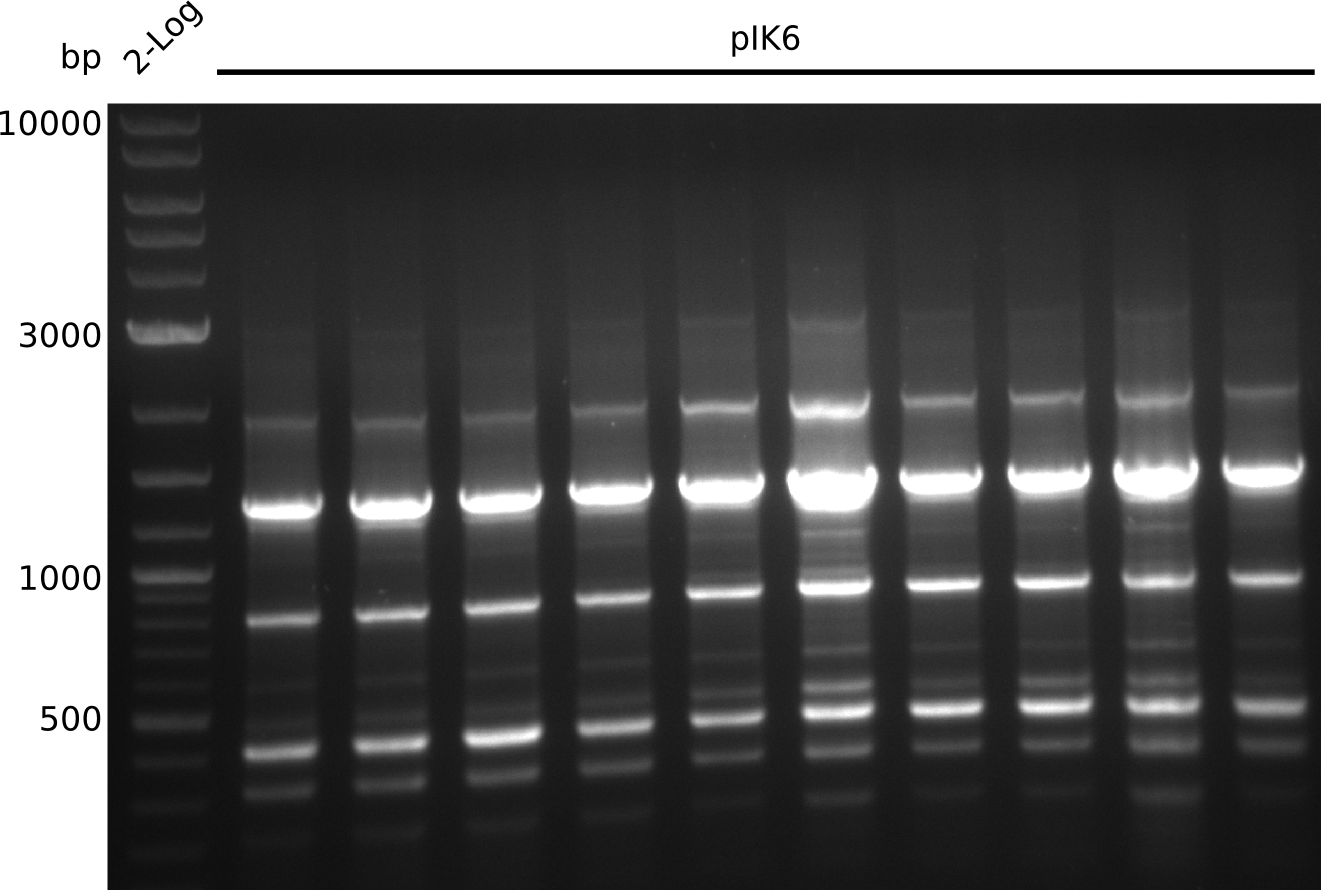
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In this week the whole procedure was repeated again from fragment amplification to extensive colony PCR. This week several different colony types were encountered and one that grew only after 3 days actually gave a band at 1.2 kb, but it turned out to be unspecific.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 145: | Line 114: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 12</h1> | <h1>Week 12</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Several possible error sources such as unselective plates were investigated, but all turned out to be fine. The colony PCR was then repeated using yet another primer pair, but it still only gave unspecific or no bands.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 159: | Line 122: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 13</h1> | <h1>Week 13</h1> | ||
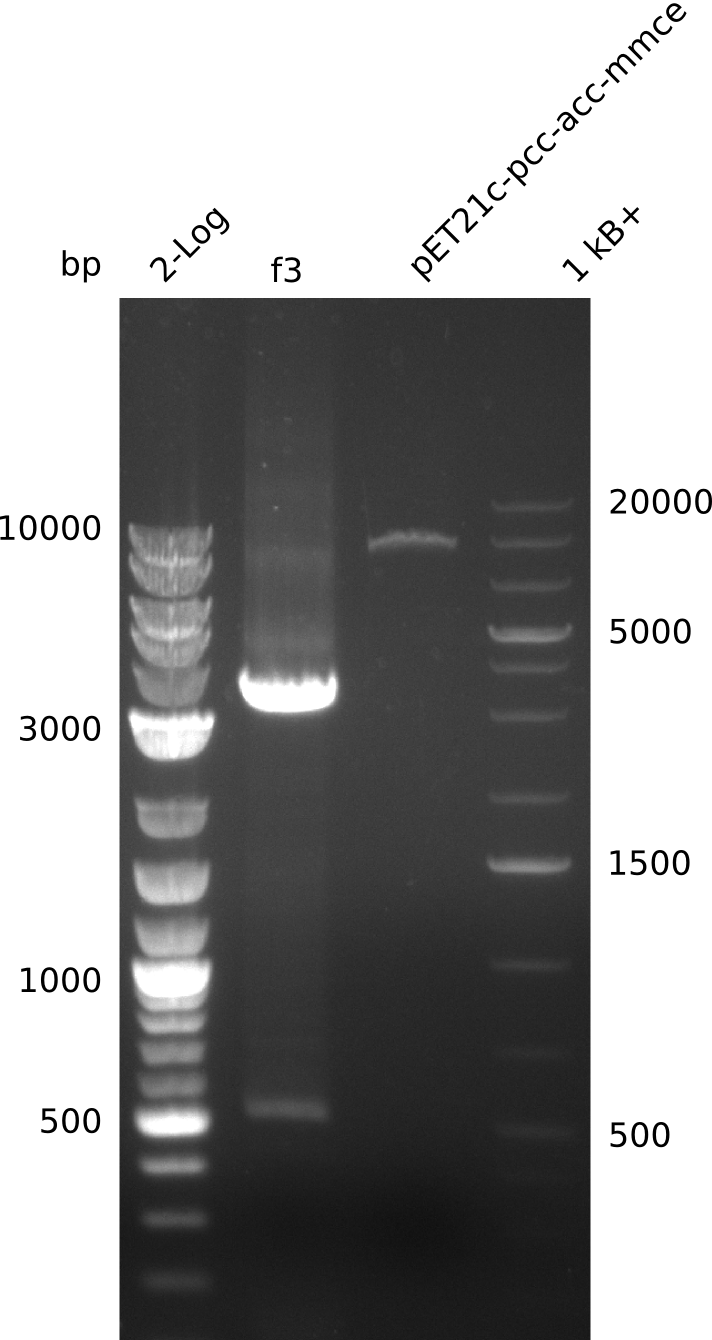
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to check whether the cell line used was actually the right one primers against sfp were used and cell from other subgroups were included. It turned out that something really weird was going onwith the cells, since sfp could not be detected. Thus the entirely new strategy of generating a plasmid instead of doing genomic integration was initialised.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 168: | Line 130: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 14</h1> | <h1>Week 14</h1> | ||
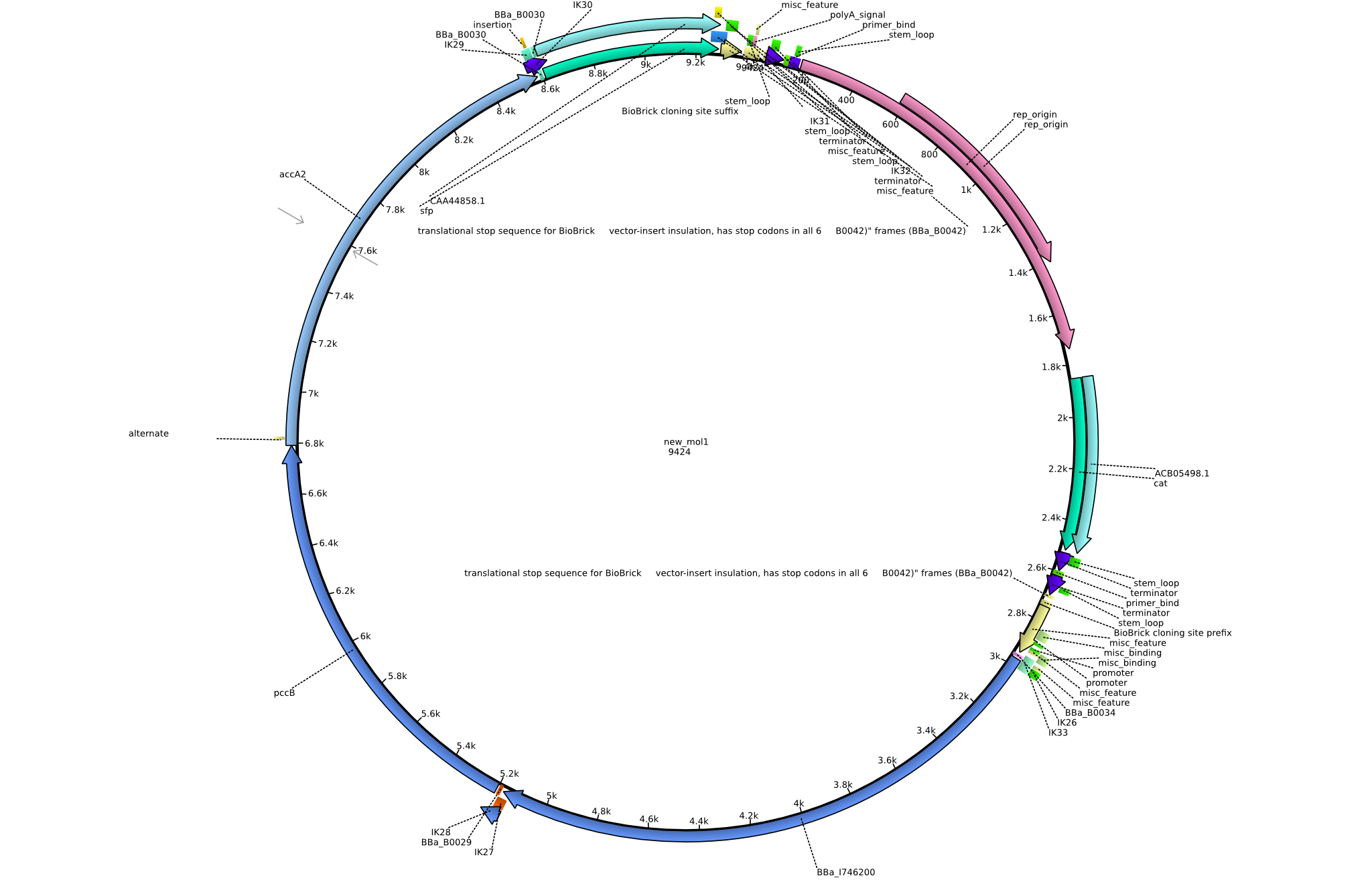
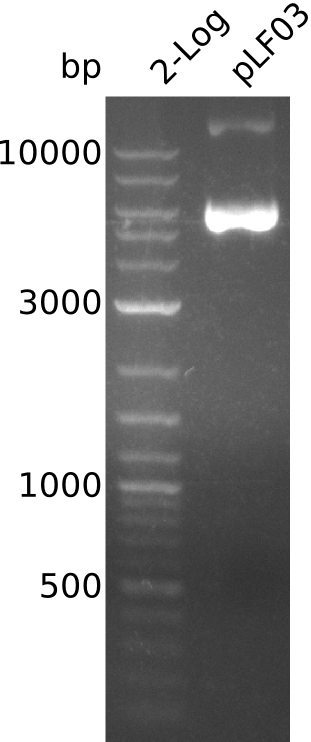
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">To be really sure about the sfp a new plasmid preparation was made and cells were tranformed again. In parallel the pLF03 was also prepared and sent for sequencing. It turned out, that there is a deletion in the sequence and thus the primers can't bind properly.</p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 177: | Line 139: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 15</h1> | <h1>Week 15</h1> | ||
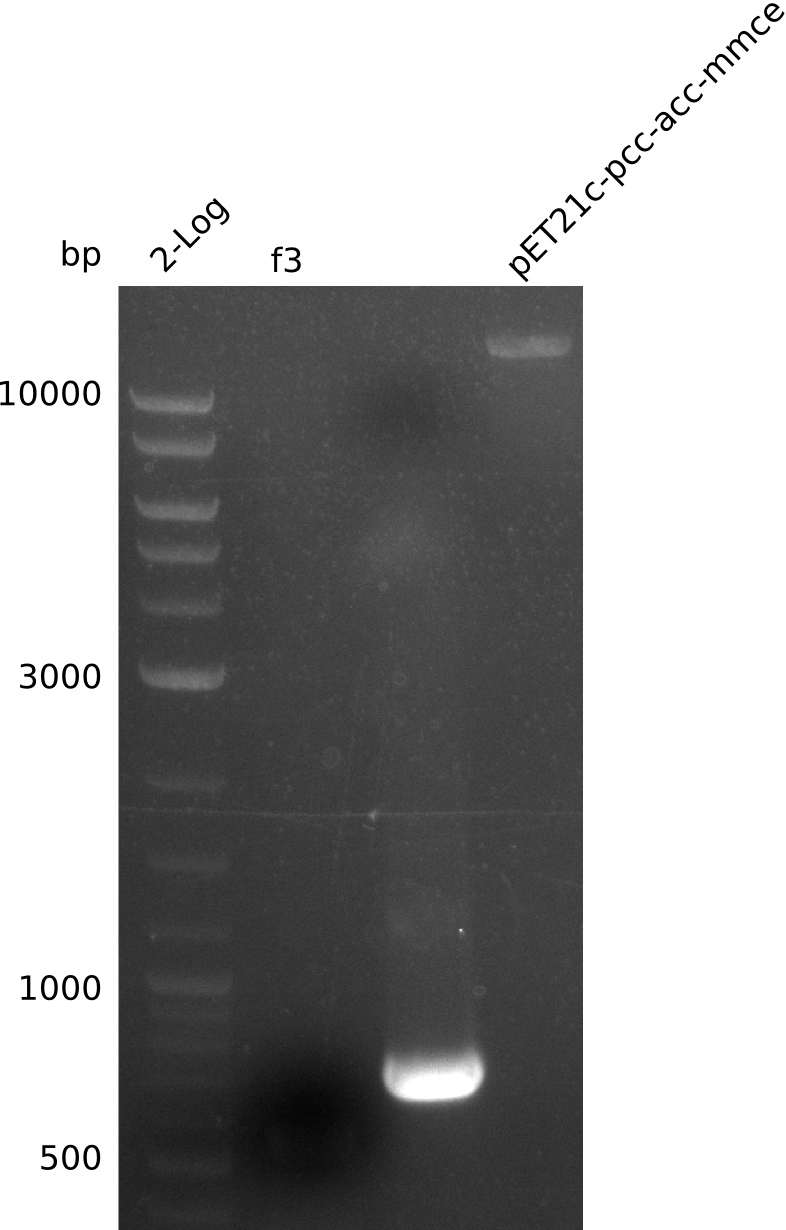
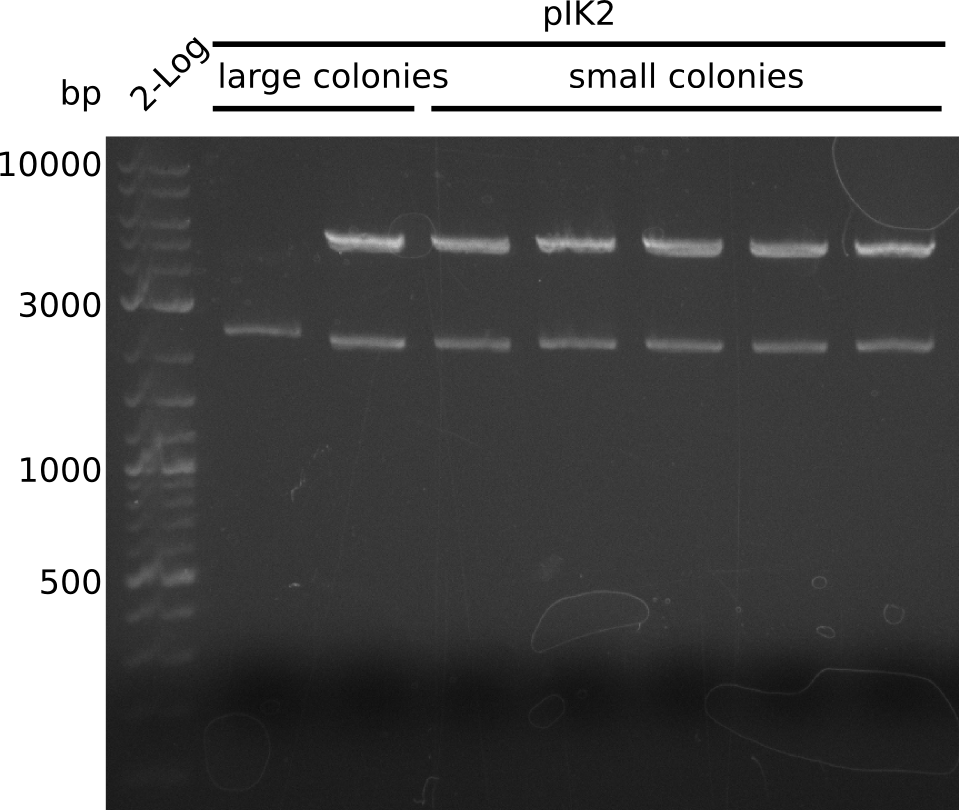
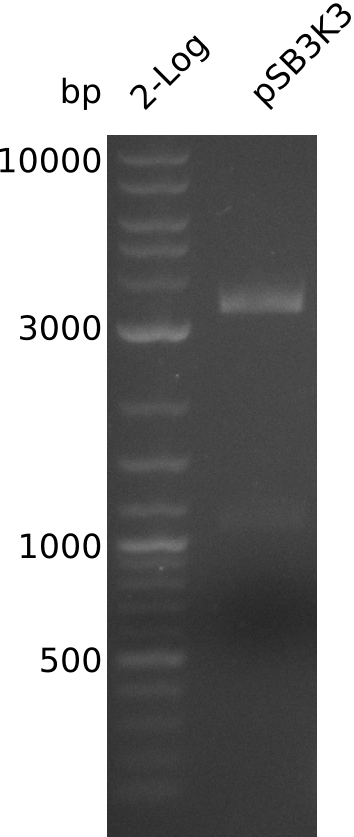
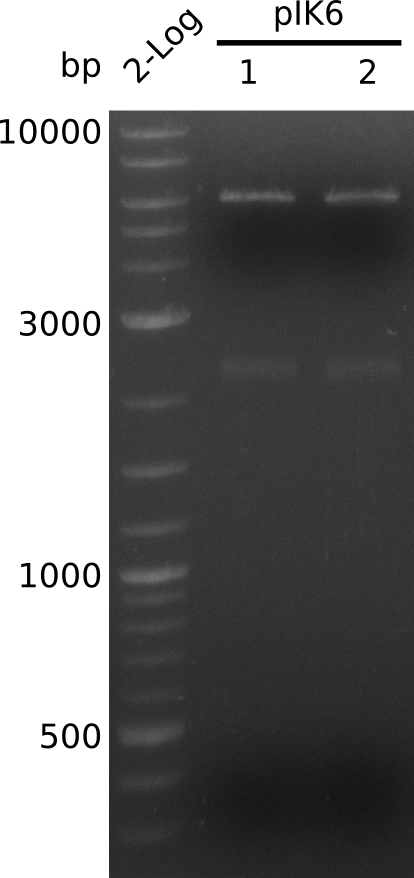
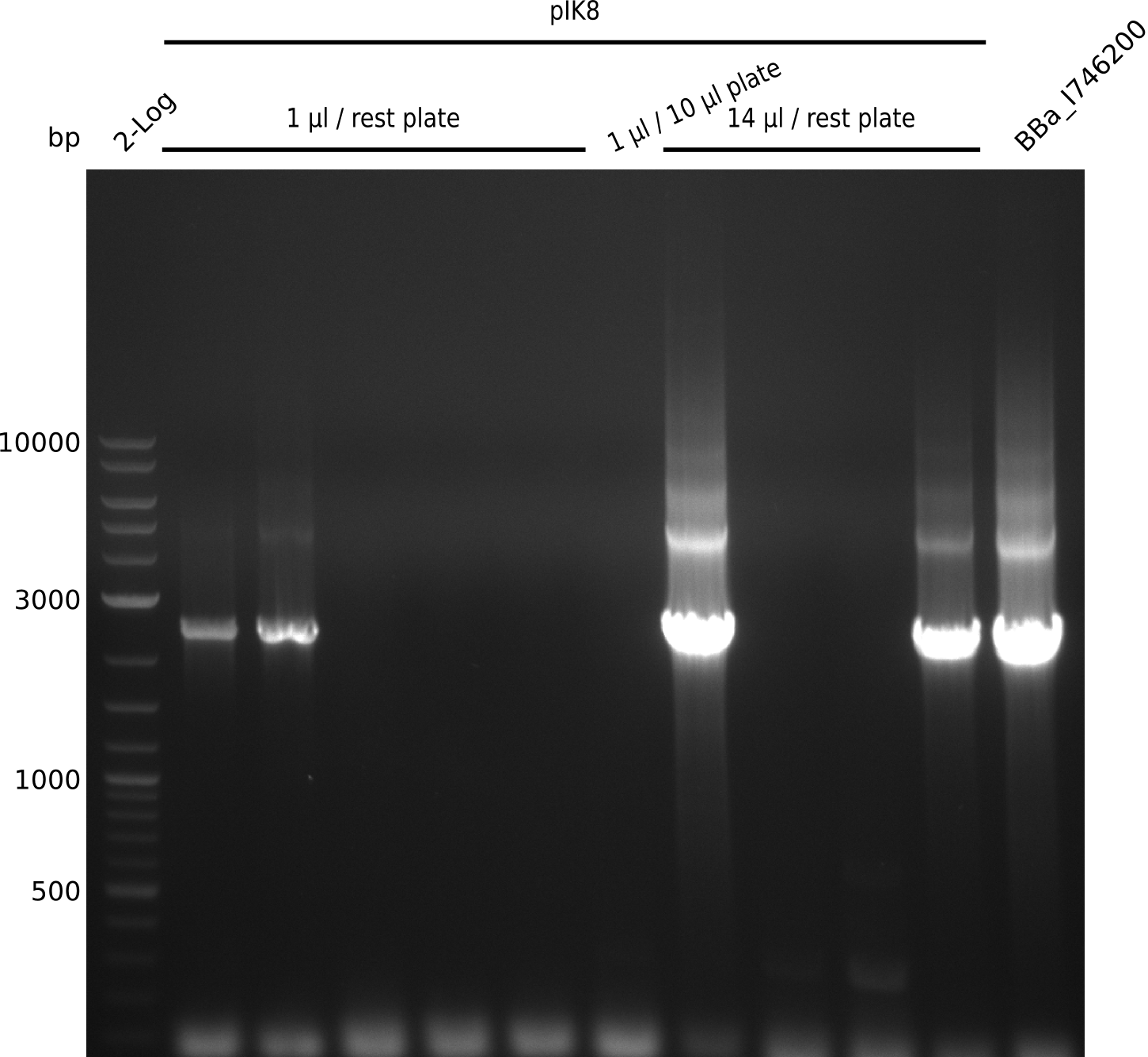
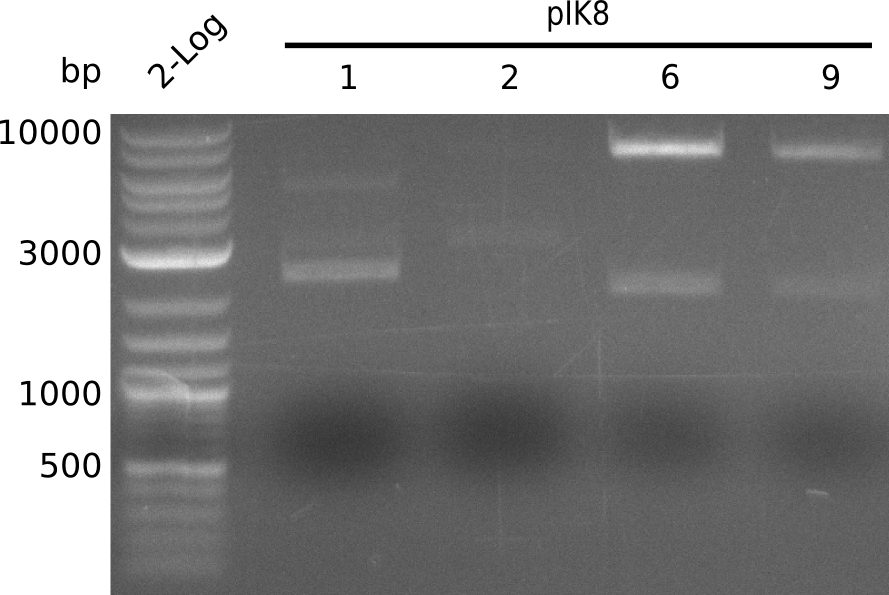
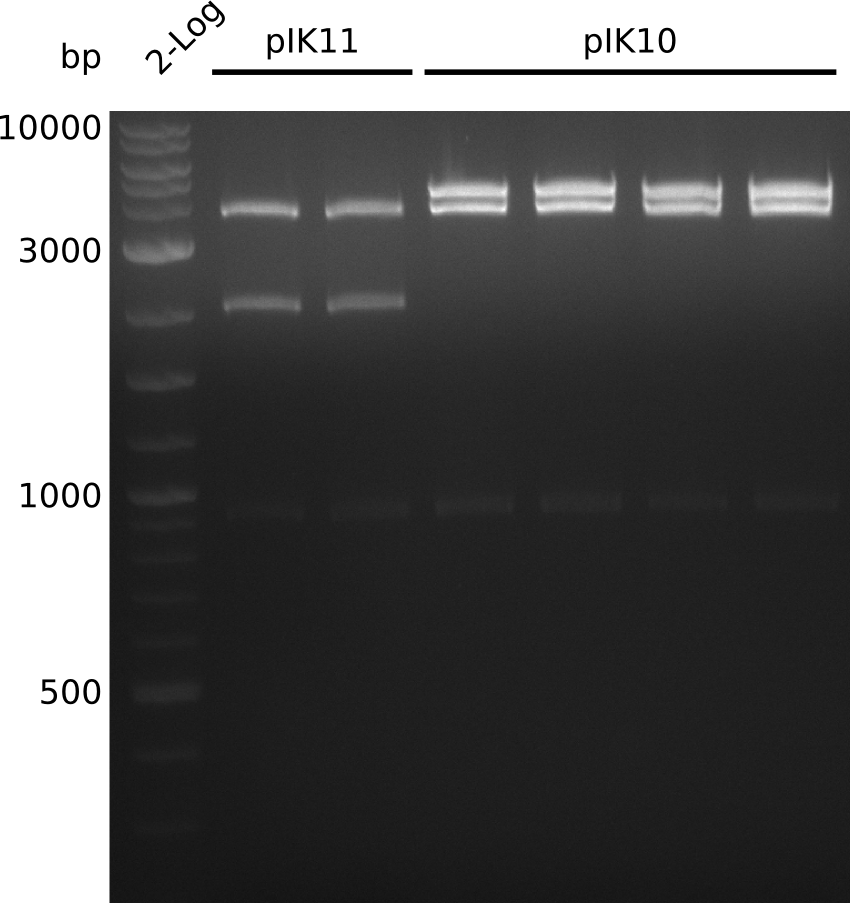
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">A final try on integrating the methyl-malonyl-CoA pathway was made. At the same time new plasmids carrying the pathway arrived and had to be validated first via PCR and restriction digestion. Several PCR reaction were run and also the first Gibson assembly and transformation was done for both plasmids. The positive colonies were sent for sequencing by the end of the week.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 186: | Line 147: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 16</h1> | <h1>Week 16</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After finally giving up on the genomic integration we focused on the plasmid strategy, where the sequencing results of the two plasmids arrived. As they were already positive glycerol stocks were prepared.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 196: | Line 156: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 17</h1> | <h1>Week 17</h1> | ||
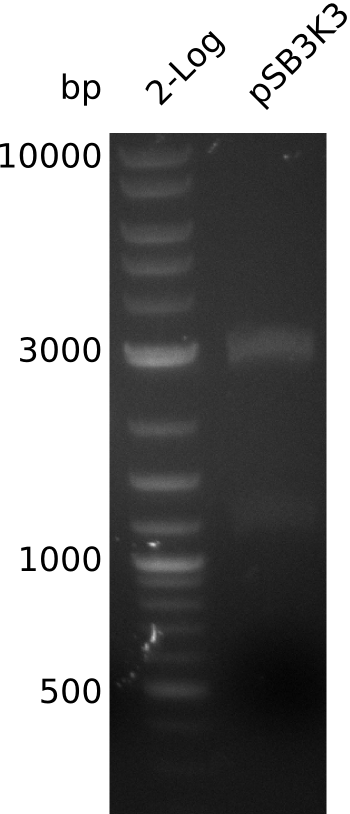
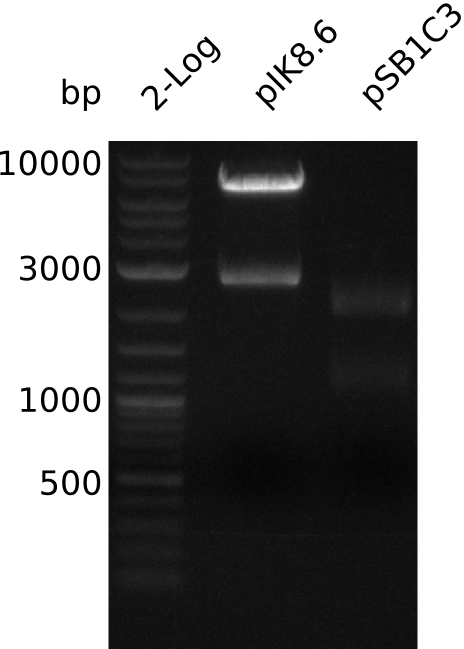
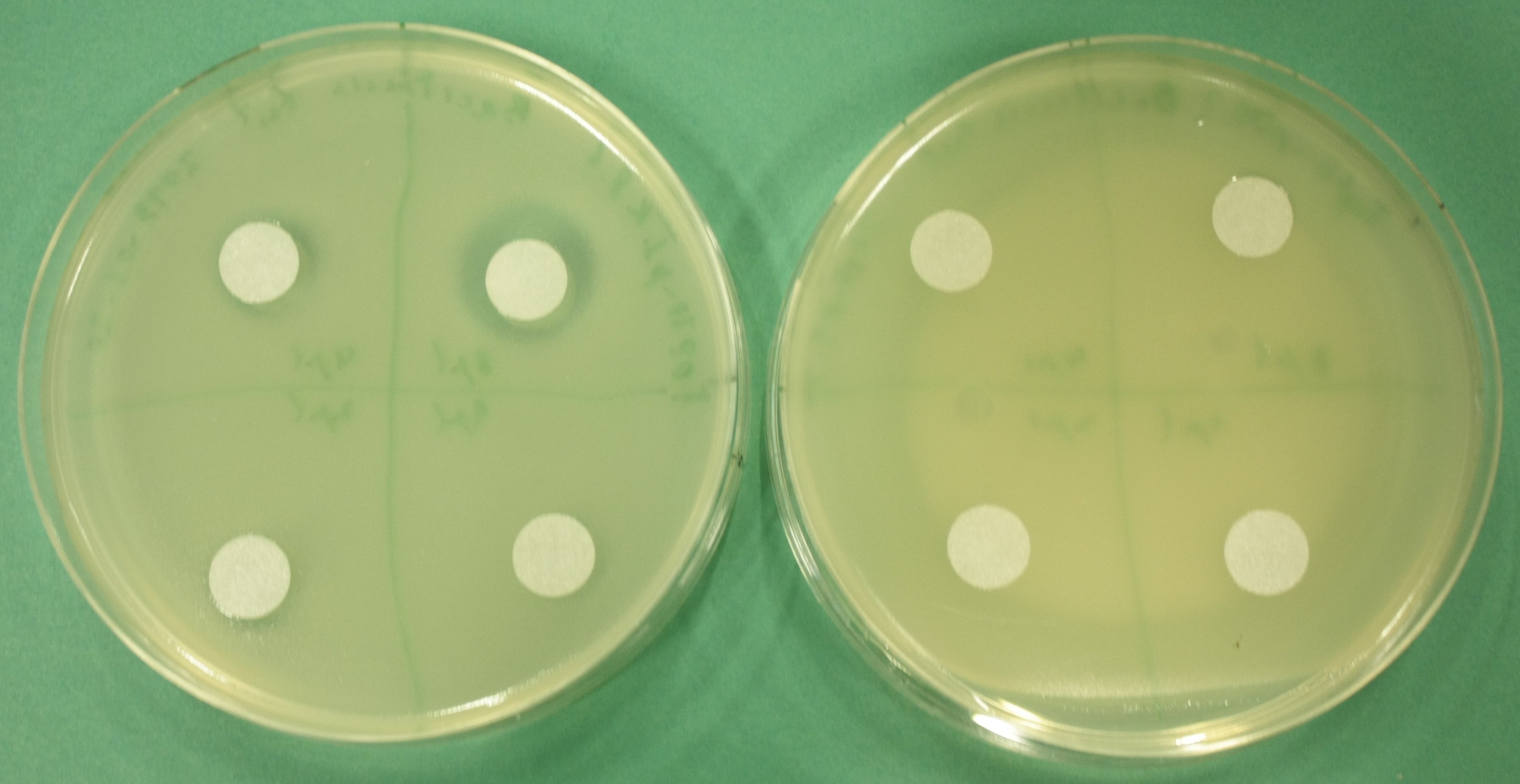
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to check for sfp expression pMM64, carrying an indigoidine synthetase was cotransformed, but didn't give any blue colonies. Thus the plasmid backbone had to be changed to allow for cotransformation with our own indigoidine construct. One of the new plasmids was sent for sequencing.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 206: | Line 165: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 18</h1> | <h1>Week 18</h1> | ||
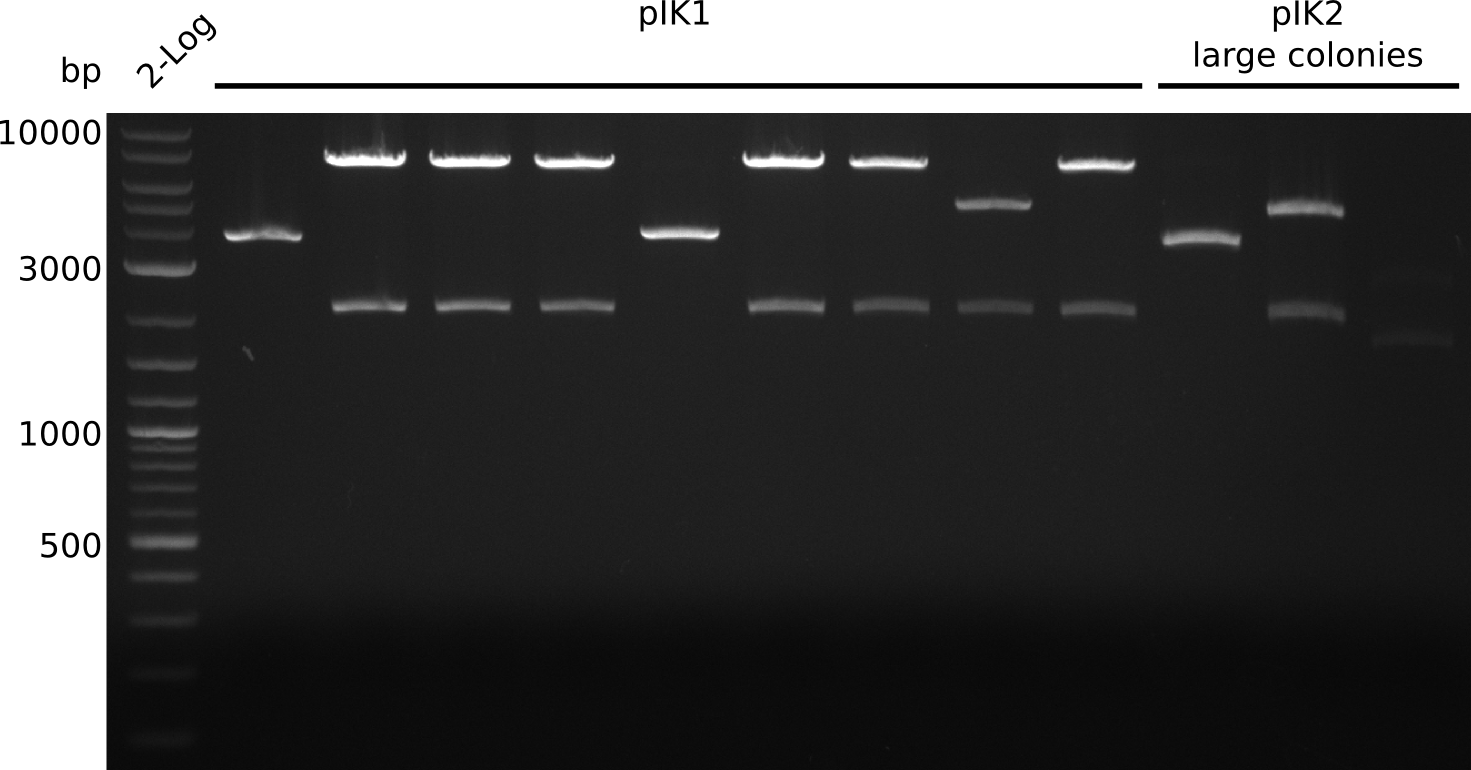
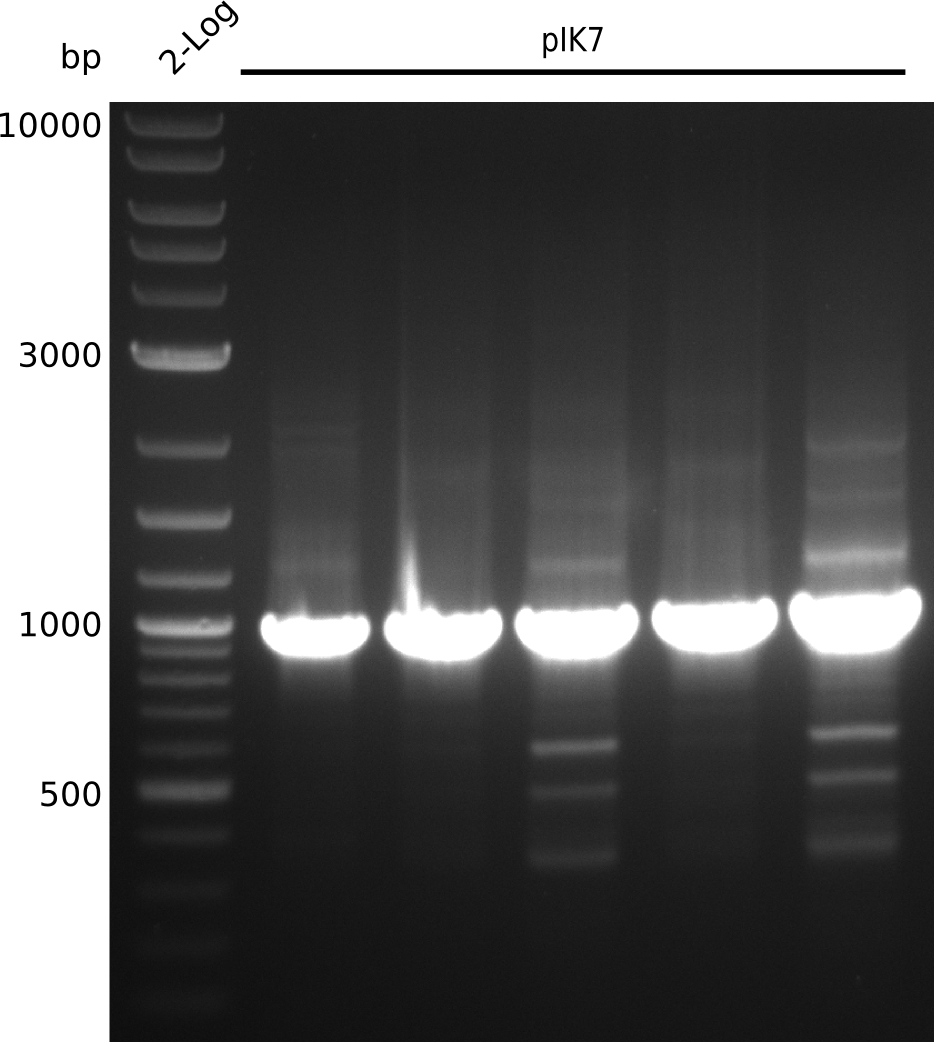
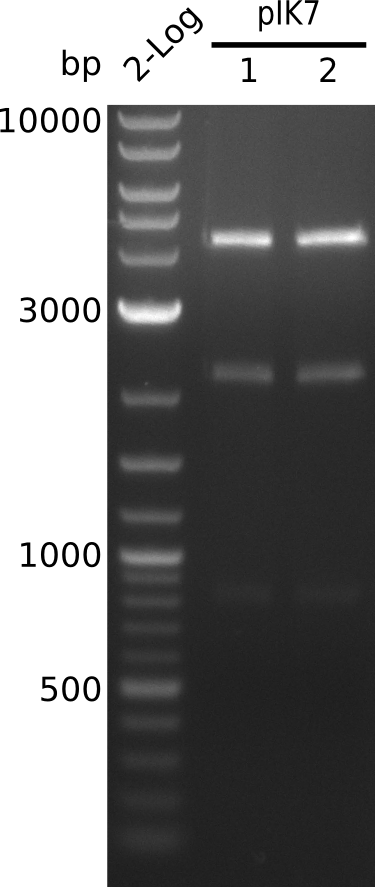
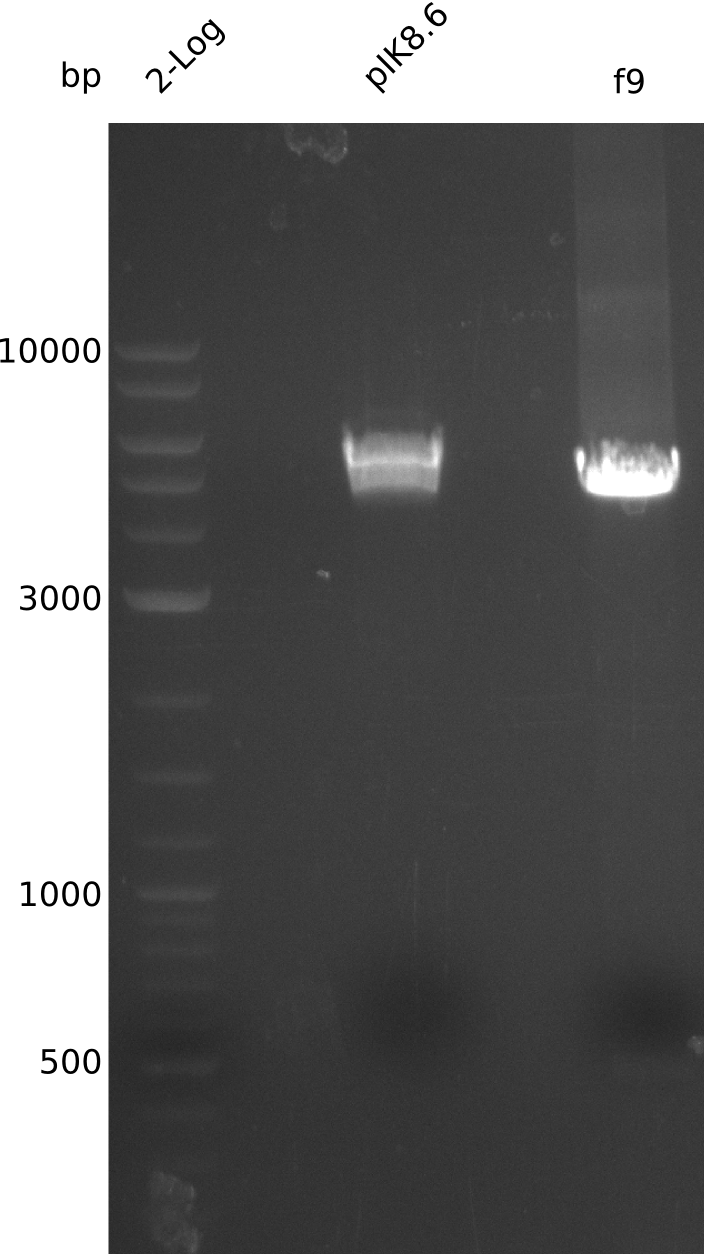
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The sequencing results of pIK1.3 revealed a point mutation in the permeability device. All the other plasmids sent for sequencing also carried some kind of mutation that introduced a stop codon within the permeability device, which is thus non-functional. Moreover there were still no blue colonies even though we now used our own construct.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 216: | Line 174: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 19</h1> | <h1>Week 19</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After doing some research in 2007's Cambridge team wiki it was found, that the permeability device is probably toxic to the cells and can thus only be succesfully cloned behind a very weak promotor.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 226: | Line 183: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 20</h1> | <h1>Week 20</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we grew the first blue colonies with an updated version of the indigoidine construct. We also put the permeability device behind a very weak promotor and prepared new plasmids for sequencing.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 236: | Line 192: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 21</h1> | <h1>Week 21</h1> | ||
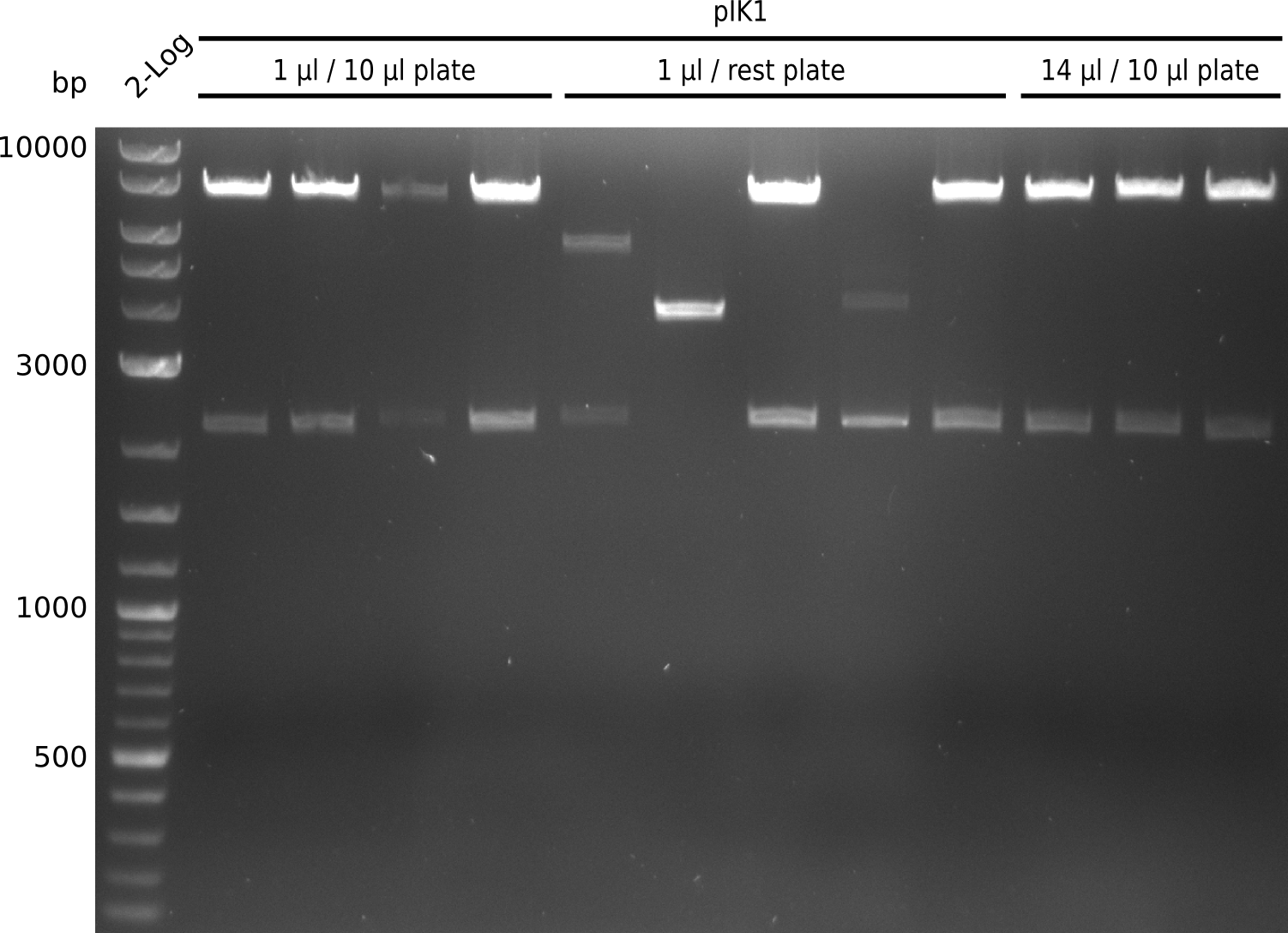
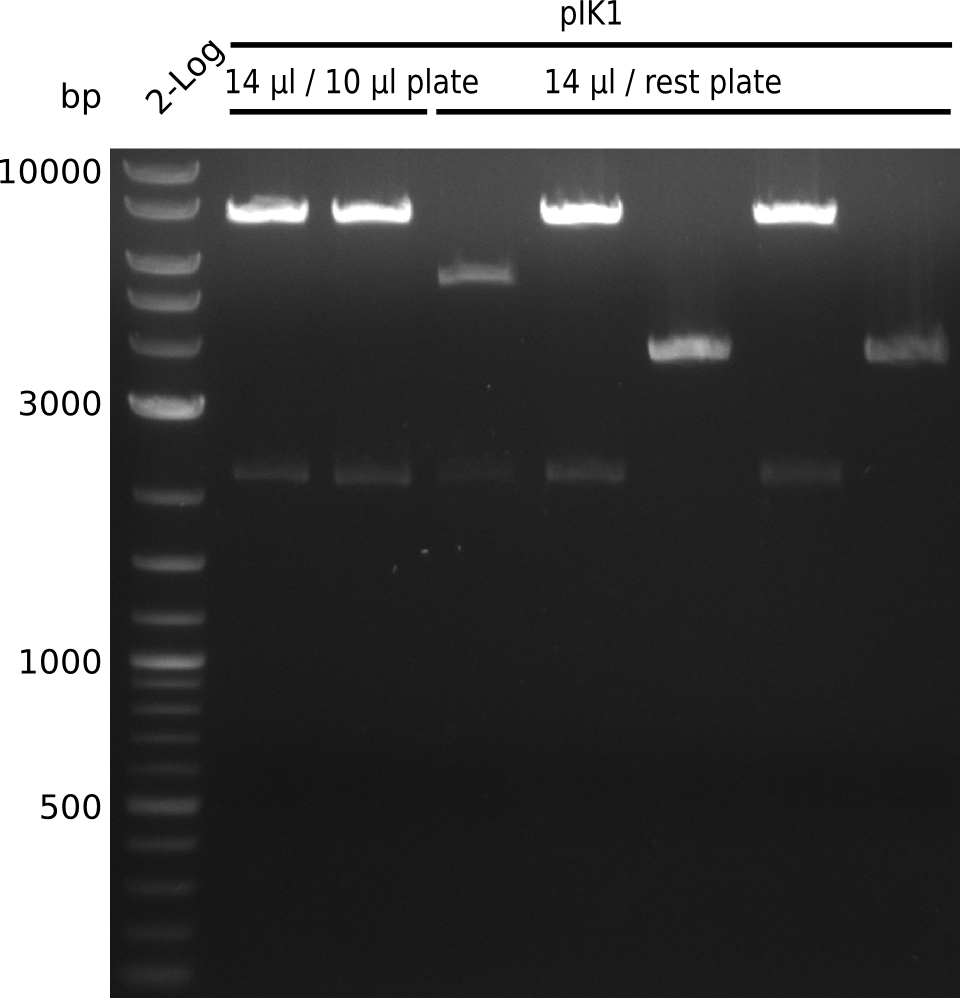
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">When the sequencing results were analysed there were no mutations in the permeability device. Now the cloning of the plasmids for the parts submission started and by the end of the week we sent them for sequening.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 246: | Line 201: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 22</h1> | <h1>Week 22</h1> | ||
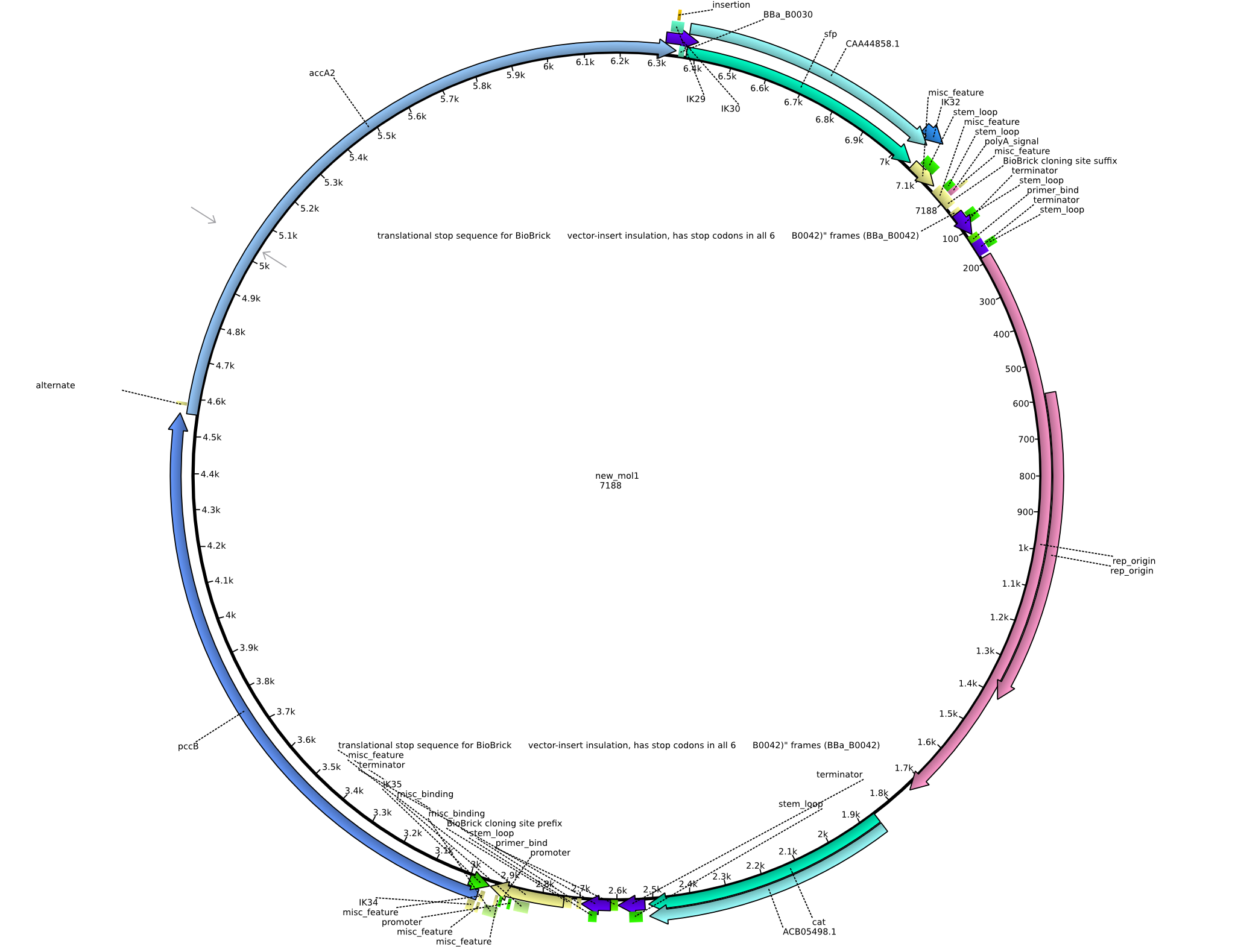
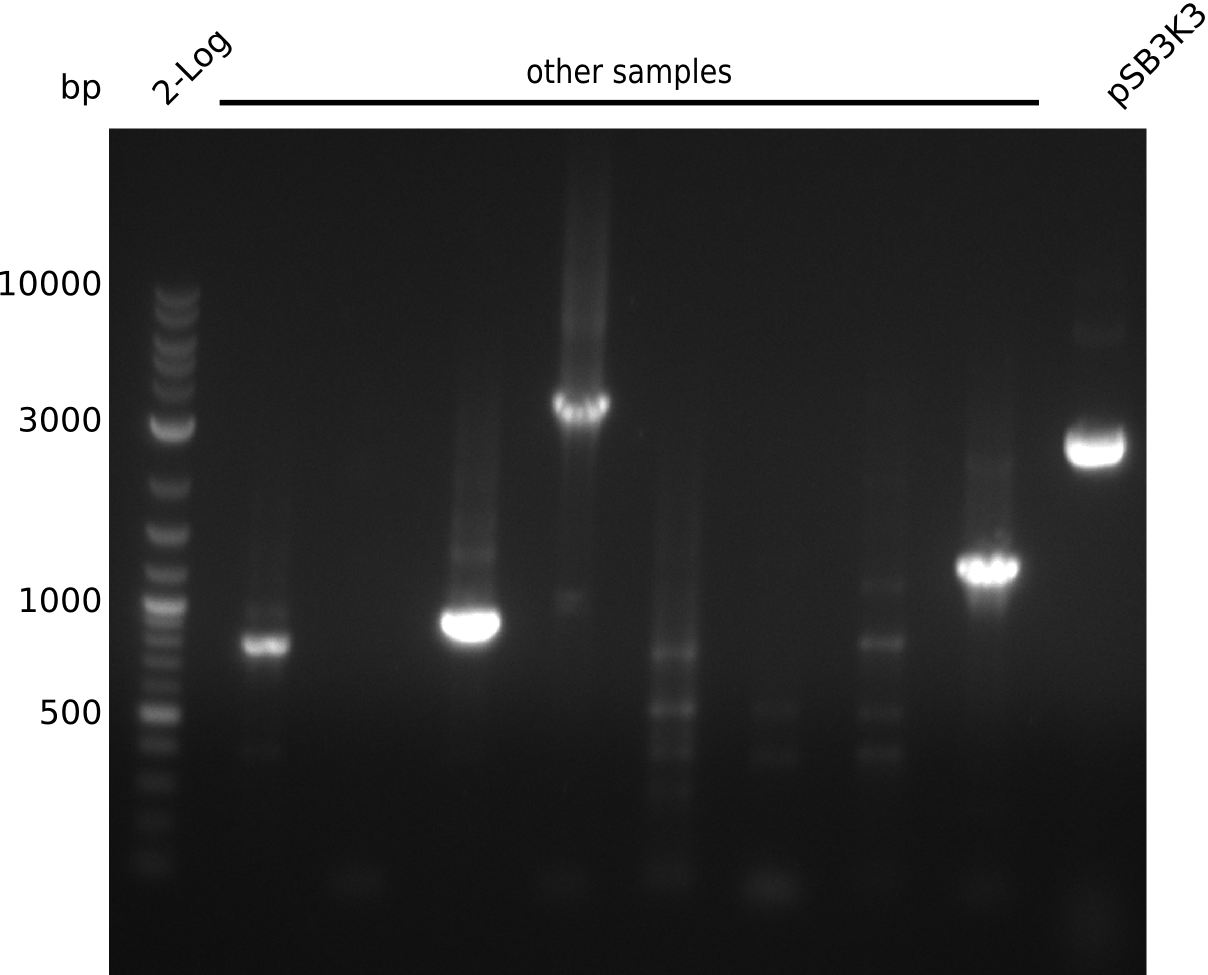
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Unfortunately we noticed that we accidentally introduced an additional restriction site and could thus not submit the part to the registry. We already removed the restriction site and validated it by restriction digestion. Anyway we are going to try different restriction enzymes and send it for sequencing after wiki freeze.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 257: | Line 211: | ||
</div> | </div> | ||
</div><!-- /.carousel --> | </div><!-- /.carousel --> | ||
| + | </div> | ||
| + | |||
| + | <div class="col-sm-12 col-md-6"> | ||
| + | <div class="jumbotron methods" data-spy="scroll" data-target="#navbarExample" data-offset="0" style="margin-top:10%"> | ||
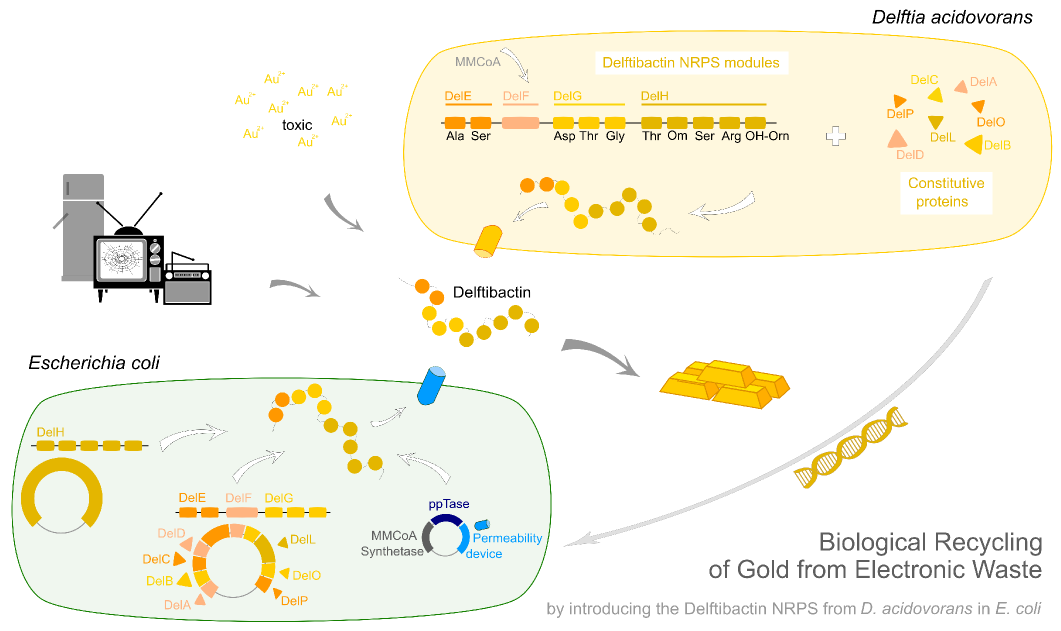
| + | <h3>Graphical Abstract</h3> | ||
| + | <div style="width:100%;"> | ||
| + | <a class="fancybox fancyGraphical" rel="group" href="https://static.igem.org/mediawiki/2013/e/e3/Heidelberg_ga_delf.png"> | ||
| + | |||
| + | <img style="width:100%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 4px; border-color: grey;" src="https://static.igem.org/mediawiki/2013/e/e3/Heidelberg_ga_delf.png"></img> | ||
| - | + | </a> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | </div> | |
</div> | </div> | ||
</div> | </div> | ||
| - | <div class="col-sm- | + | <div class="container"> |
| + | <div class="row"> | ||
| + | <div class="col-sm-12"> | ||
<!--Start Weekly Labjournal--> | <!--Start Weekly Labjournal--> | ||
| Line 277: | Line 242: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 307: | Line 272: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 333: | Line 298: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 359: | Line 324: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 384: | Line 349: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 410: | Line 375: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 435: | Line 400: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 461: | Line 426: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 498: | Line 463: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 531: | Line 496: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 564: | Line 529: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 594: | Line 559: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 617: | Line 582: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 640: | Line 605: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 663: | Line 628: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 686: | Line 651: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 709: | Line 674: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 717: | Line 682: | ||
</p> | </p> | ||
</div> | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| Line 725: | Line 691: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
{{:Team:Heidelberg/Templates/Footer-Nav}} | {{:Team:Heidelberg/Templates/Footer-Nav}} | ||
{{:Team:Heidelberg/Templates/Footer-MMCoA}} | {{:Team:Heidelberg/Templates/Footer-MMCoA}} | ||
Latest revision as of 09:42, 18 October 2013


 "
"