Team:Heidelberg/Delftibactin/DelRest
From 2013.igem.org
(Difference between revisions)
Nils.kurzawa (Talk | contribs) |
|||
| (39 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:Heidelberg/Templates/Navigation}} | {{:Team:Heidelberg/Templates/Navigation}} | ||
| + | {{:Team:Heidelberg/Templates/scrollbar-css}} | ||
| + | |||
<html> | <html> | ||
<style type="text/css"> | <style type="text/css"> | ||
| Line 11: | Line 9: | ||
p { | p { | ||
font-size:14px; | font-size:14px; | ||
| + | text-align:justify; | ||
| + | } | ||
| + | .carousel-inner { | ||
| + | margin-top:20%; | ||
| + | } | ||
| + | .tab-content .wikitable { | ||
| + | font-size:9pt; | ||
} | } | ||
</style> | </style> | ||
<div class="container"> | <div class="container"> | ||
<!--Project Description--> | <!--Project Description--> | ||
| - | <div | + | <div> |
| - | <h1><span style="font-size:180%;color:#FFCC00;">Del Rest.</span><span class="text-muted" style="font-size:120%"> Creating a 32 | + | <h1><span style="font-size:180%;color:#FFCC00;">Del Rest.</span><span class="text-muted" style="font-size:120%"> Creating a 32 kb plasmid.</span></h1> |
| - | + | ||
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col-sm- | + | <div class="col-sm-12 col-md-6"> |
<!--Months--> | <!--Months--> | ||
| - | <ul class="pagination" style="margin-bottom:2%; margin-left: | + | <ul class="pagination" style="margin-bottom:2%; margin-left:17%;"> |
<!--<button type="button" class="btn btn-default">May</button> | <!--<button type="button" class="btn btn-default">May</button> | ||
| Line 65: | Line 70: | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
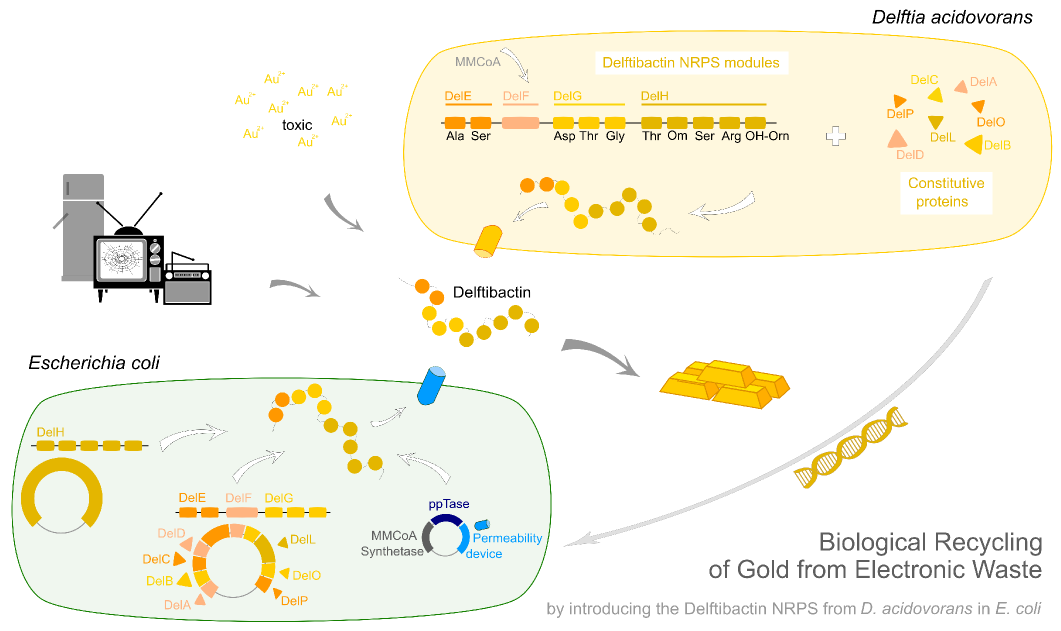
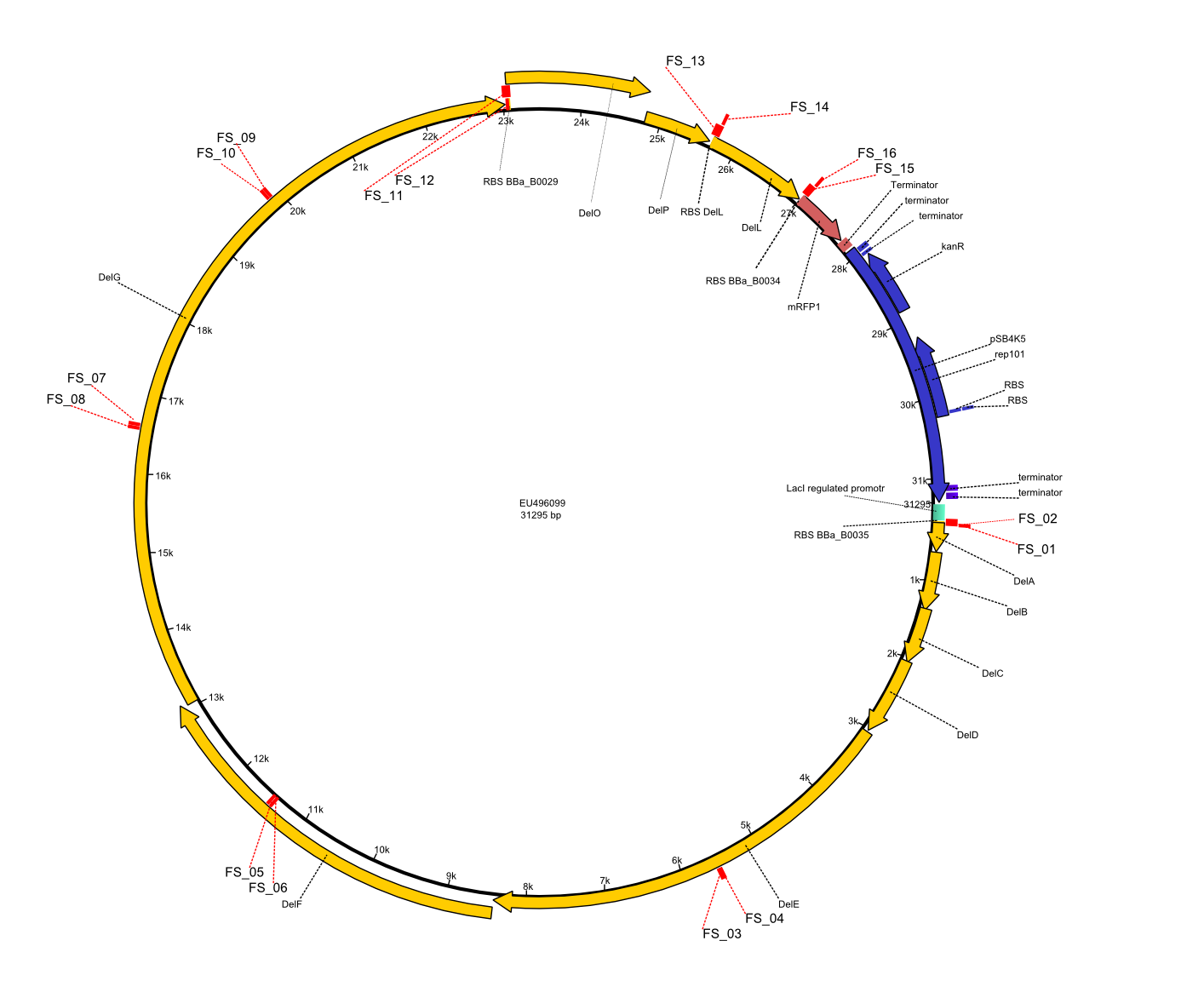
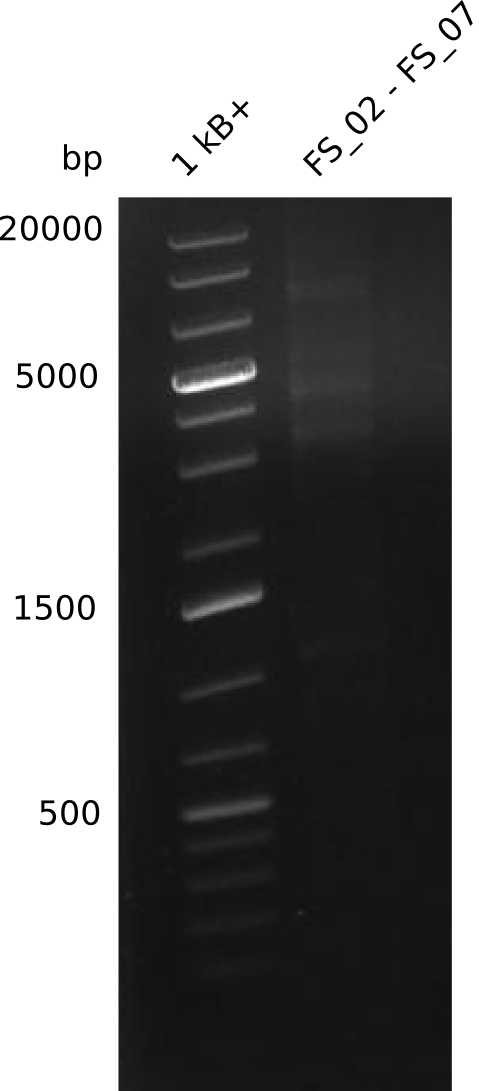
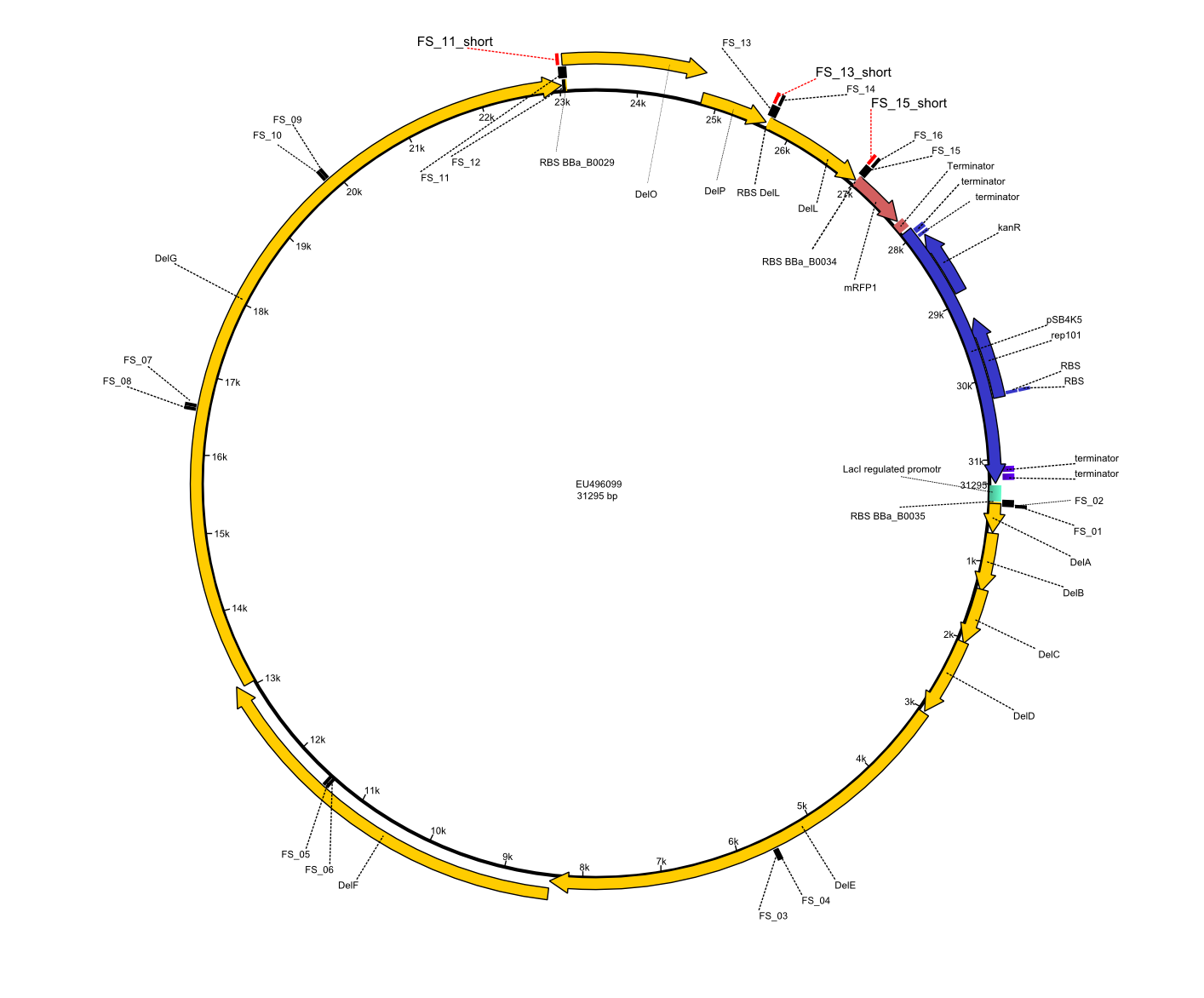
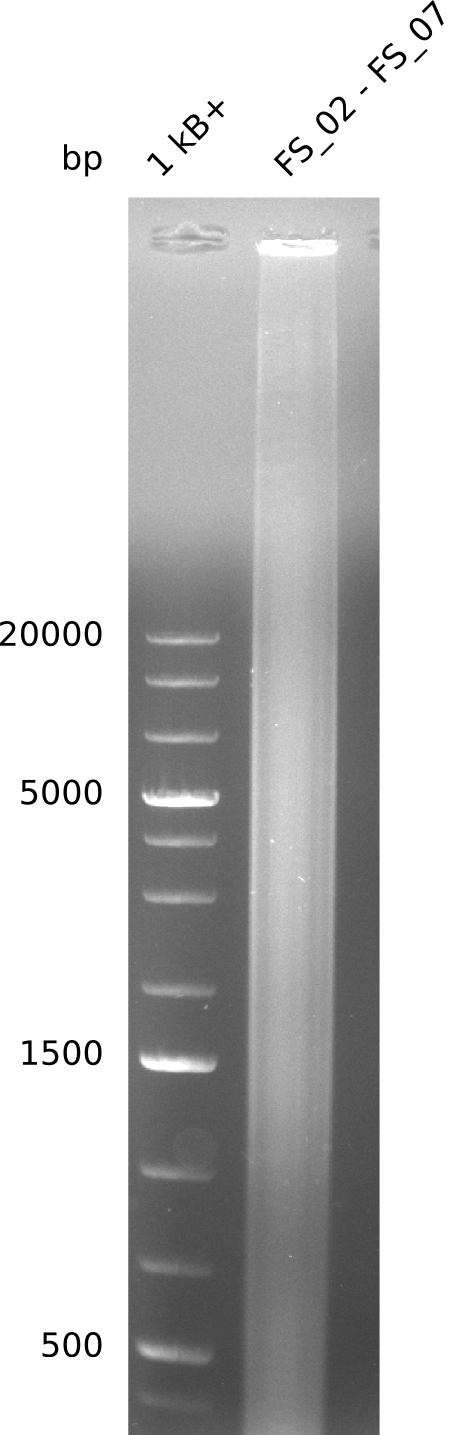
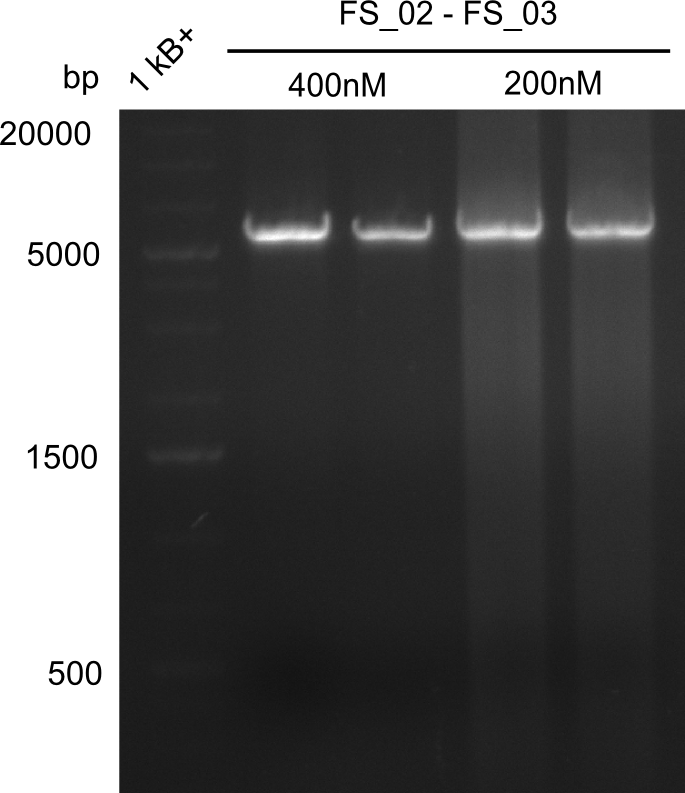
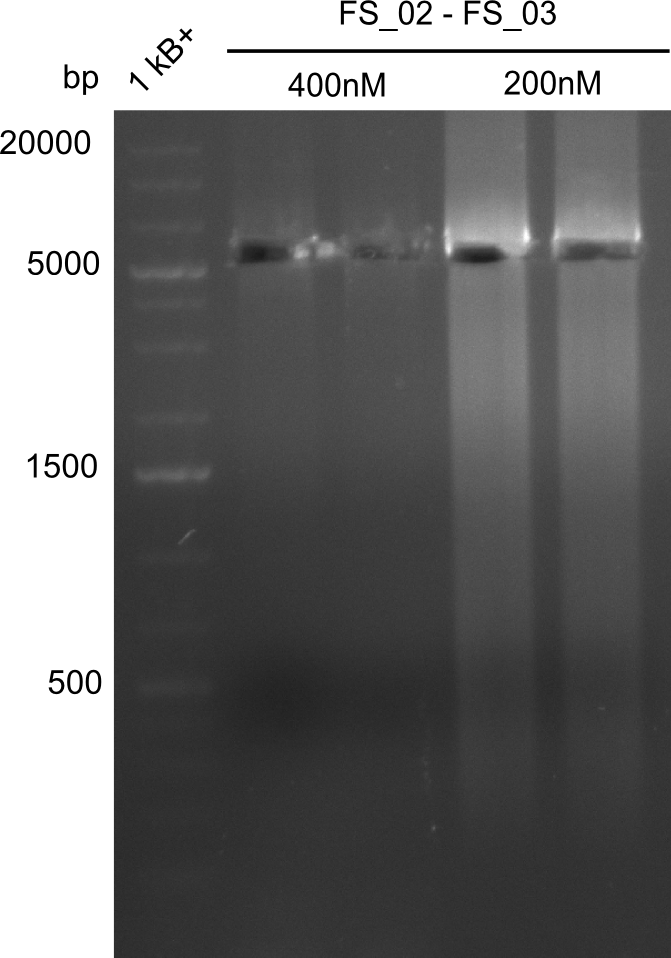
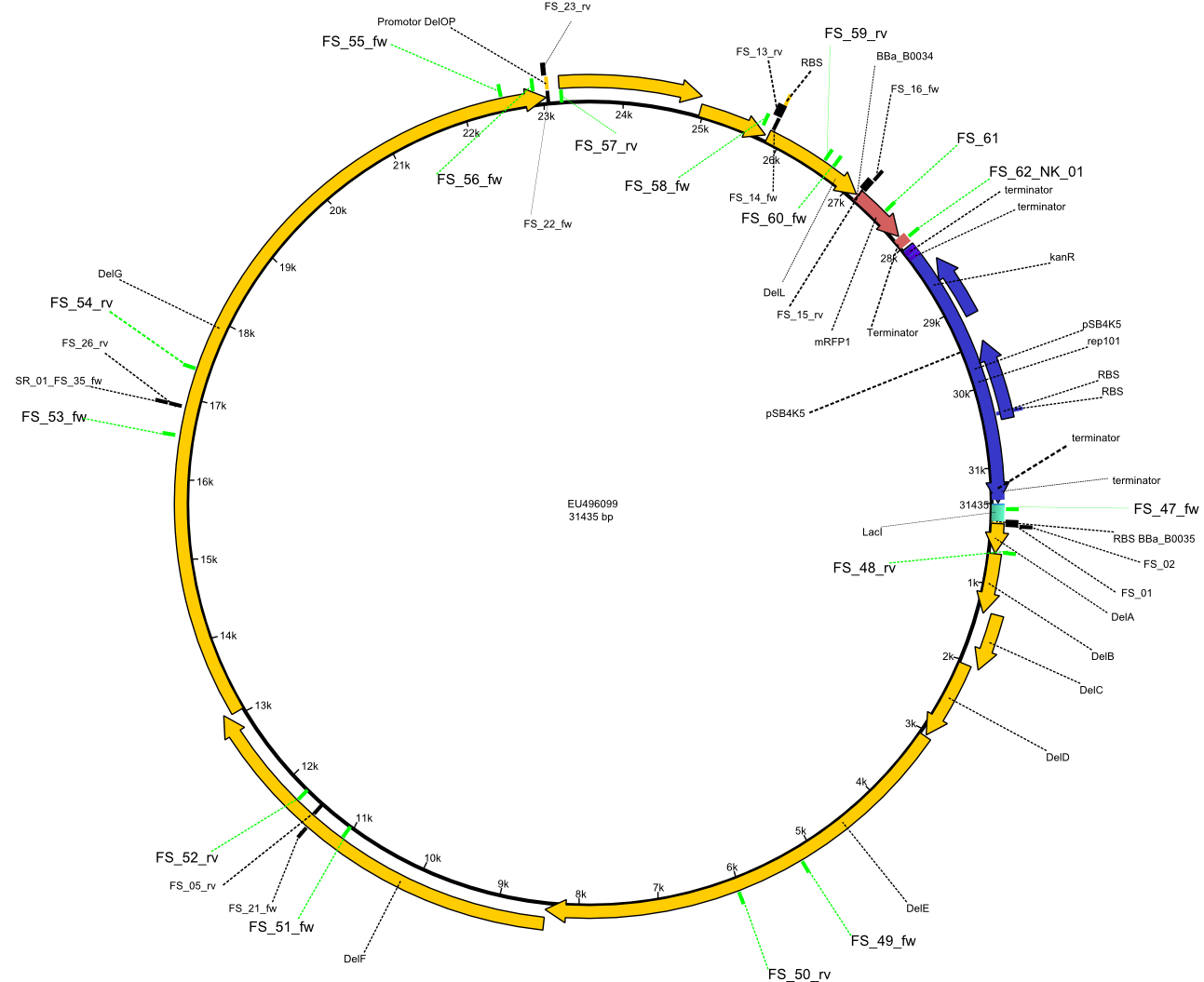
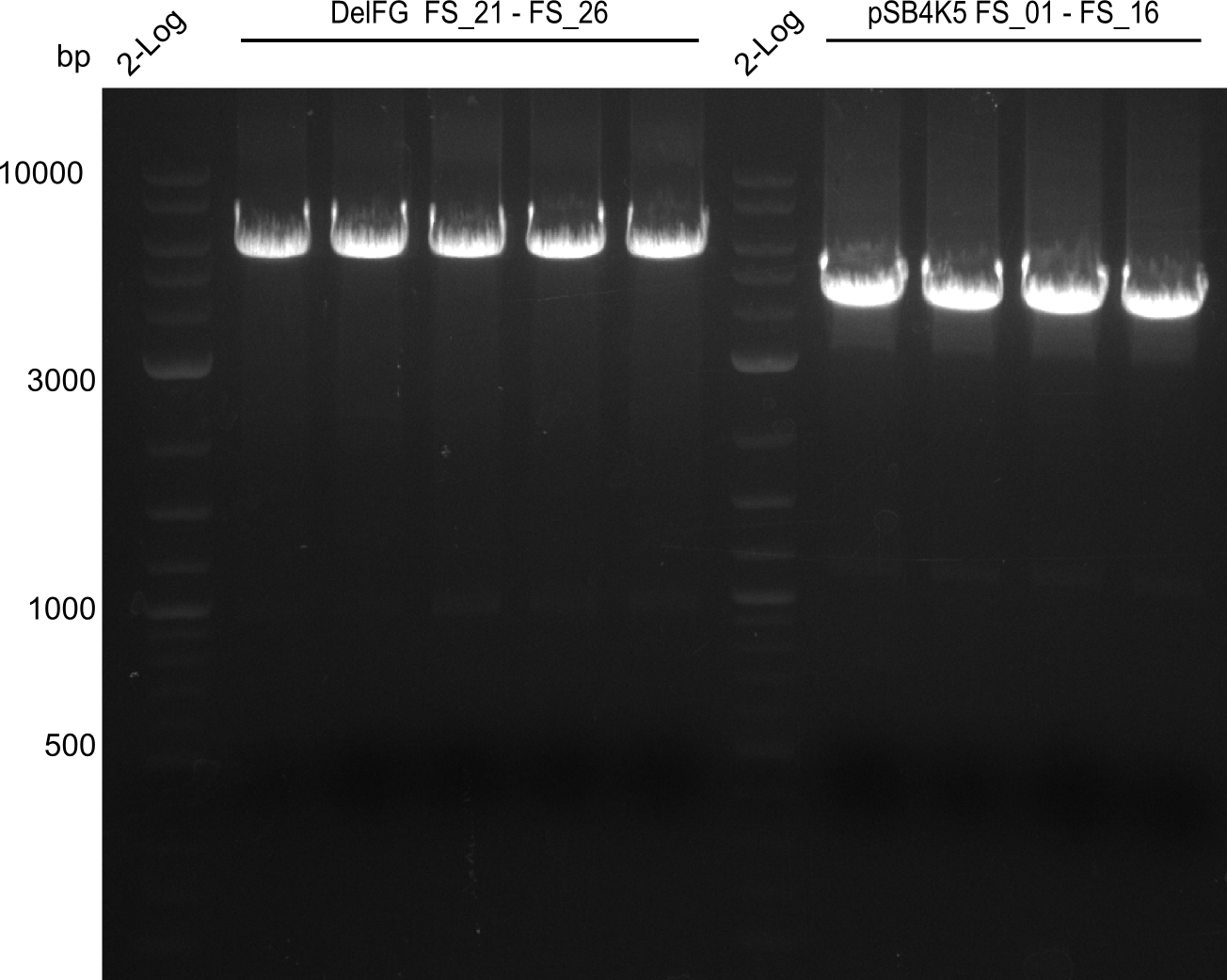
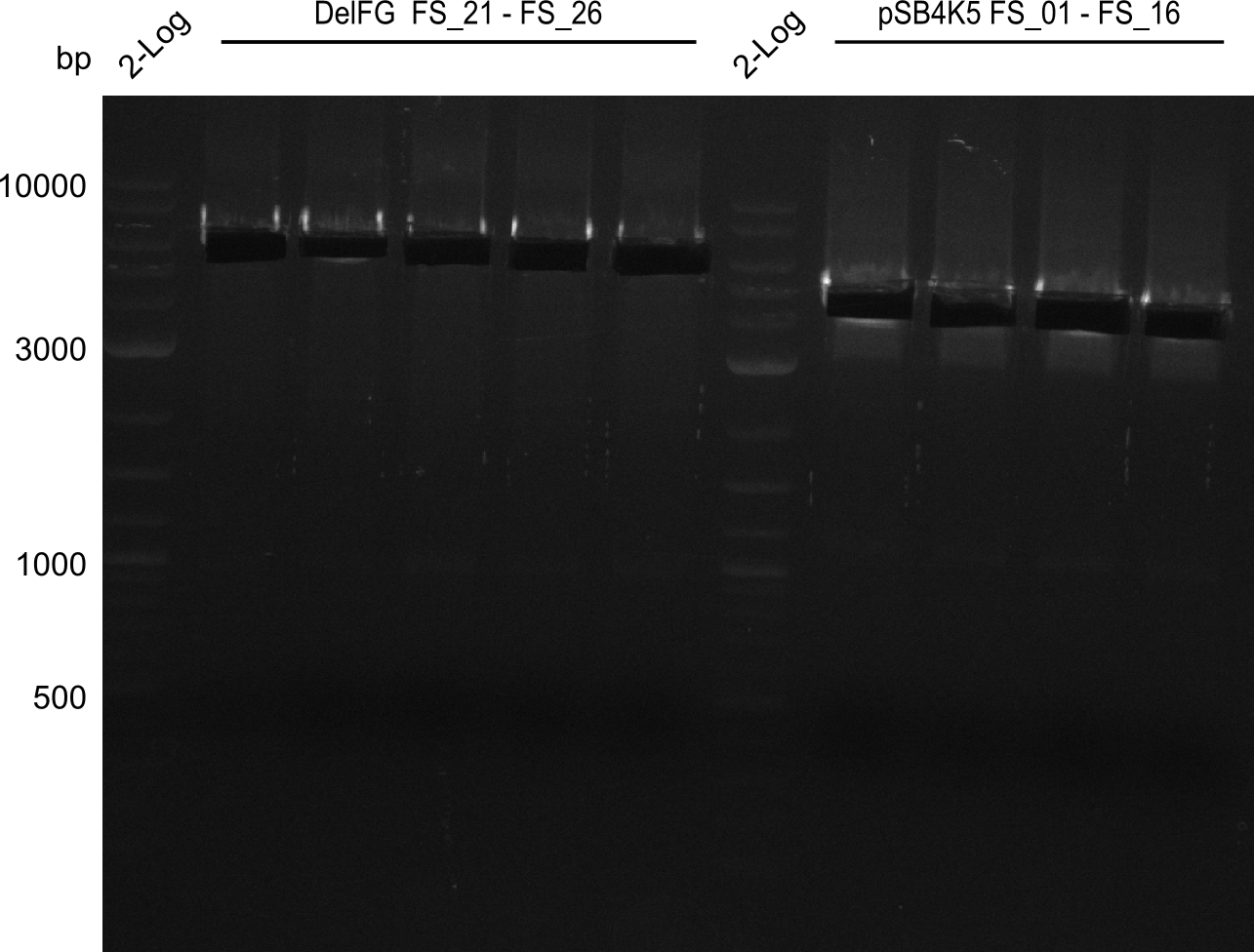
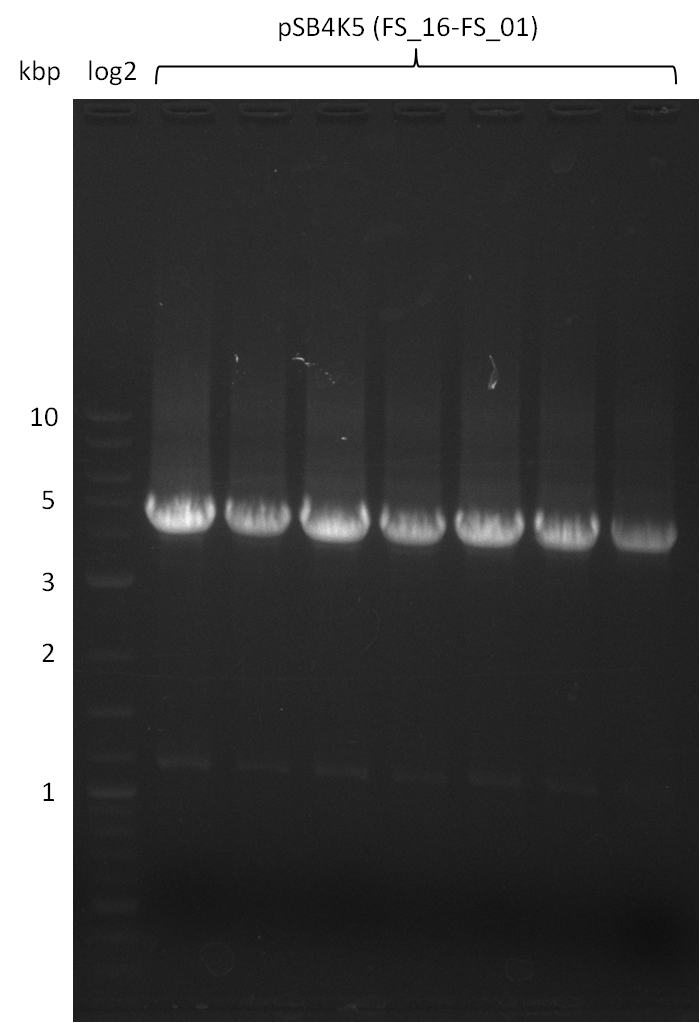
| - | This week the project "DelRest" was launched, which aims | + | This week the project "DelRest" was launched, which aims at creating a single, 32 kb plasmid enabling the expression of most genes from the <i>D. Acidovorans </i> Del cluster, namely DelA-G and DelL-P (note: the 18 kbp gene encoding DelH is cloned onto a seperate plasmid). Due to the shere size and complexity of the DelRest construct, we decided to use Gibson cloning. |
| - | + | This weeks goal was amplify the pSB4K5 backbone from the partsregistry with primers giving the intended overlaps as well as the desired genes from our host organism <i>D. Acidovorans </i>. | |
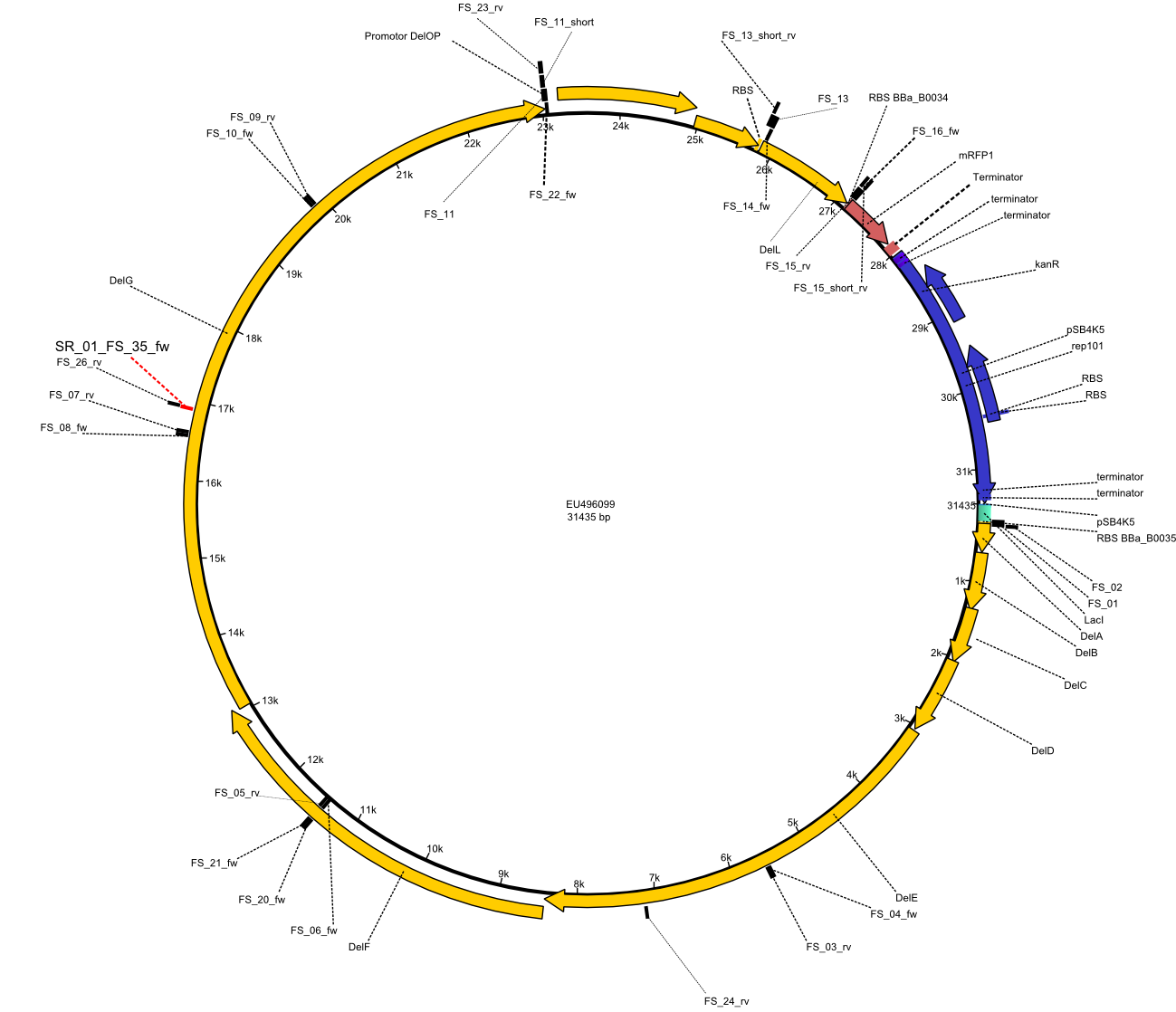
| - | + | Therefore, Gibson primers for the amplification of the target backbone pSB4K5 were designed, which also introduce a Gibson-overlap to DelA, the first gene present in the Del cluster. Furthermore Gibson primers for amplifying the fragments DelA-G and DelO-P were ordered. The last Gibson primer pair used to amplify DelL consequently carries the required overlap to the beginning of the mRFP reporter present the pSB4K5 insert part BBa_J04450. Check out our vectormap if you are curious about the detailed cloning strategy and primer design. | |
| - | + | ||
| - | Therefore Gibson primers for the amplification of the target backbone pSB4K5 were designed, which also introduce a | + | |
</p> | </p> | ||
| Line 80: | Line 83: | ||
<h1>Week 11</h1> | <h1>Week 11</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
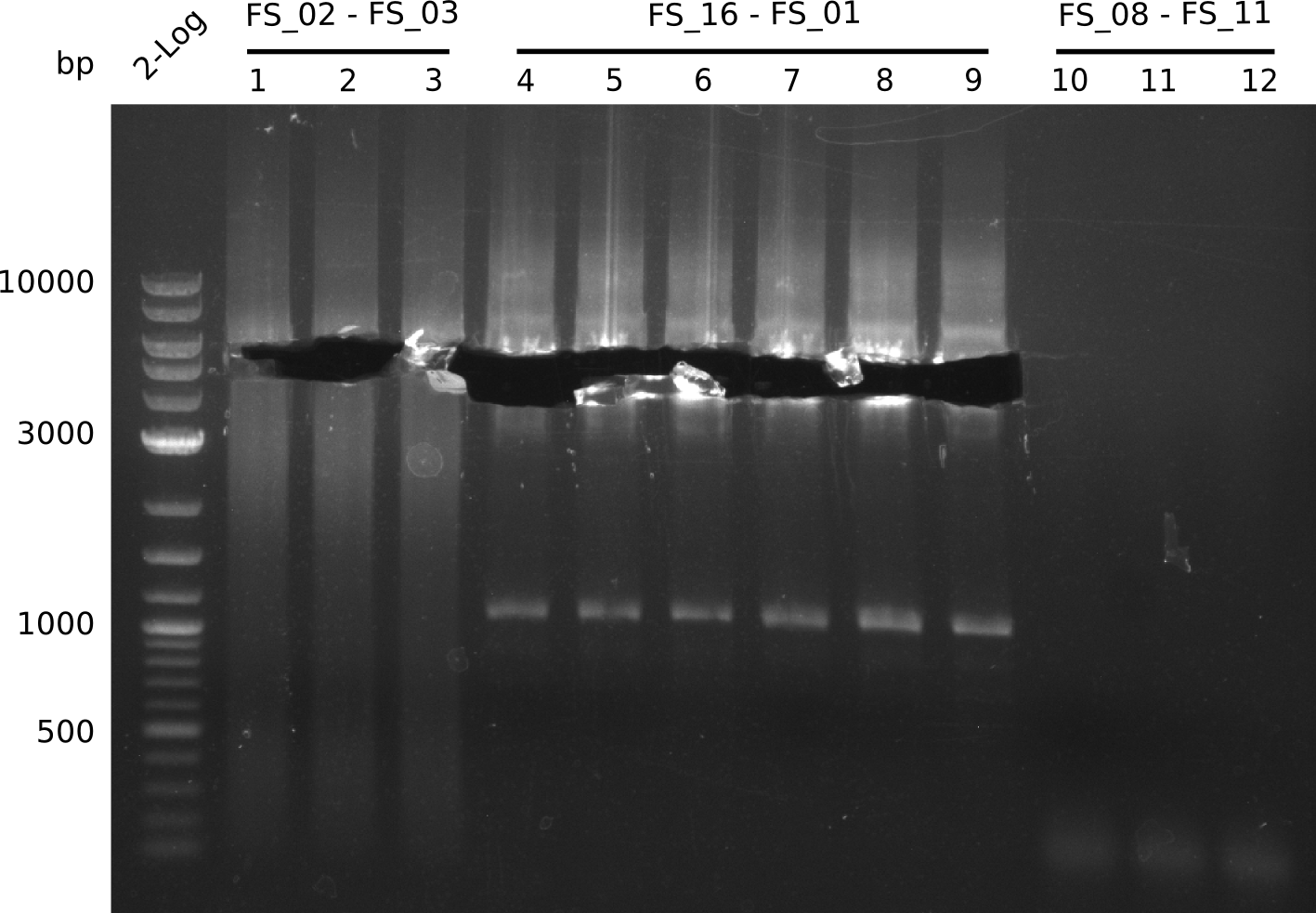
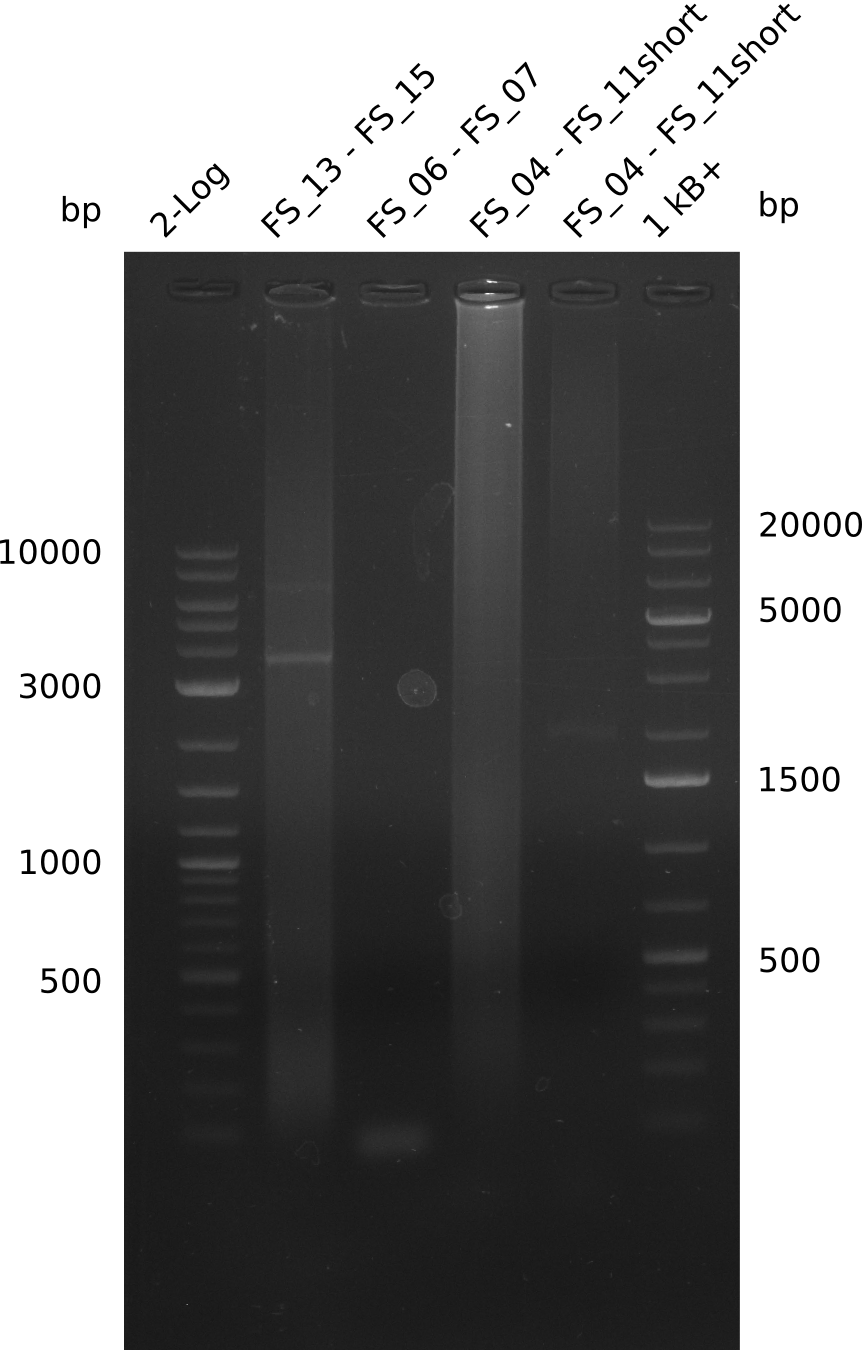
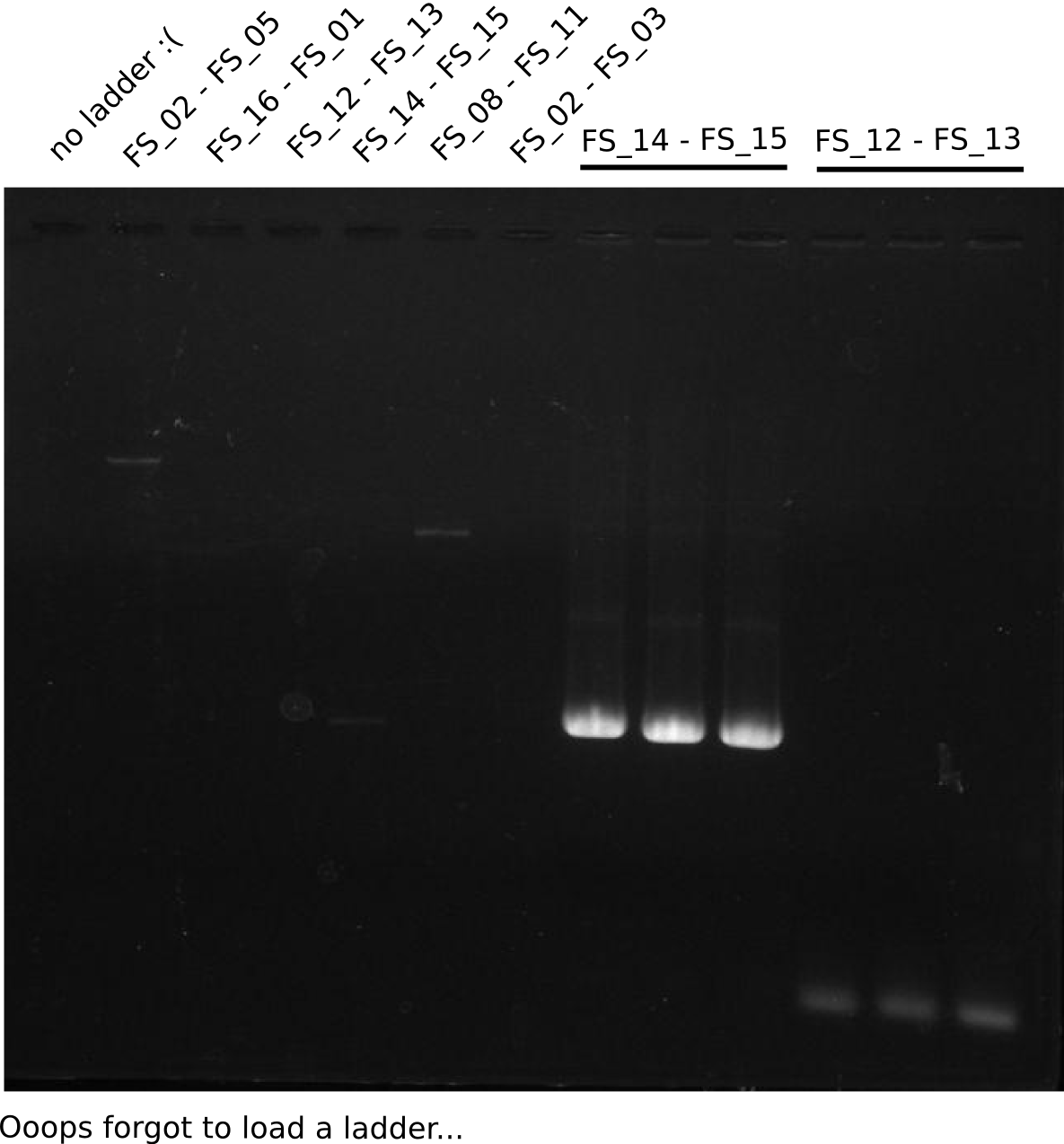
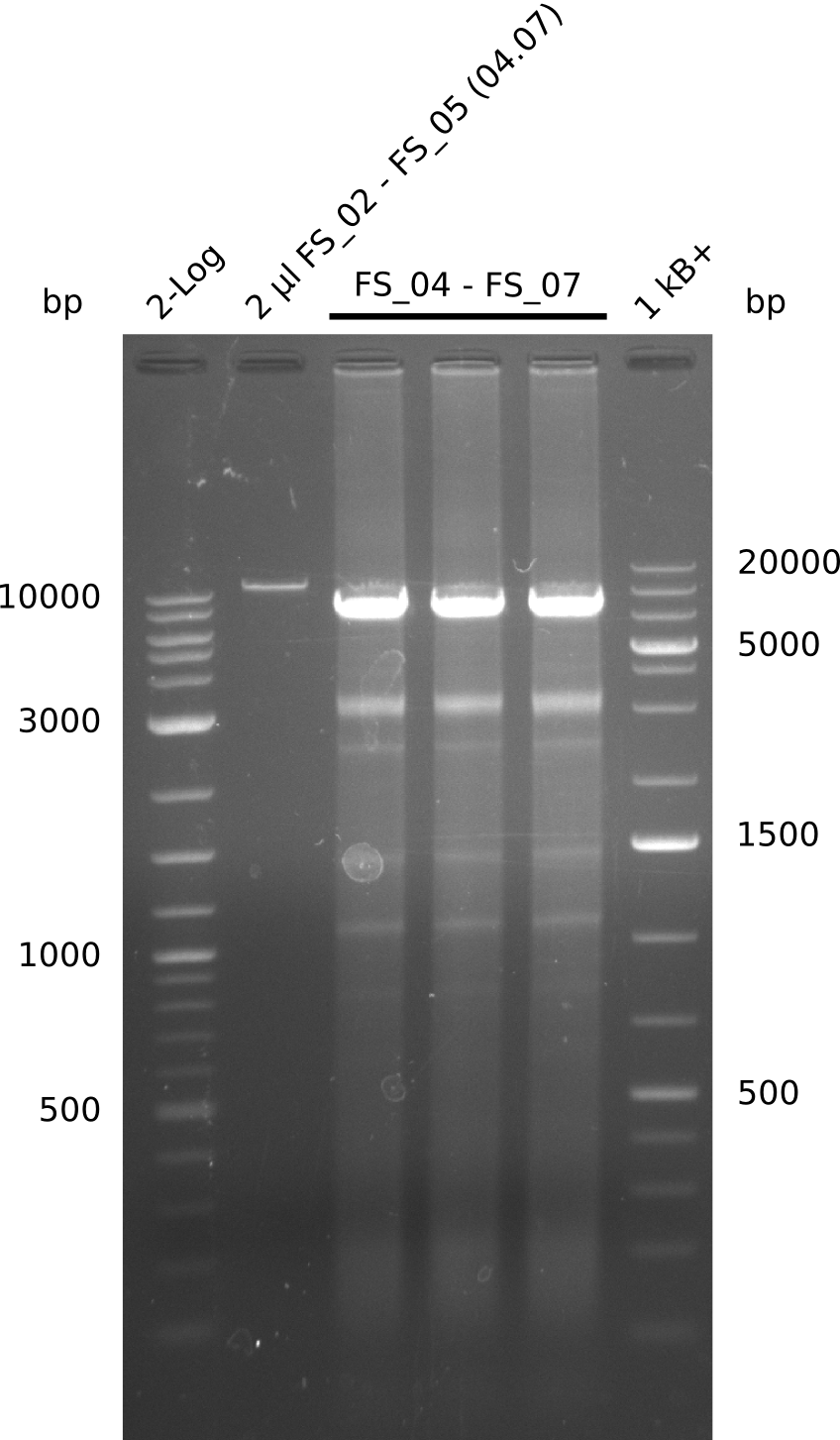
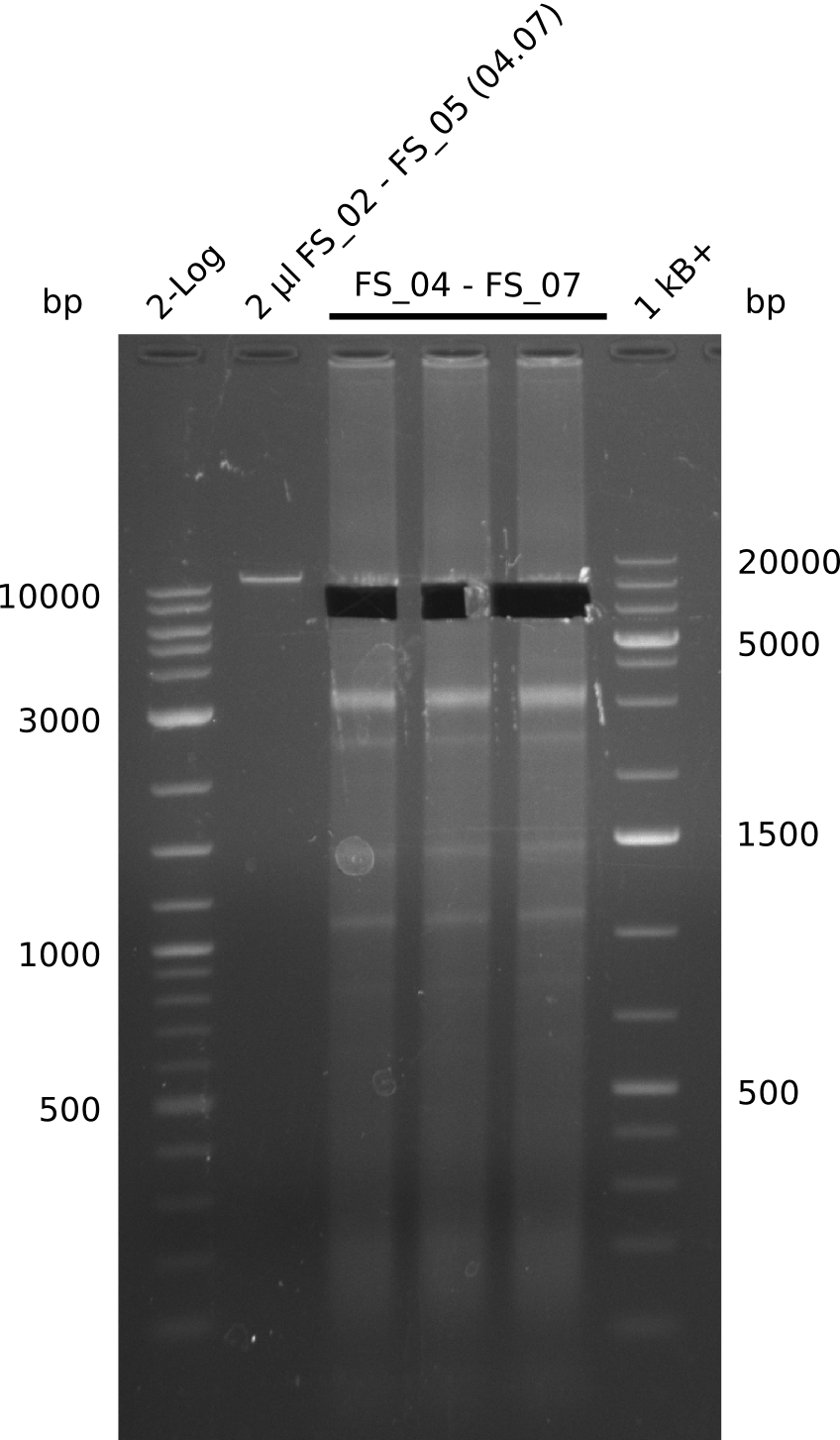
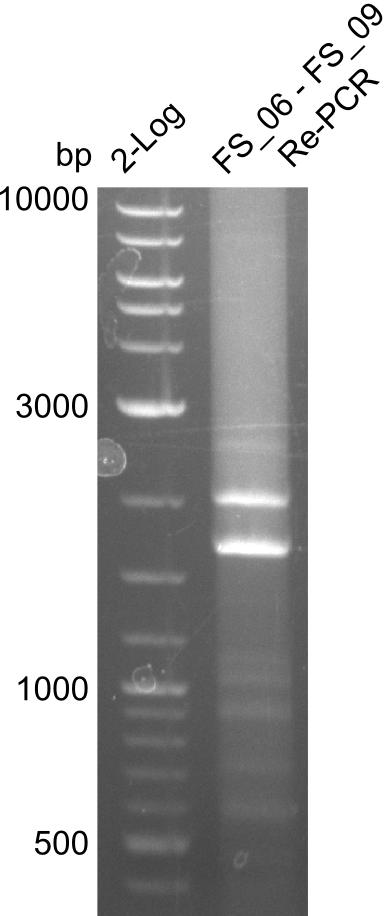
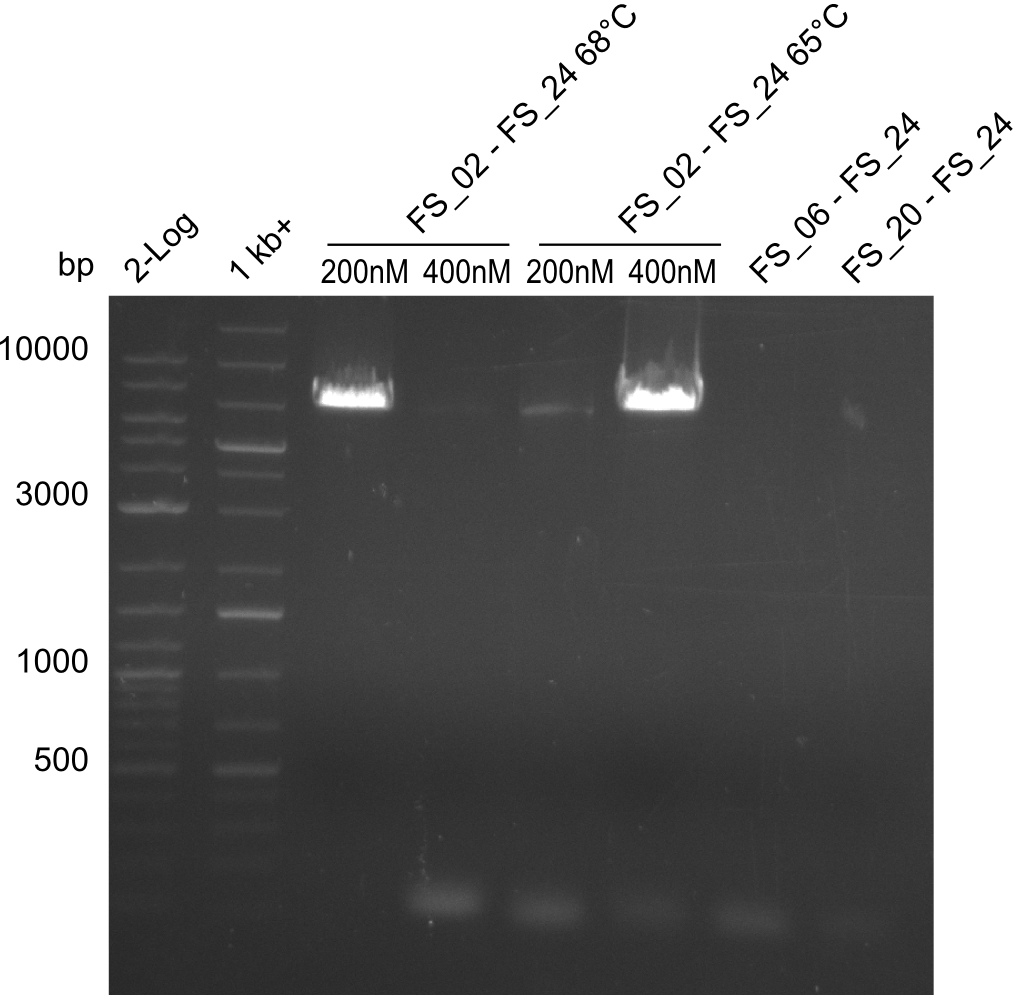
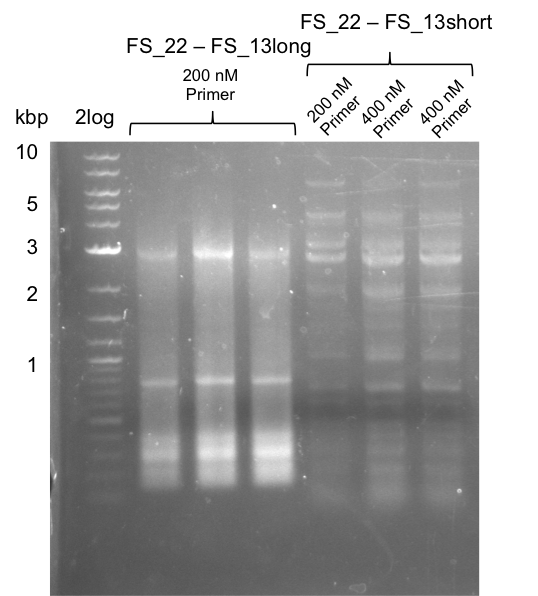
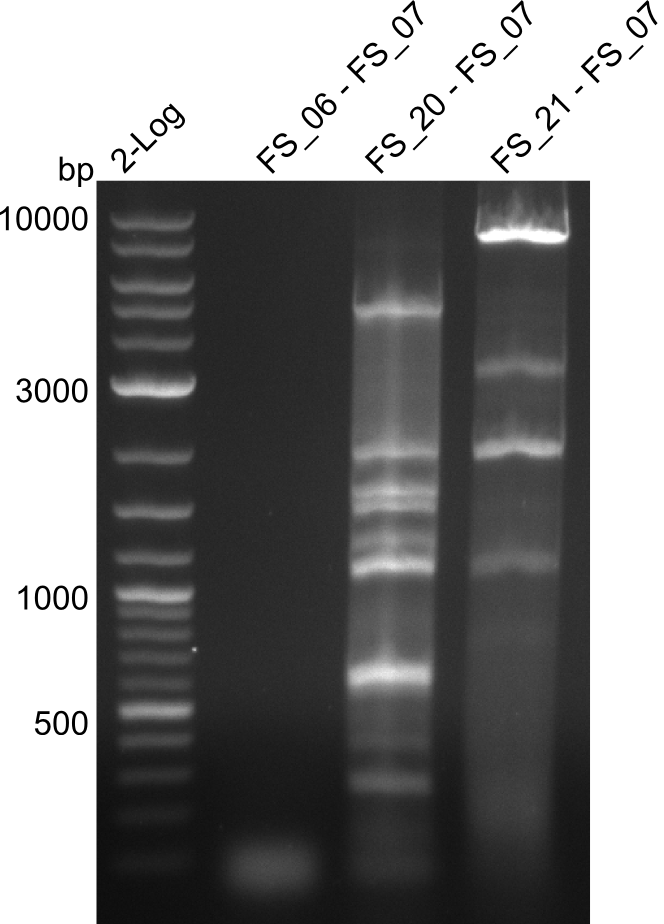
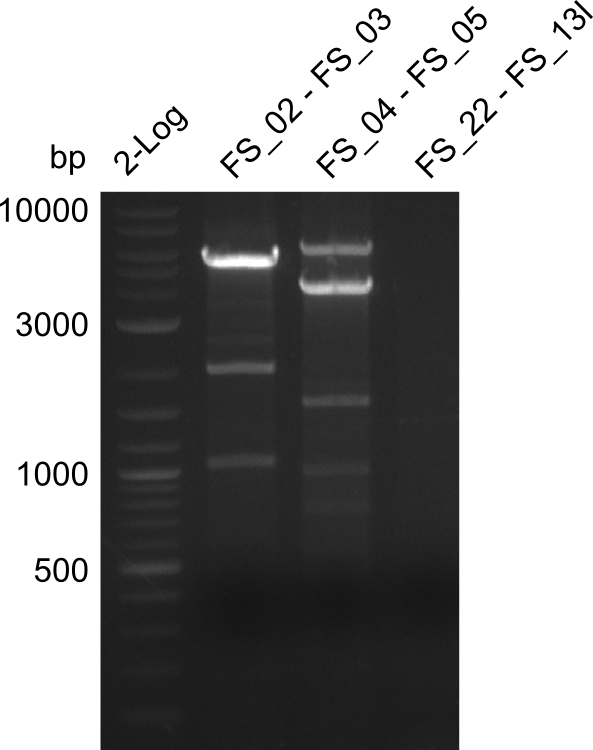
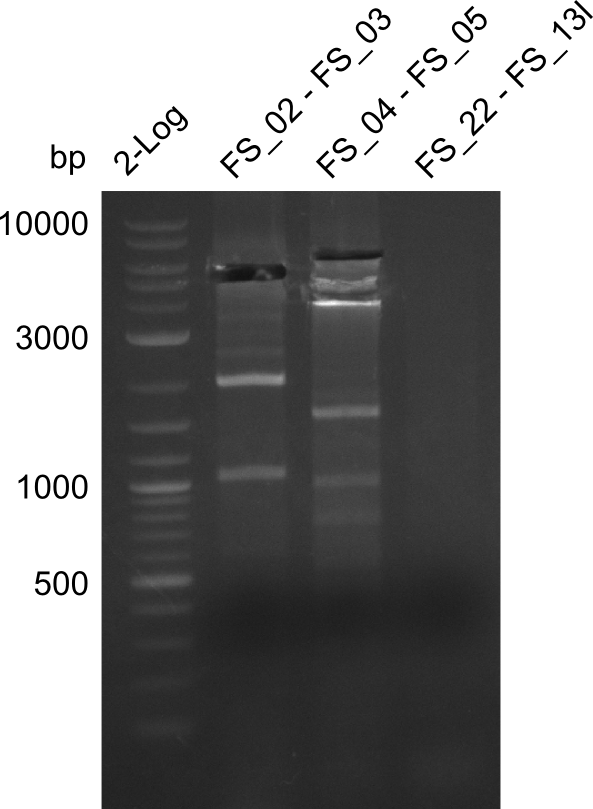
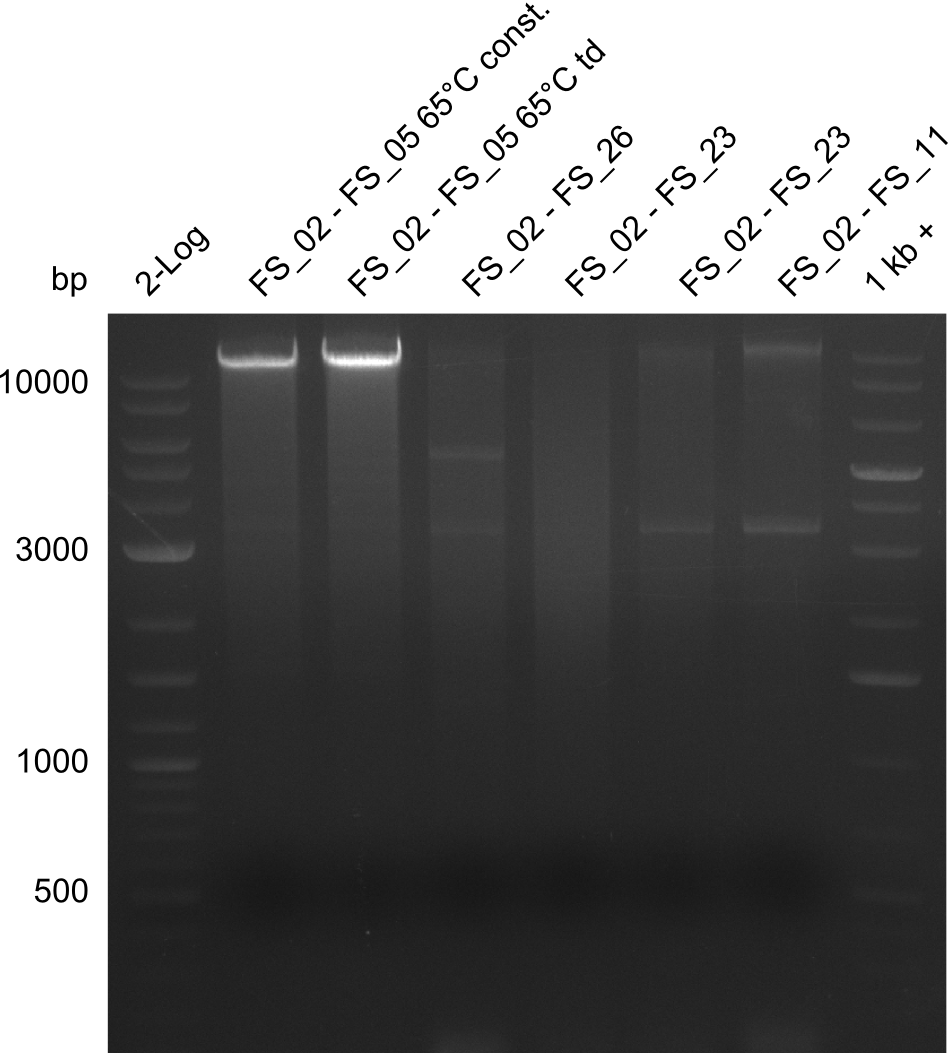
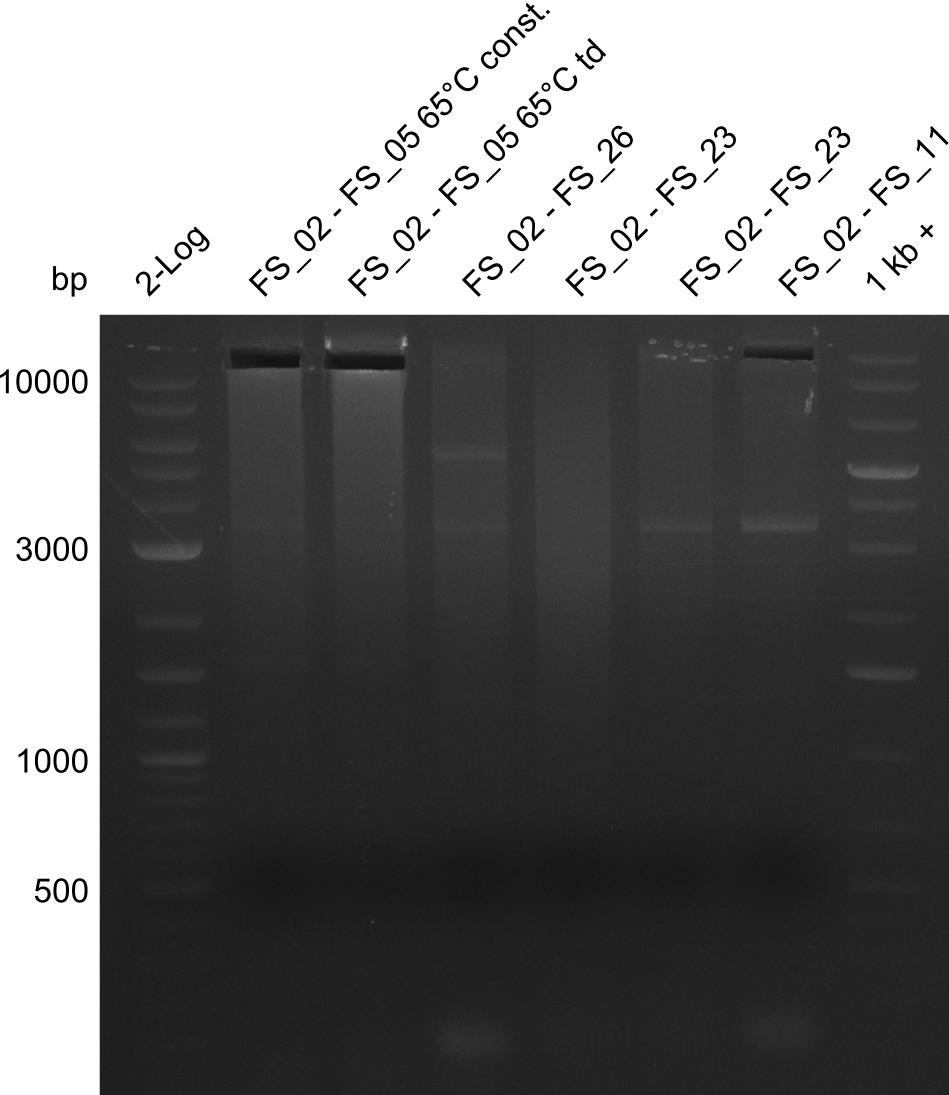
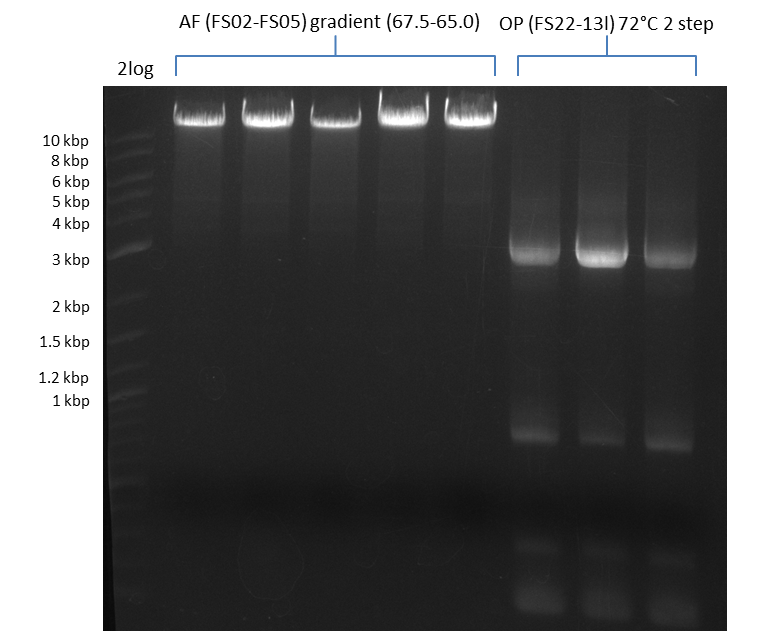
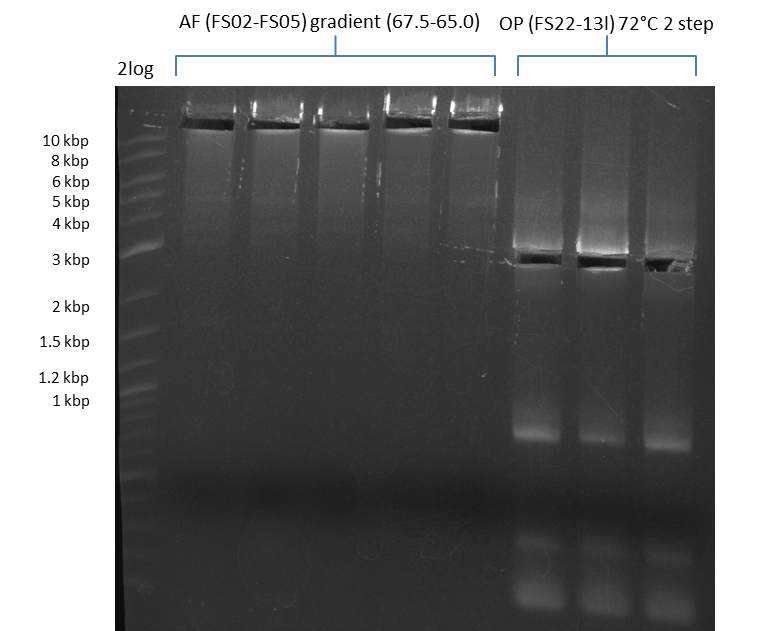
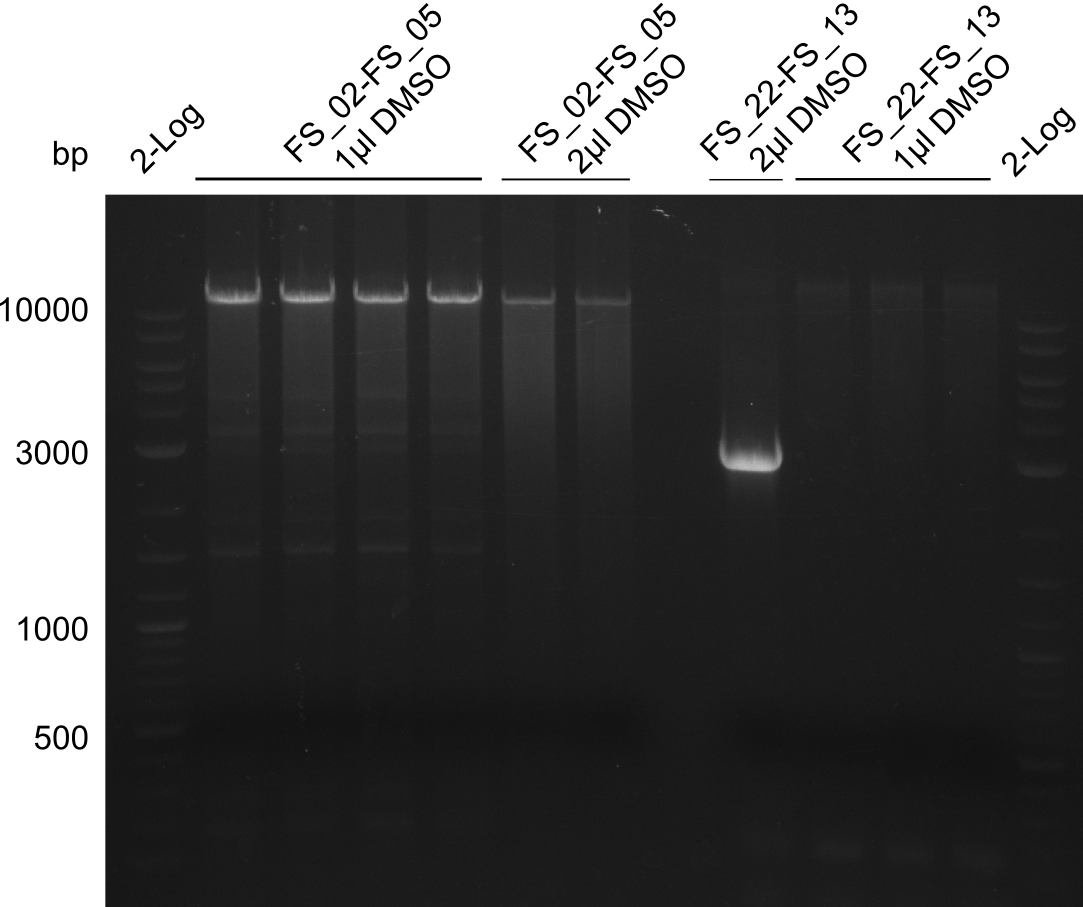
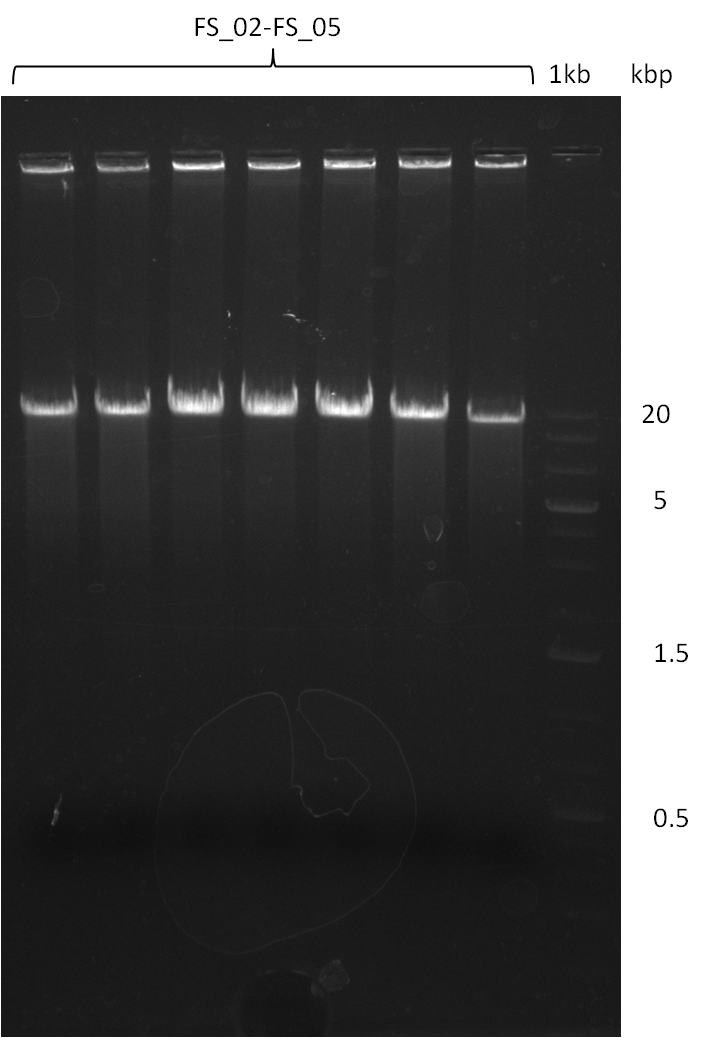
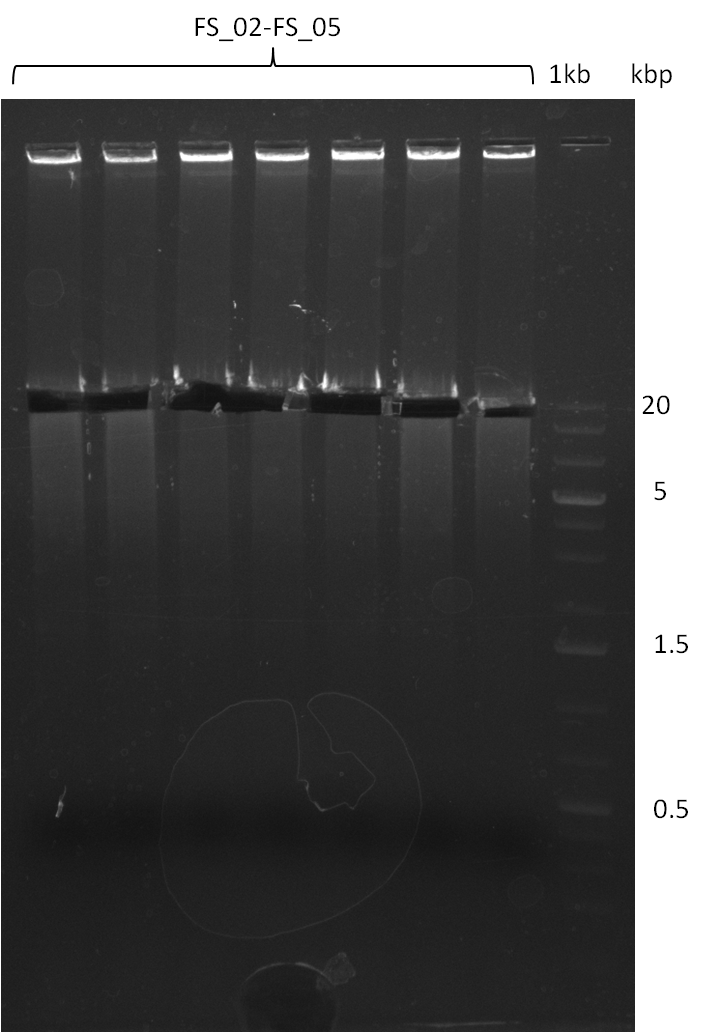
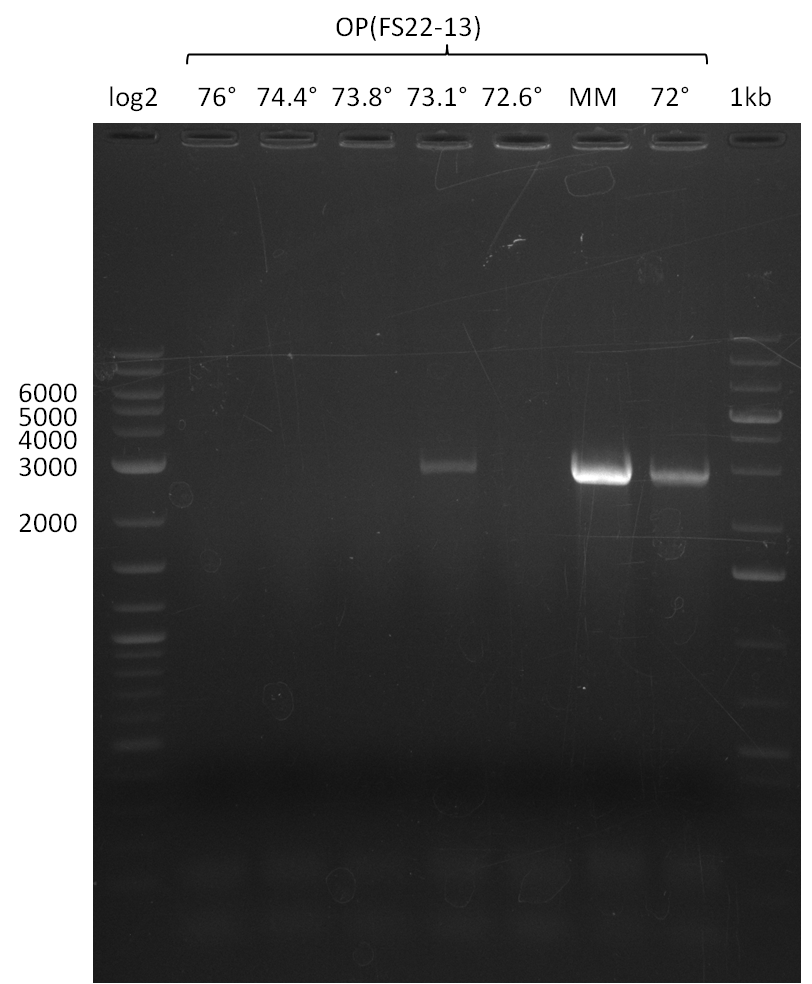
| - | During the past week we managed to amplify a fragment of 11 kbp encoding the genes DelA | + | During the past week we managed to amplify a fragment of 11 kbp encoding the genes DelA-F from genomic DNA of <i>D. Acidovorans</i>. We were further able to amplify the backbone fragment from pSB4K5 carrying the desired overlaps to the corresponding fragments to be amplified from the Del cluster. We also succeeded in the amplification of the very last gene in our construct, DelL. Unfortunately, the amplification of DelF-G and DelO-P turned out to be more difficult than expected. Since our initial plan, which required the amplification of DelO-P using a Gibson primer which introduces an overlap to DelL and furthermore introduces an artificial ribosom binding site did not work out, we changed our strategy. We orderd shorter versions of all our Gibson primers. Using these shorter primers not bearing any overlaps to other fragments, we will try to amplify the desired genes in order to obtain a specific template for the reamplification with the primers carrying the needed overlaps. Furthermore, we will optimize the PCR conditions for the amplification of the DelF-G fragment. |
</p> | </p> | ||
| Line 92: | Line 95: | ||
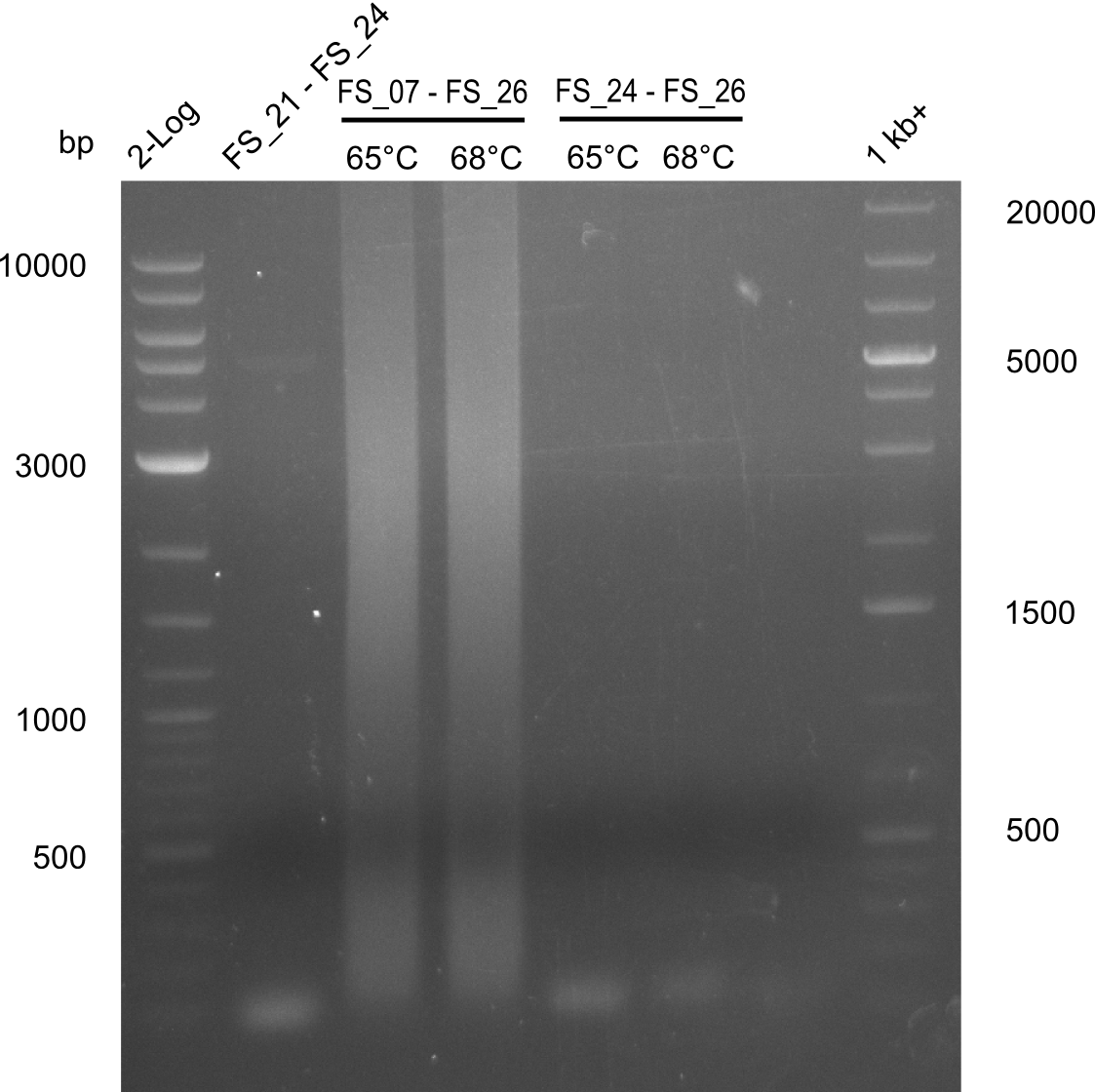
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
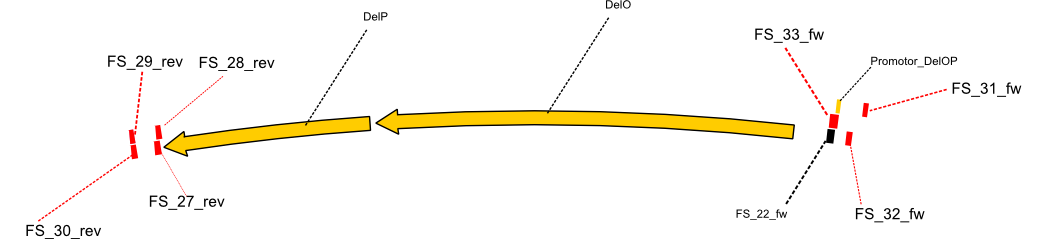
| - | By the end of last week we | + | By the end of last week we found out, that the short primers we had ordered did not improve amplification of the missing fragments from <i>D. Acidovorans. </i> Additionly, further analysis of the Delftibactin cluster, led to the discovery of a predicted promoter present in front of DelO-P potentially essential for DelO-P expression. Therefore, we not only decided to design new primers for the amplyfication of DelF-G, but also modifie the entire strategy concerning the DelO-P fragment. To ensure the expression of DelO-P in our target organism <i> E.coli </i> we decided to amplify DelO-P together with its putative promoter. In consequence, we ordered new primers for DelO-P and also for the last DelA-G fragment in order to introduce the required new overhang to the new DelO-P fragment. The correlating primers can be found in our new vector map. |
</p> | </p> | ||
| Line 103: | Line 106: | ||
<h1>Week 13</h1> | <h1>Week 13</h1> | ||
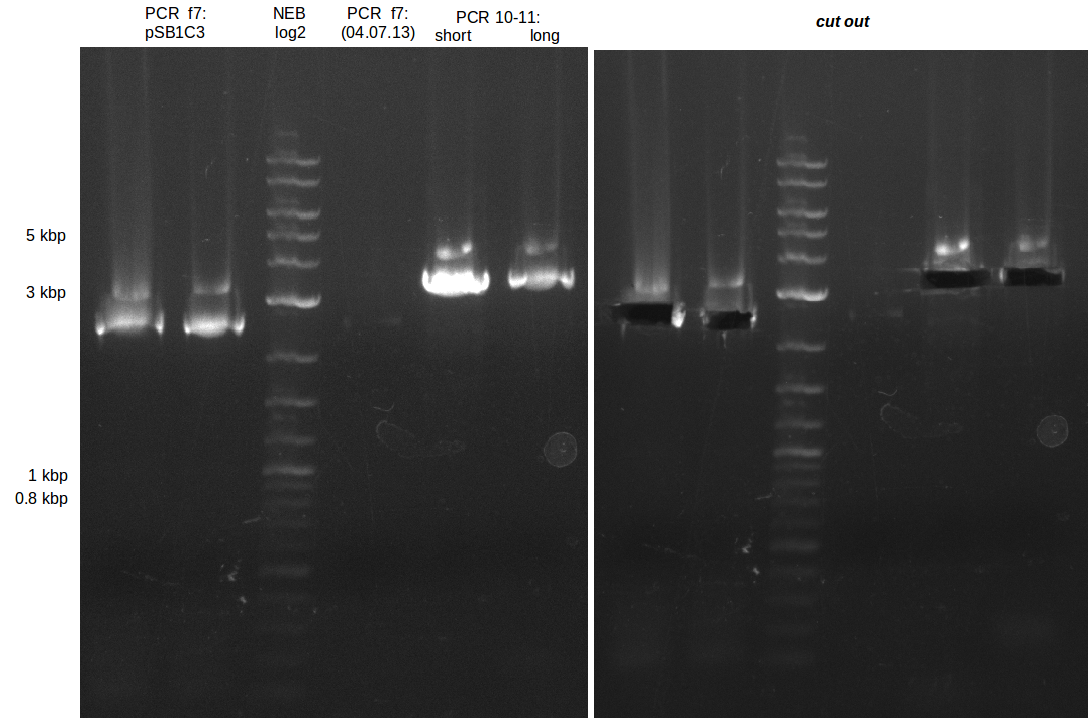
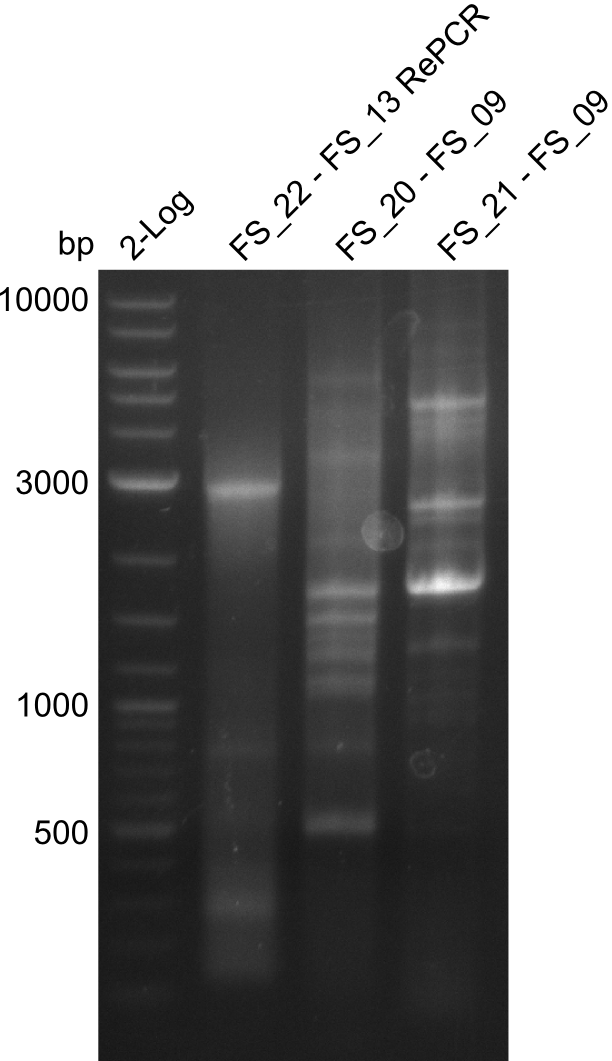
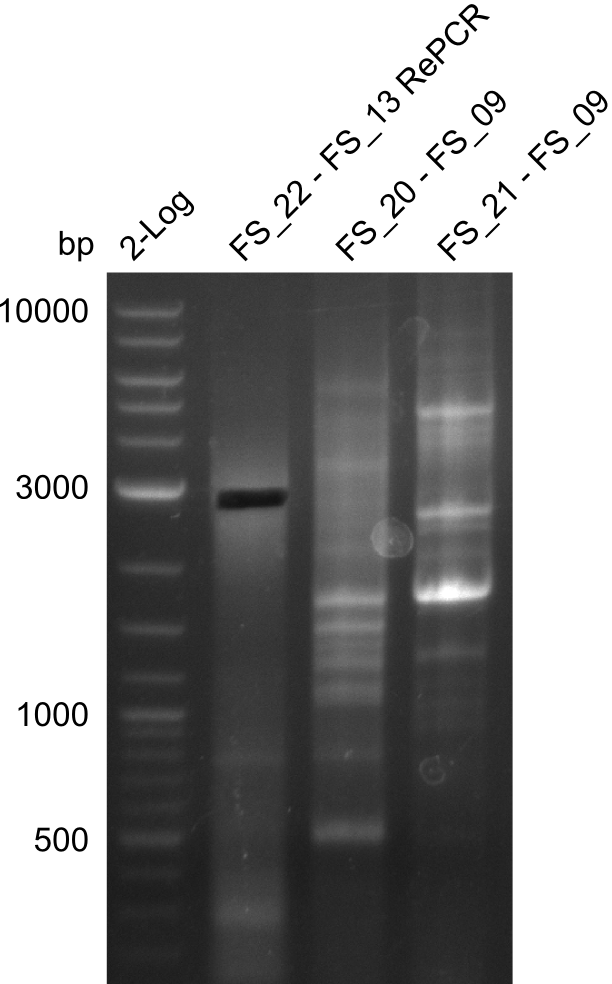
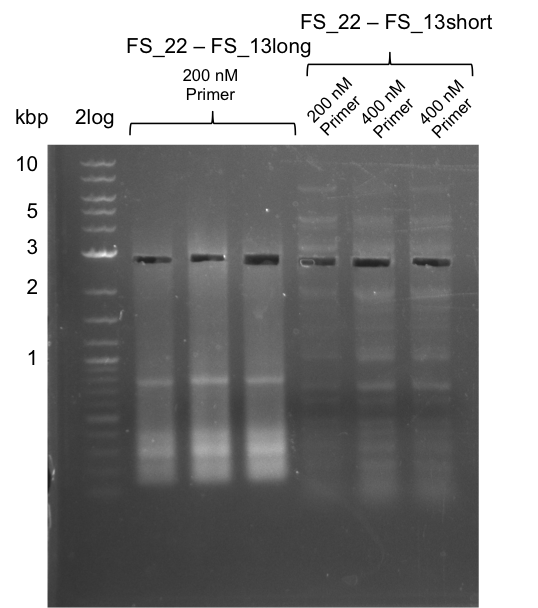
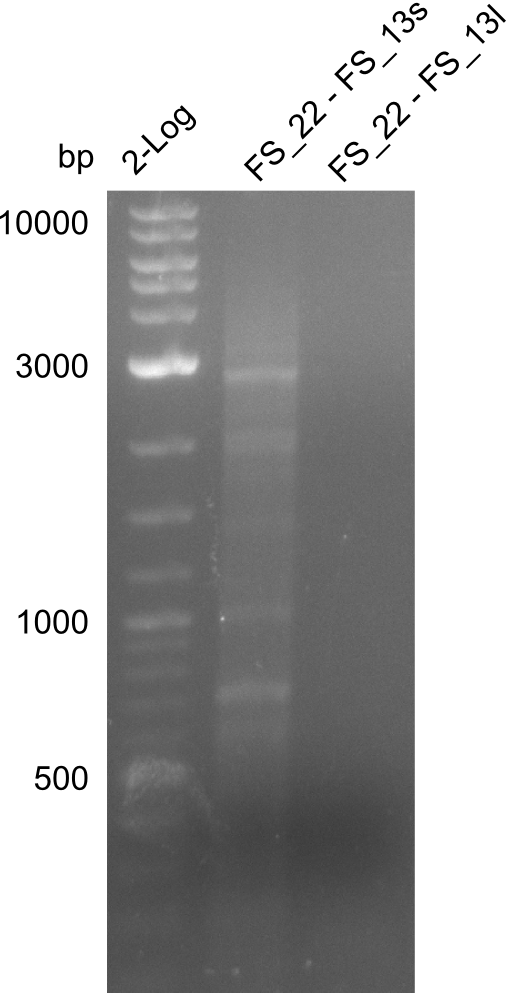
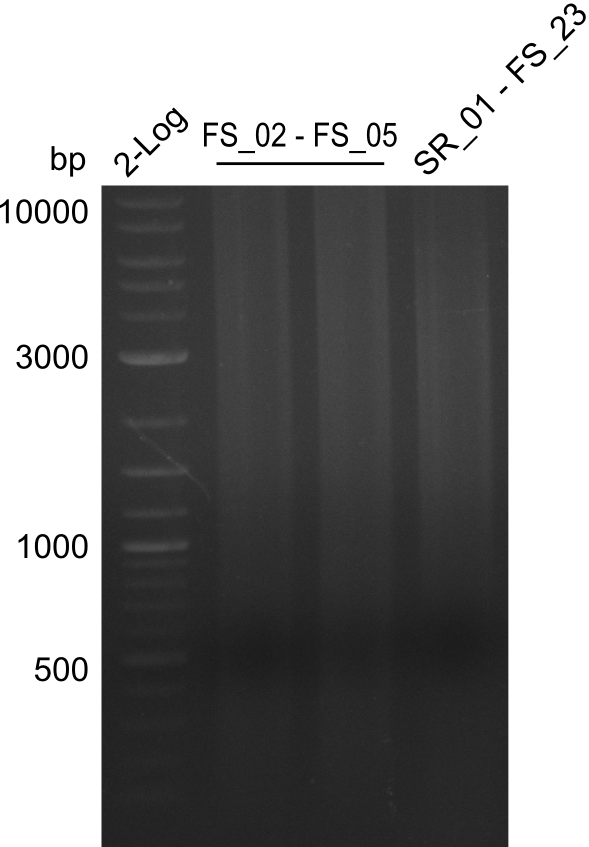
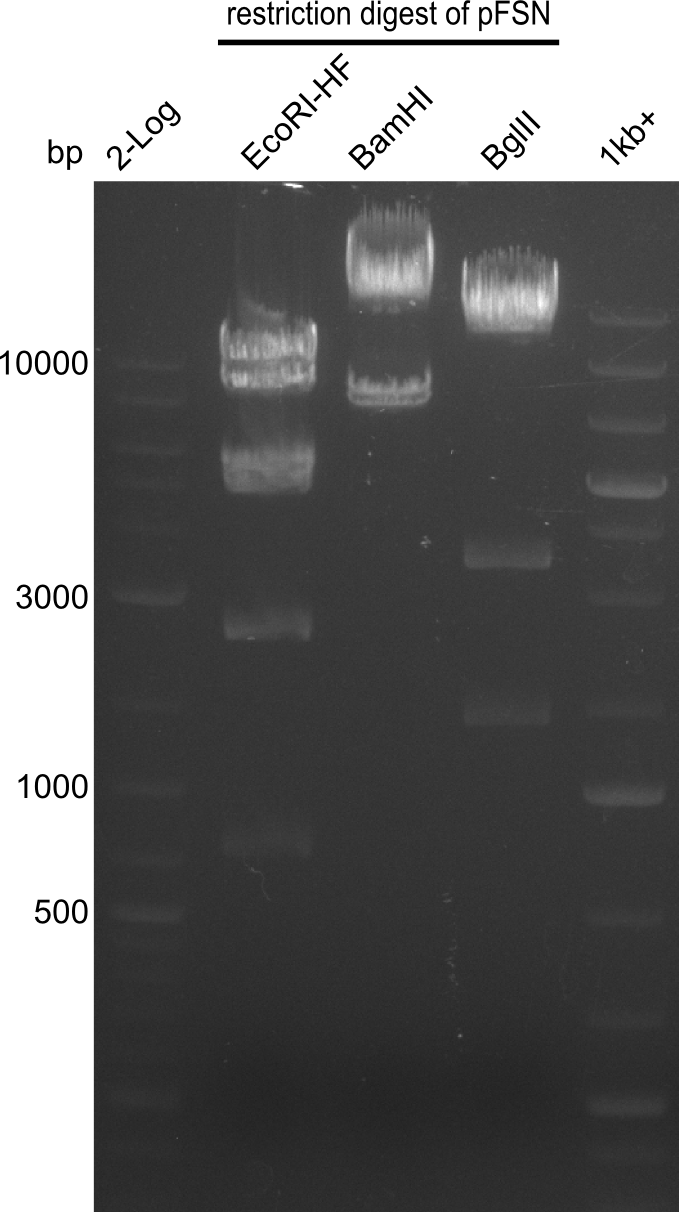
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
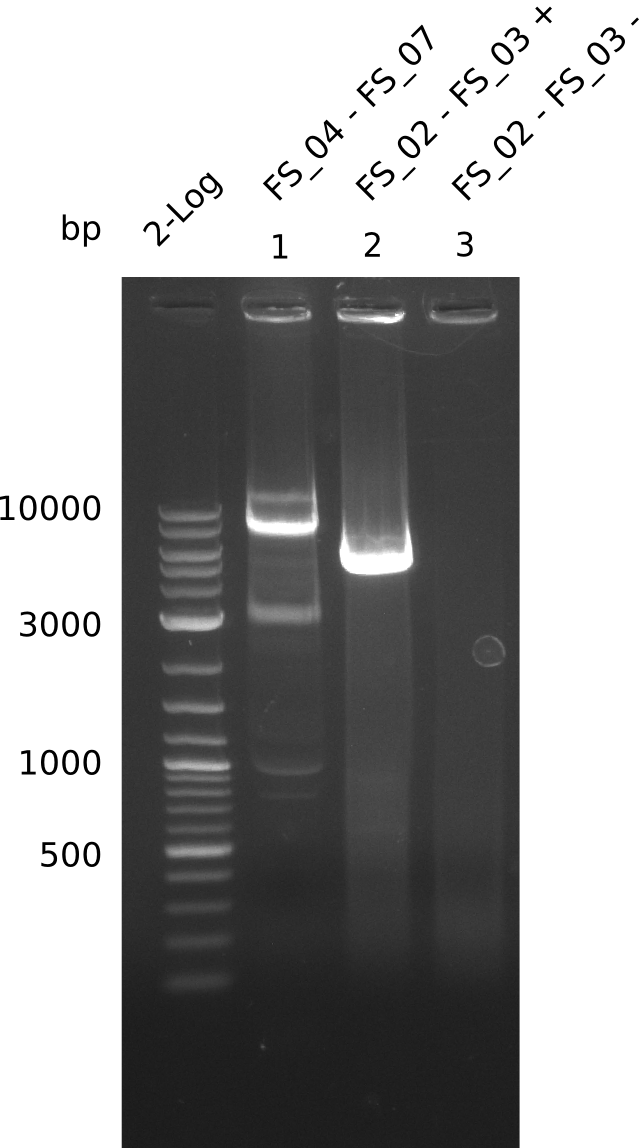
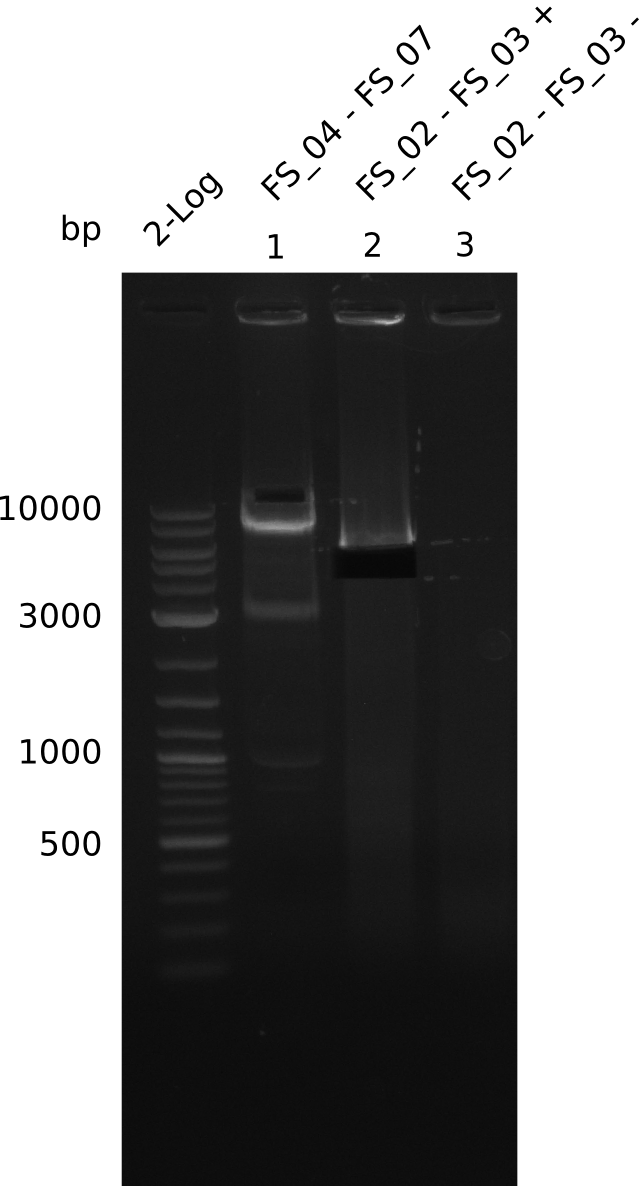
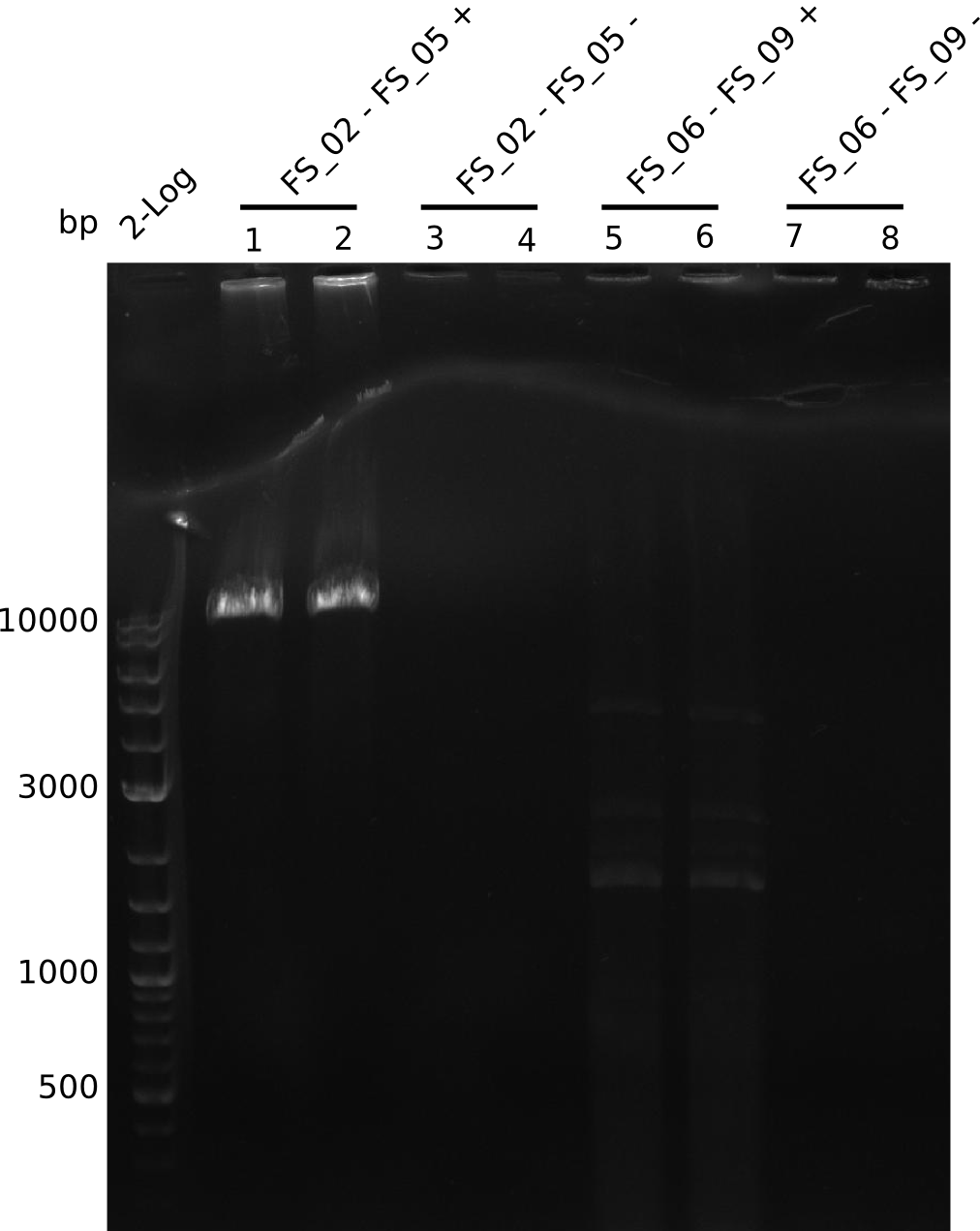
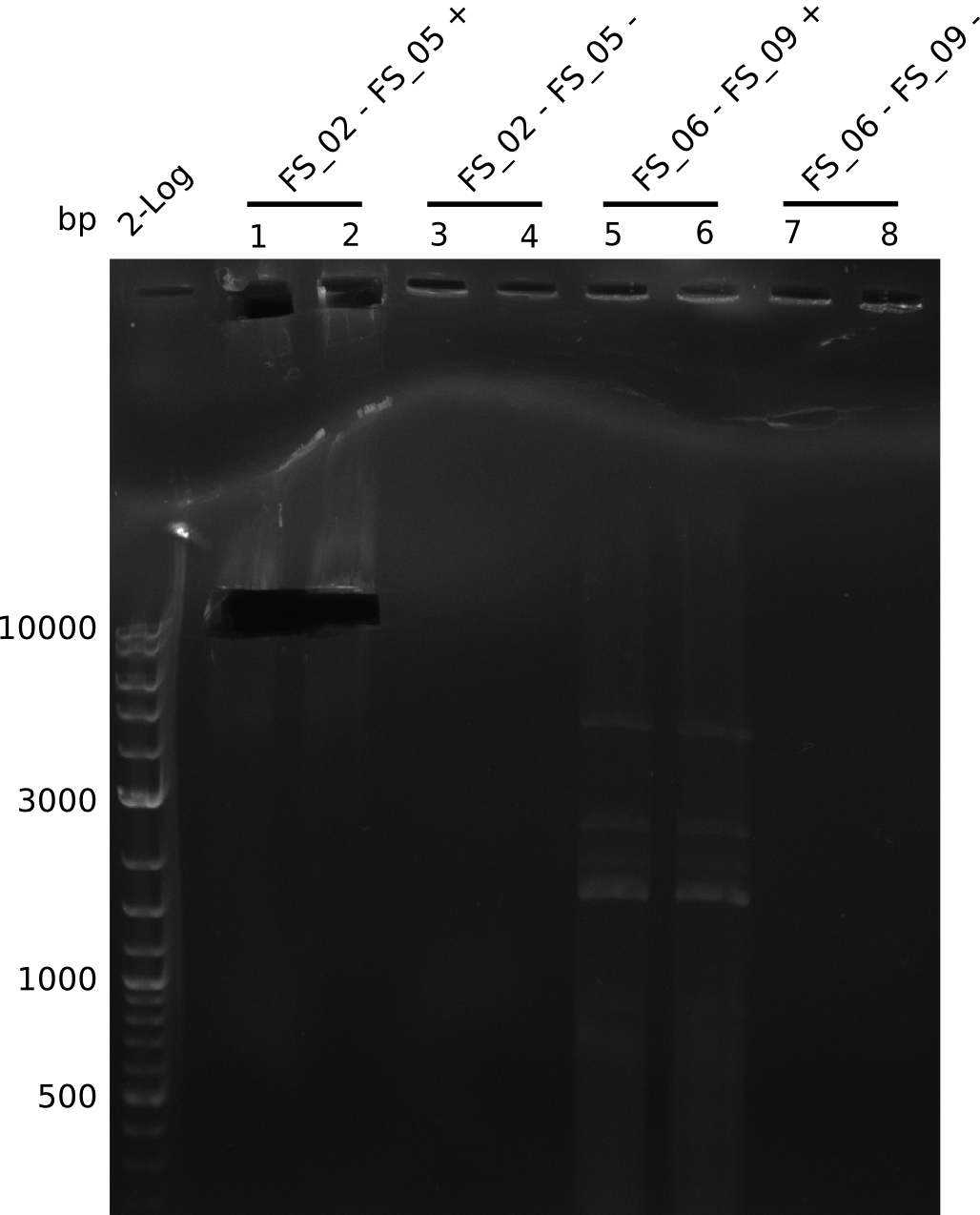
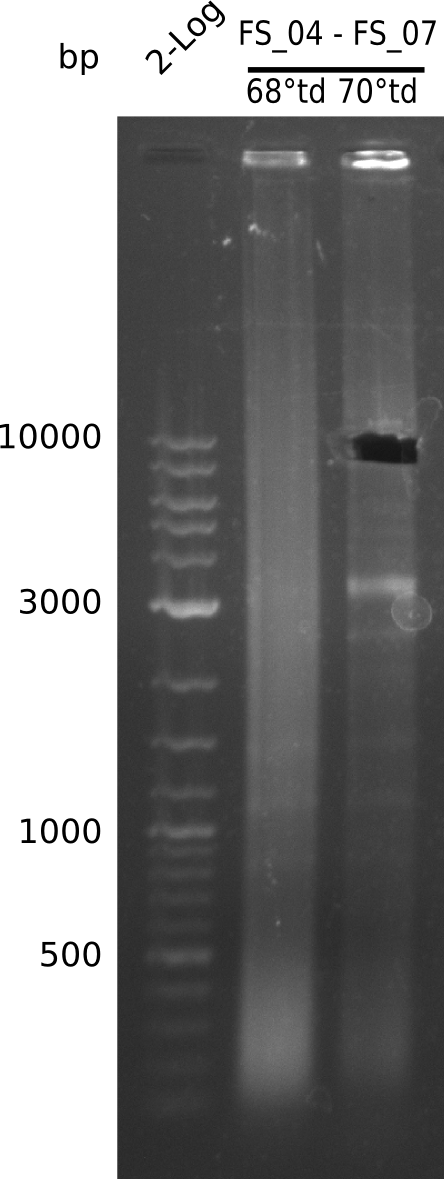
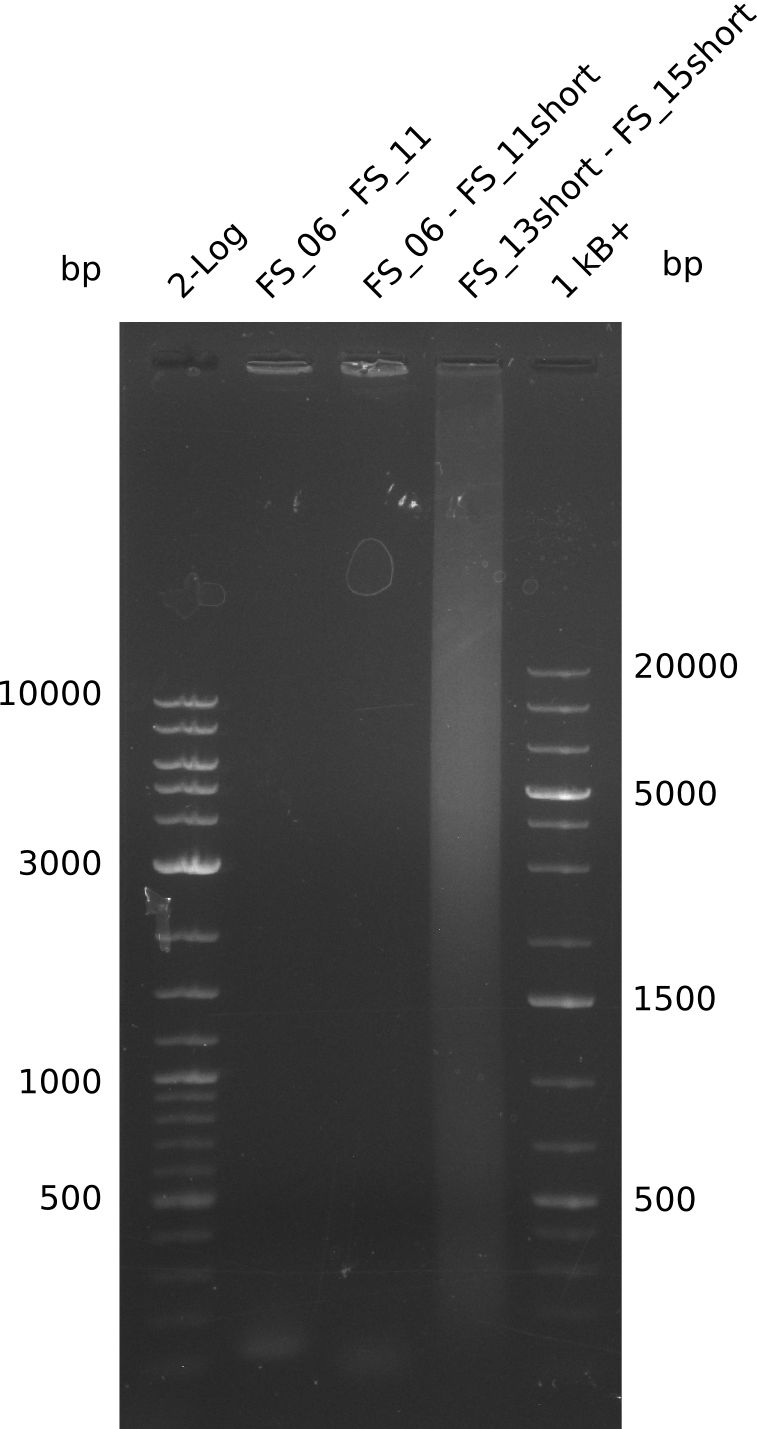
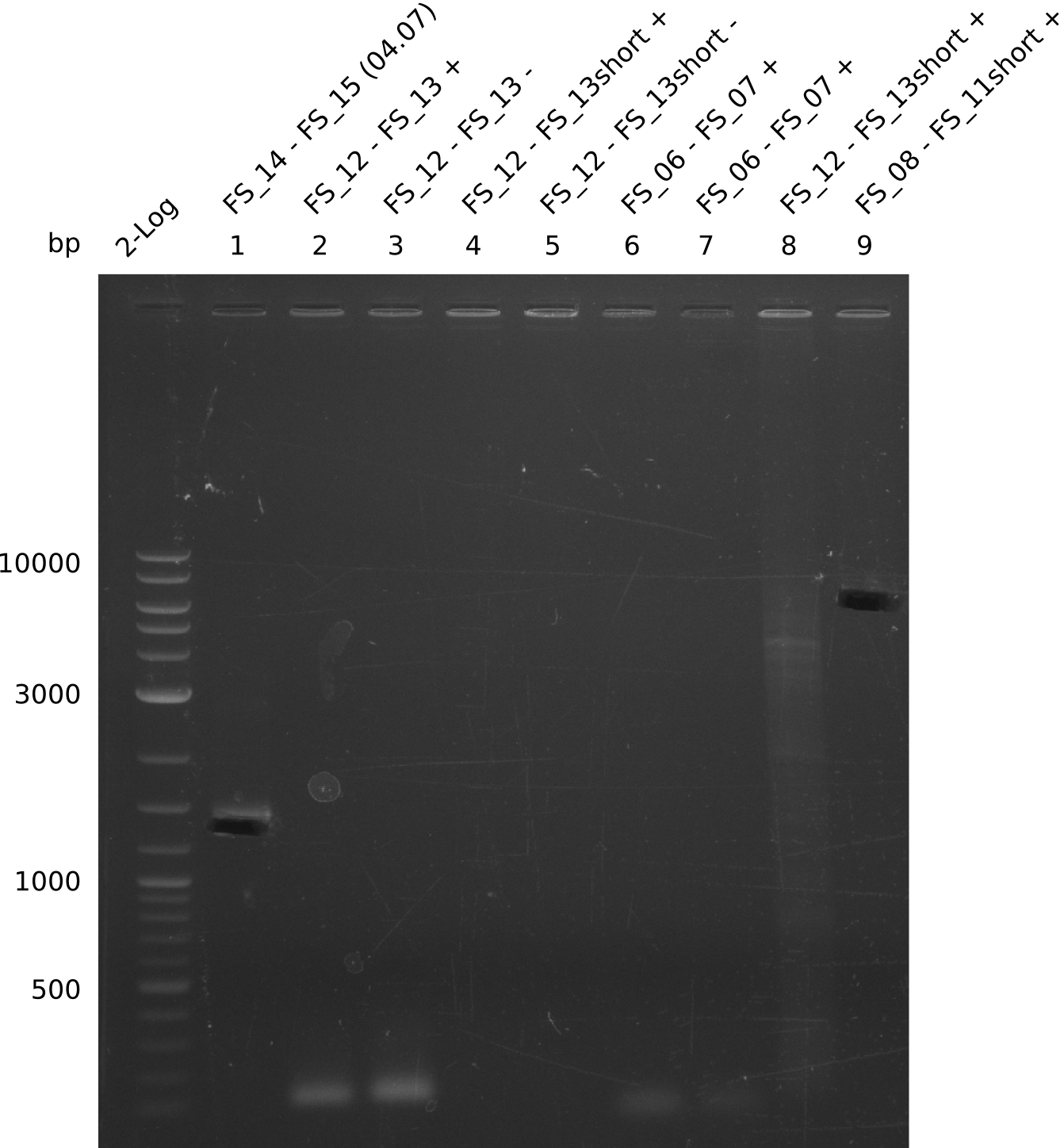
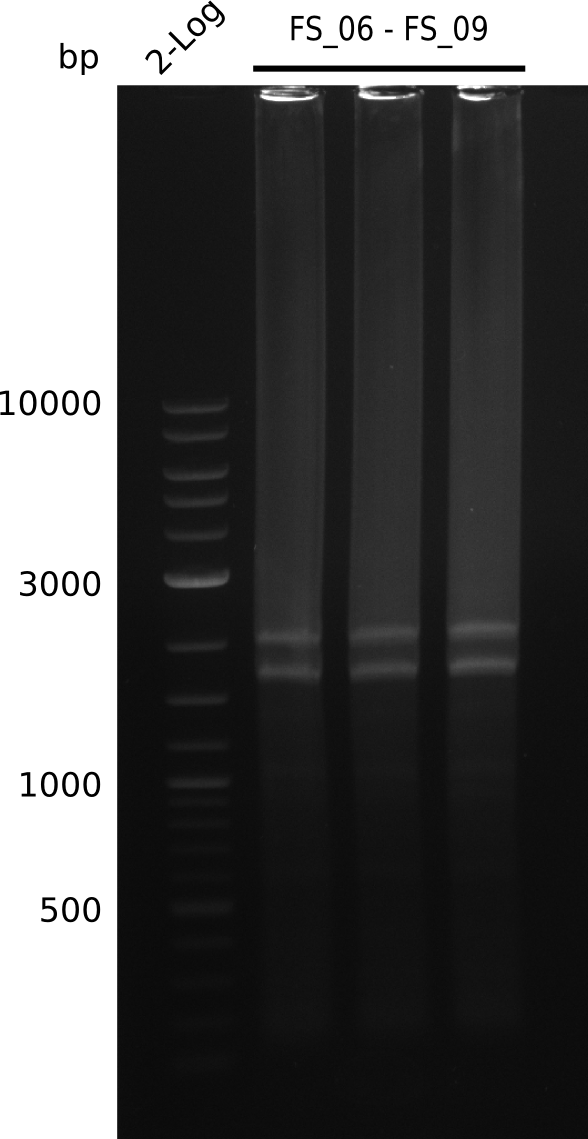
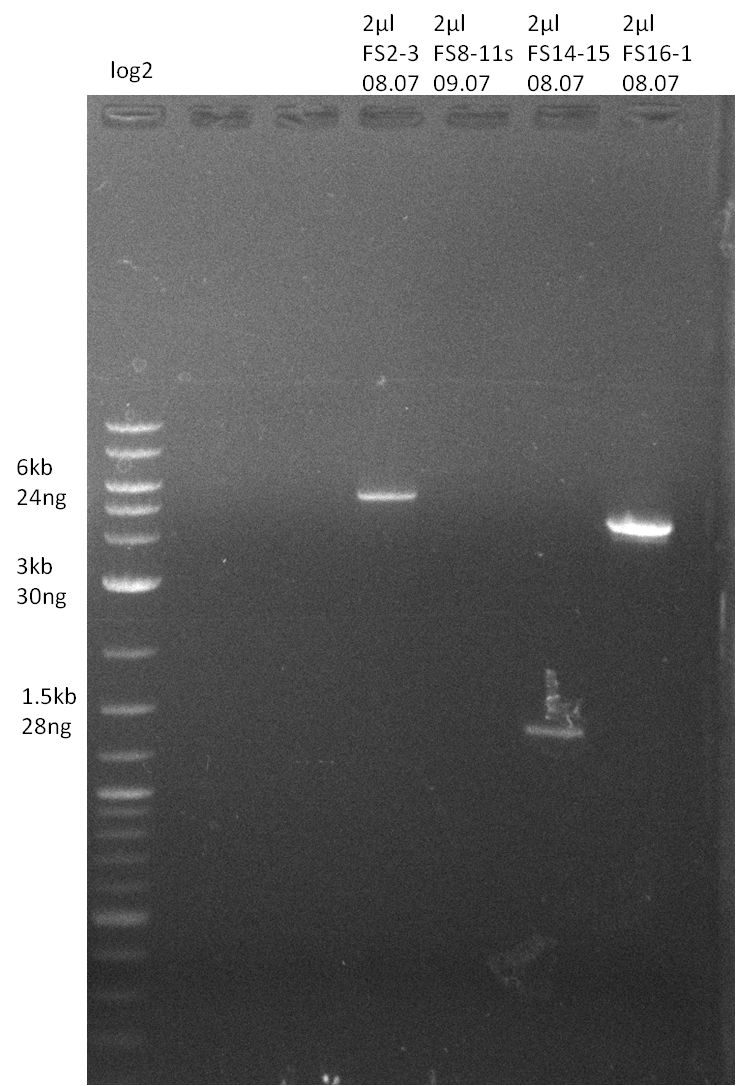
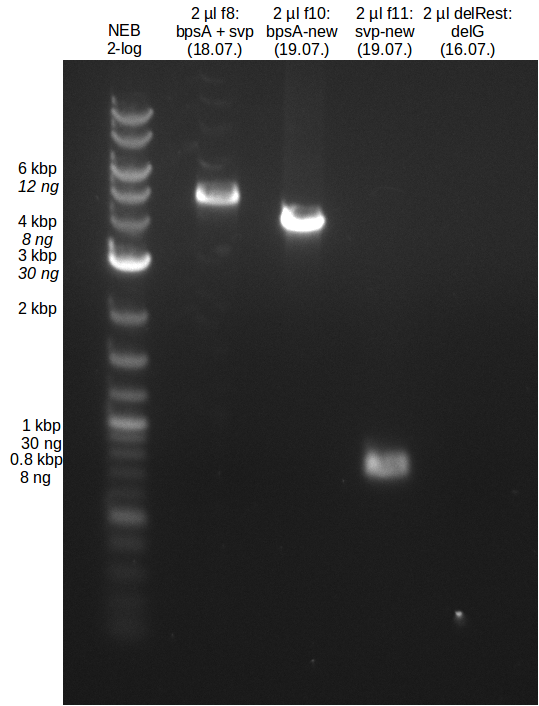
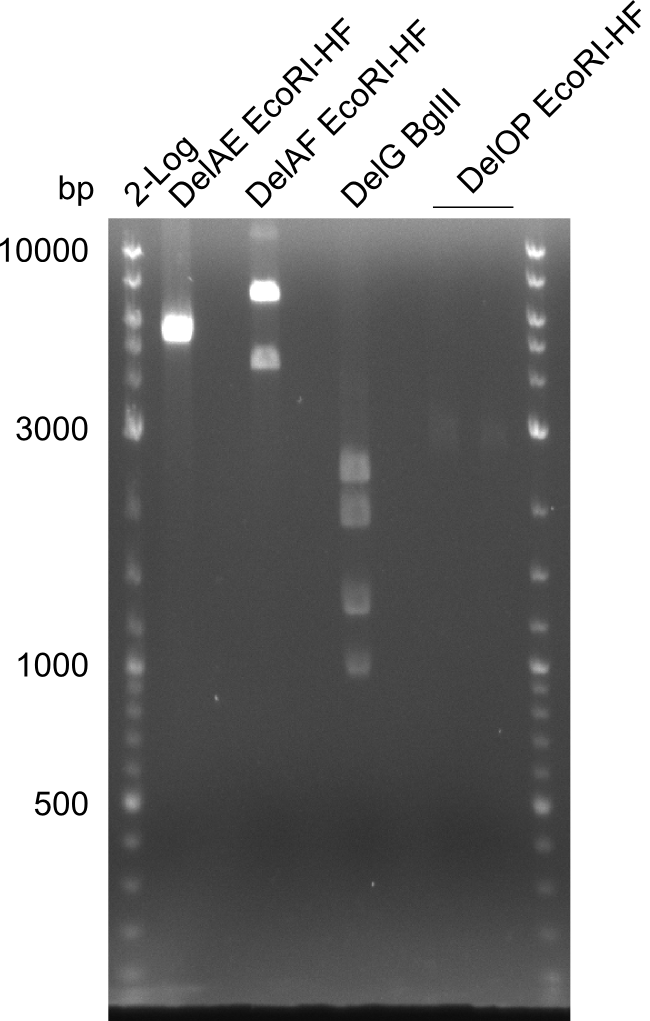
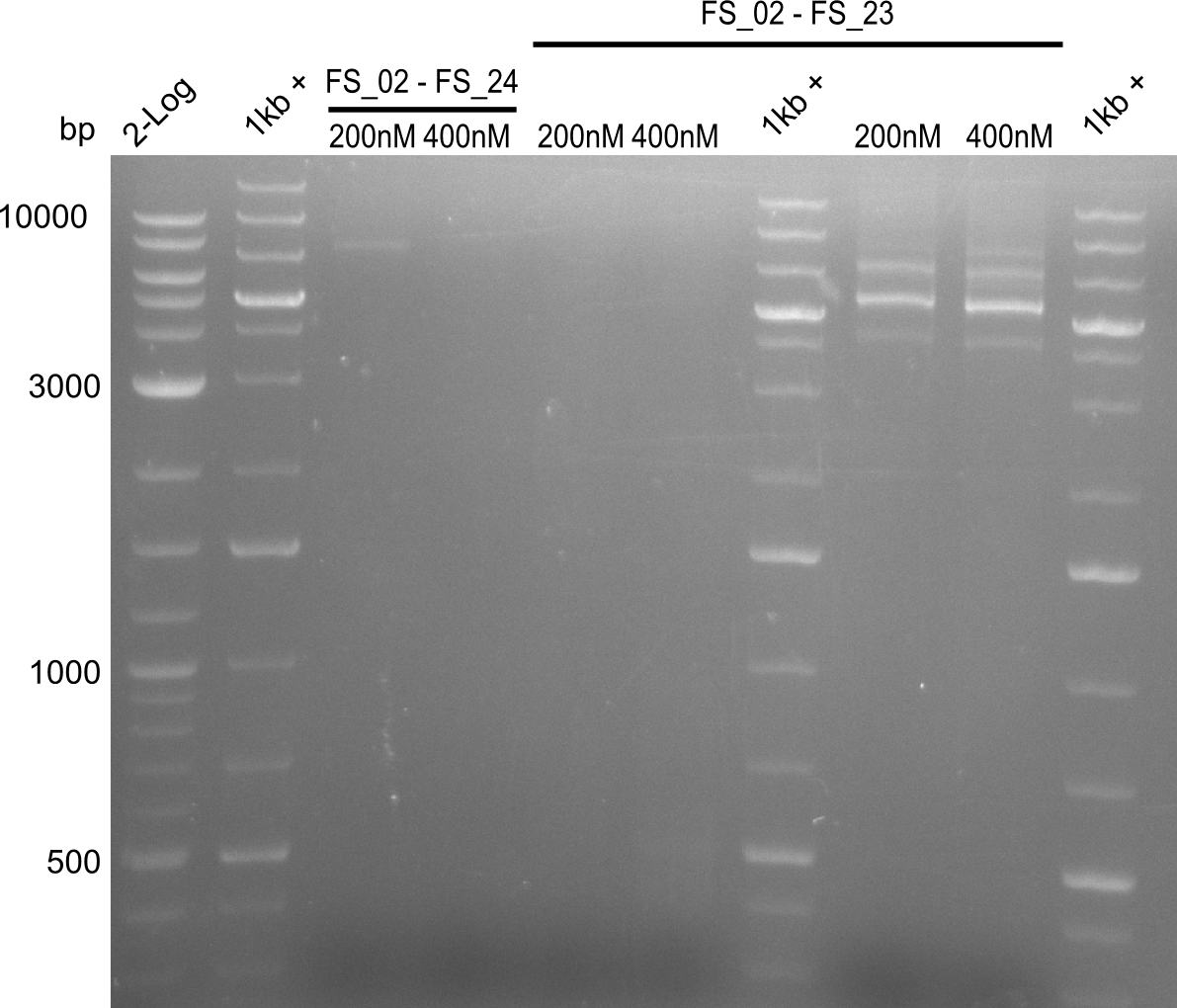
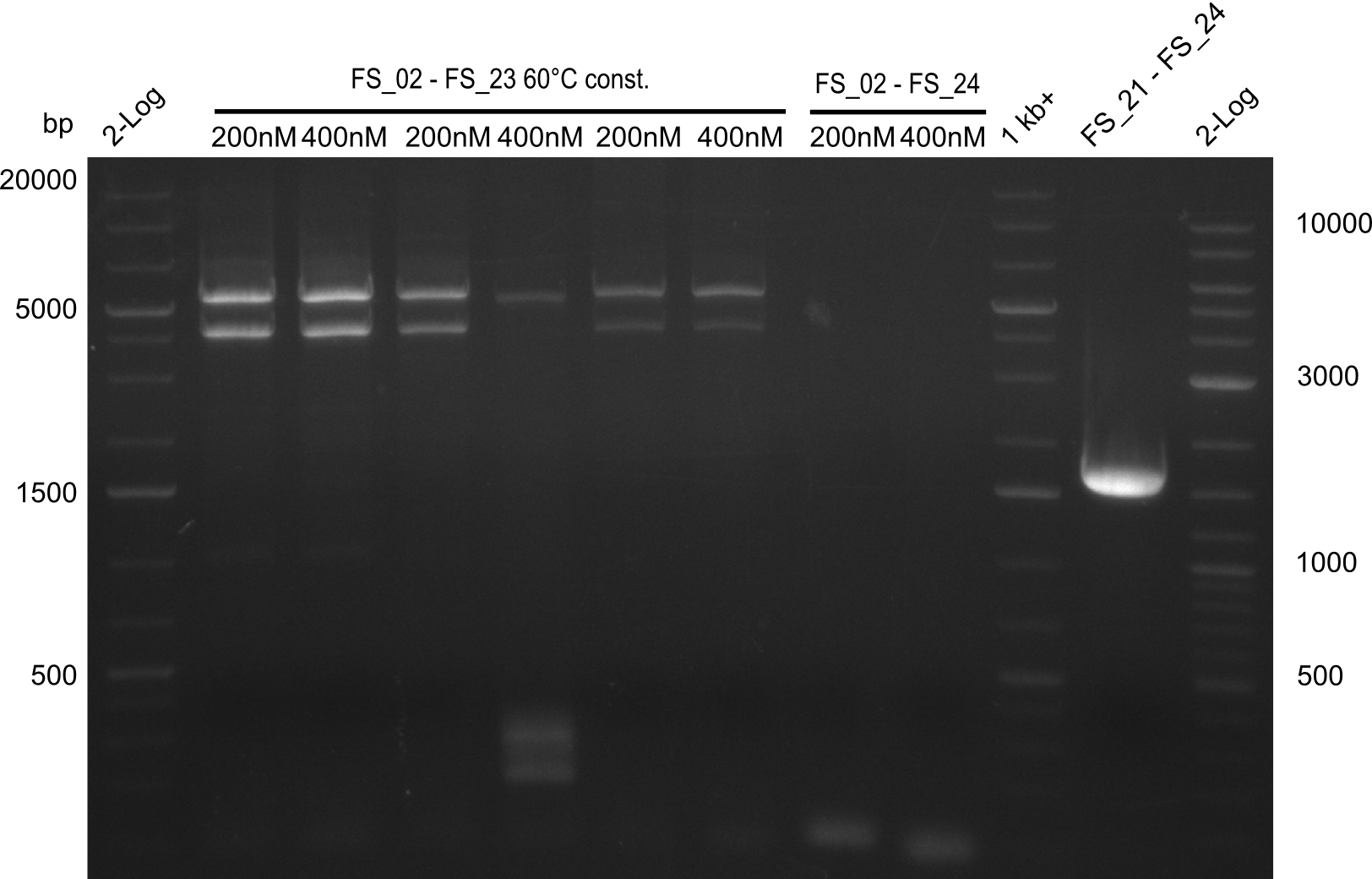
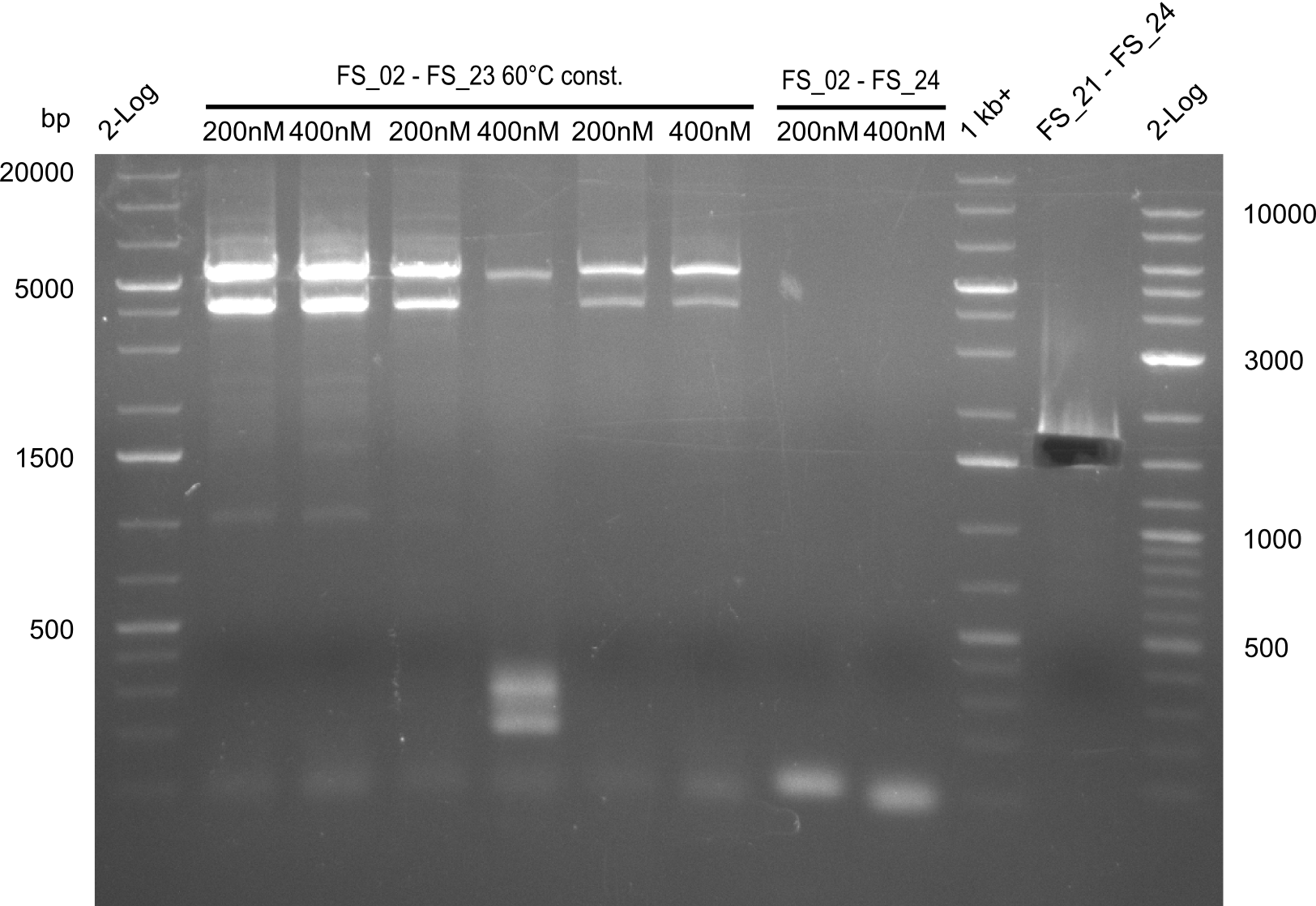
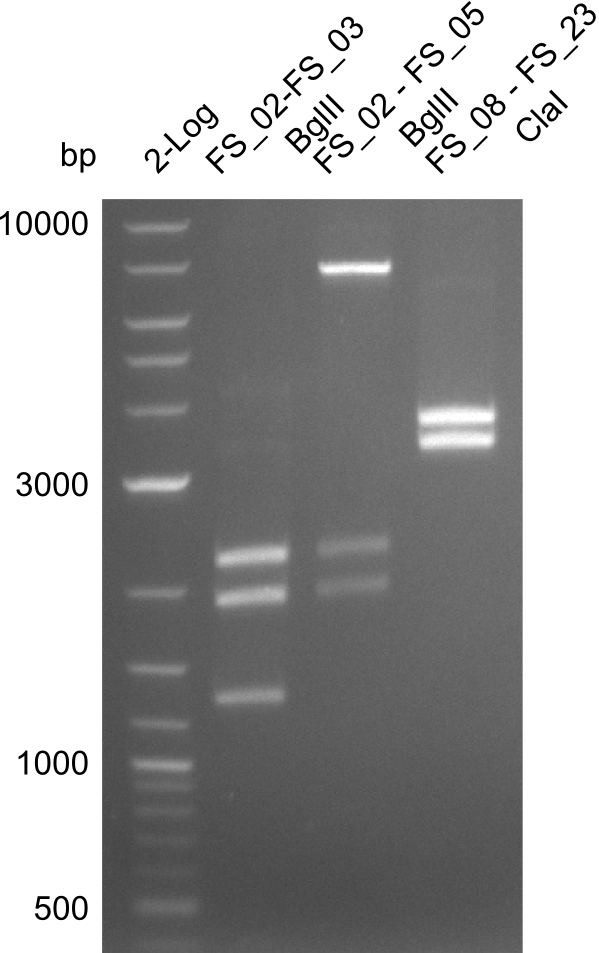
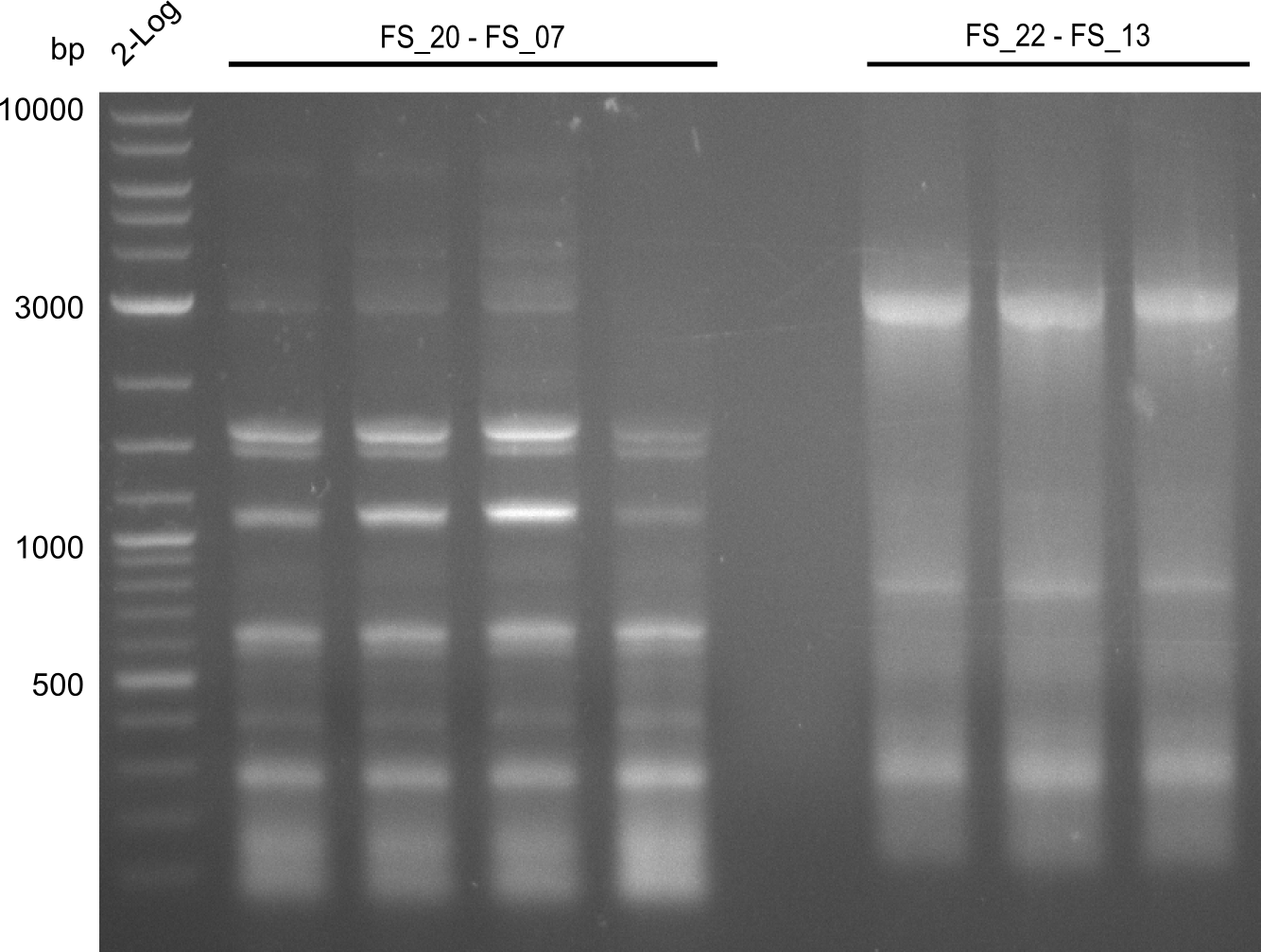
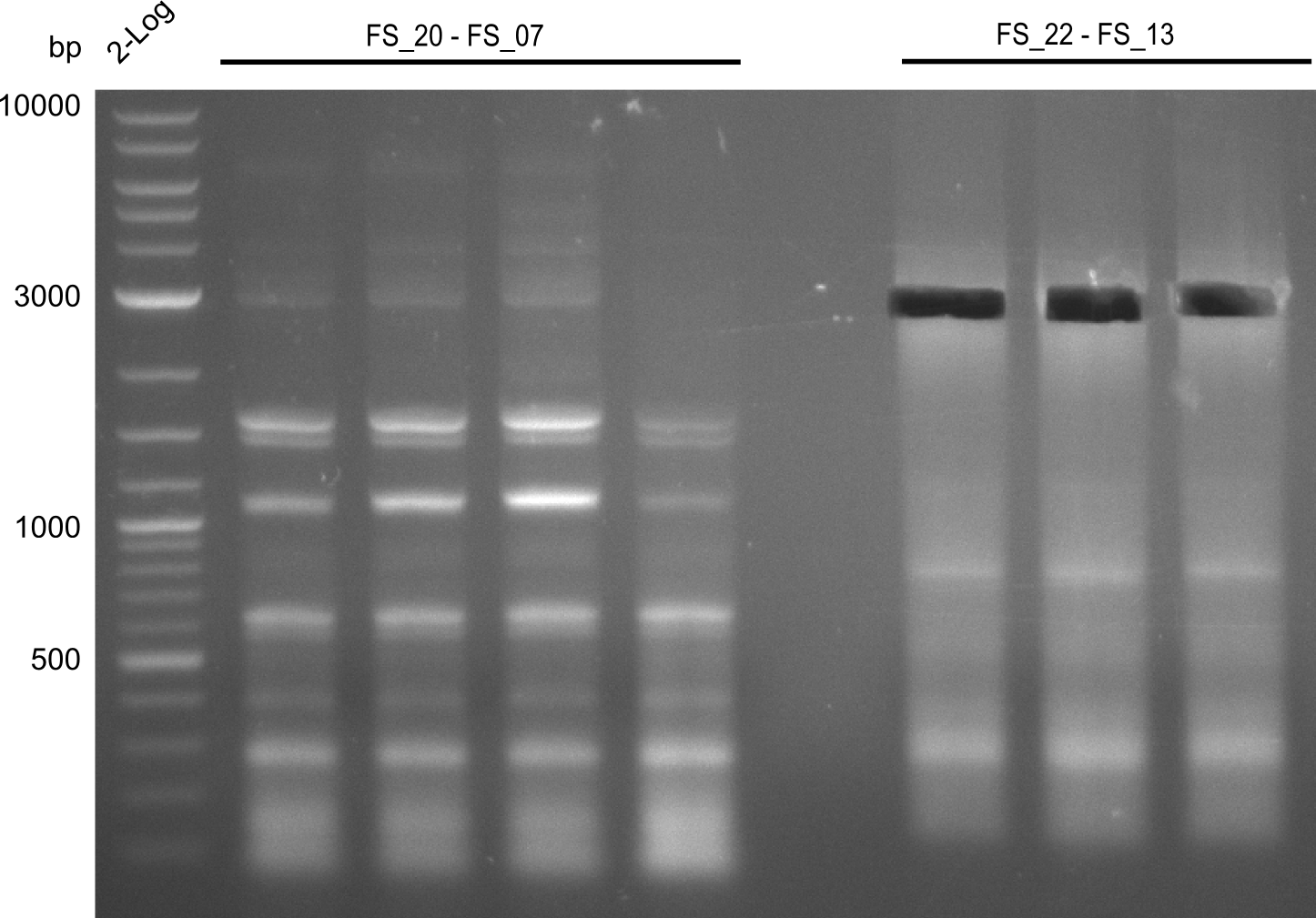
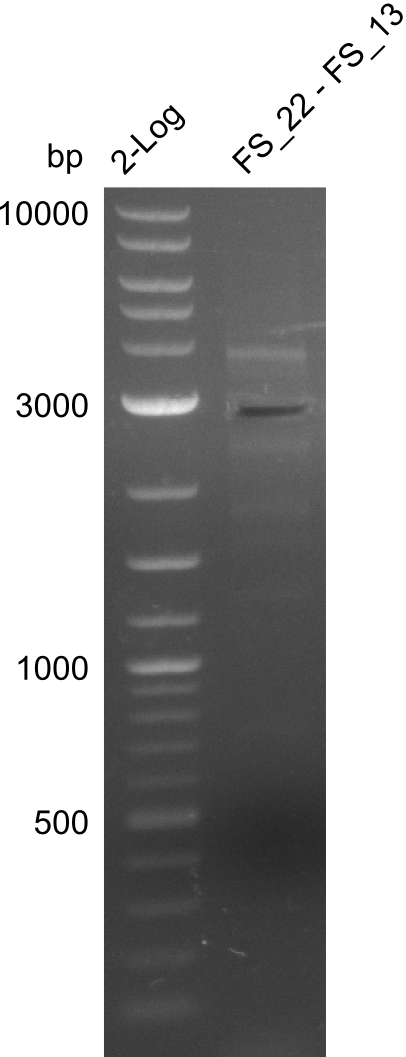
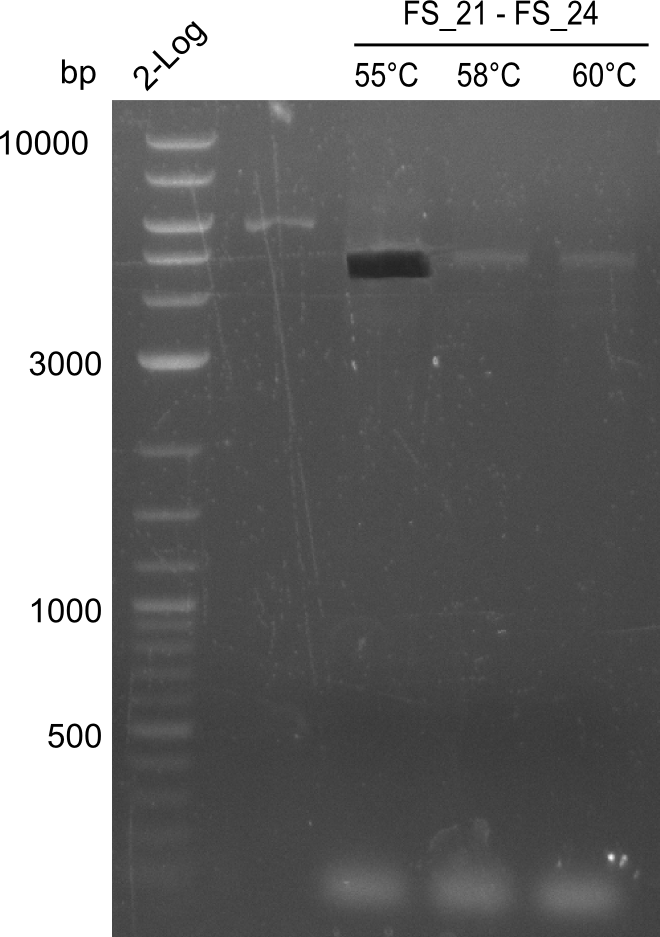
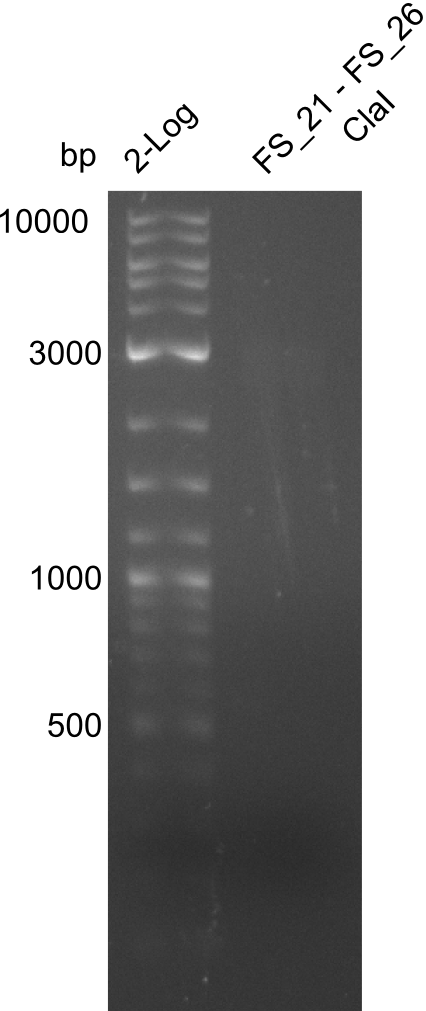
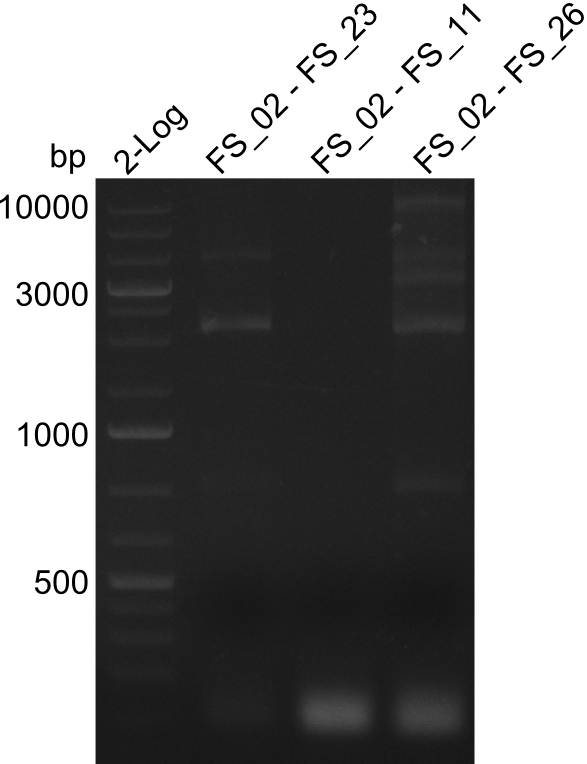
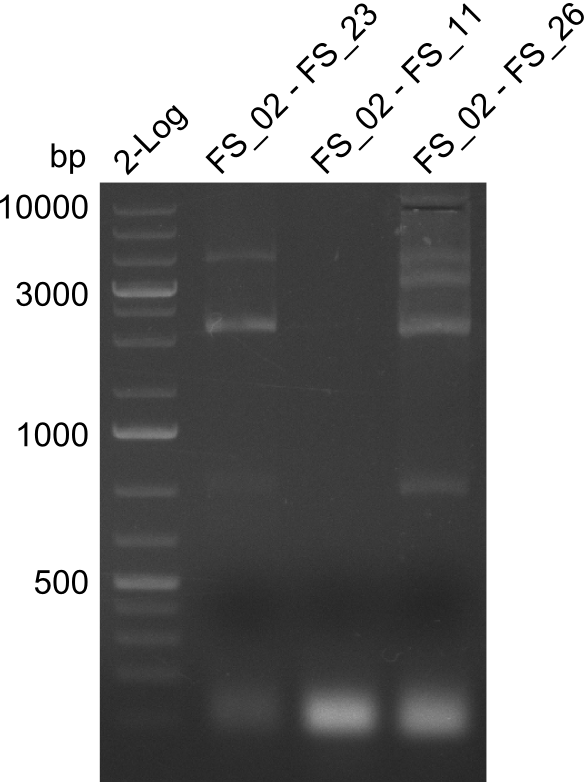
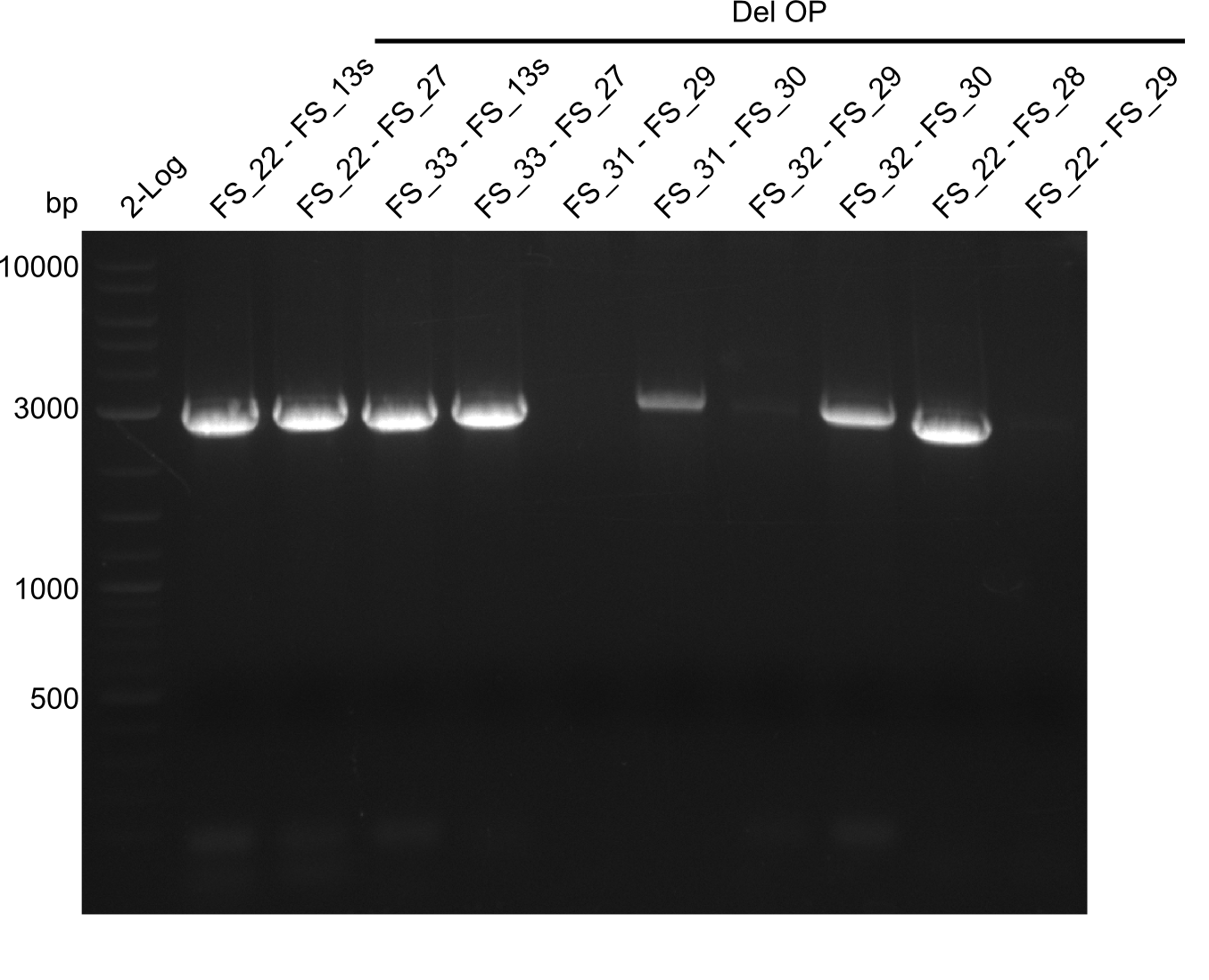
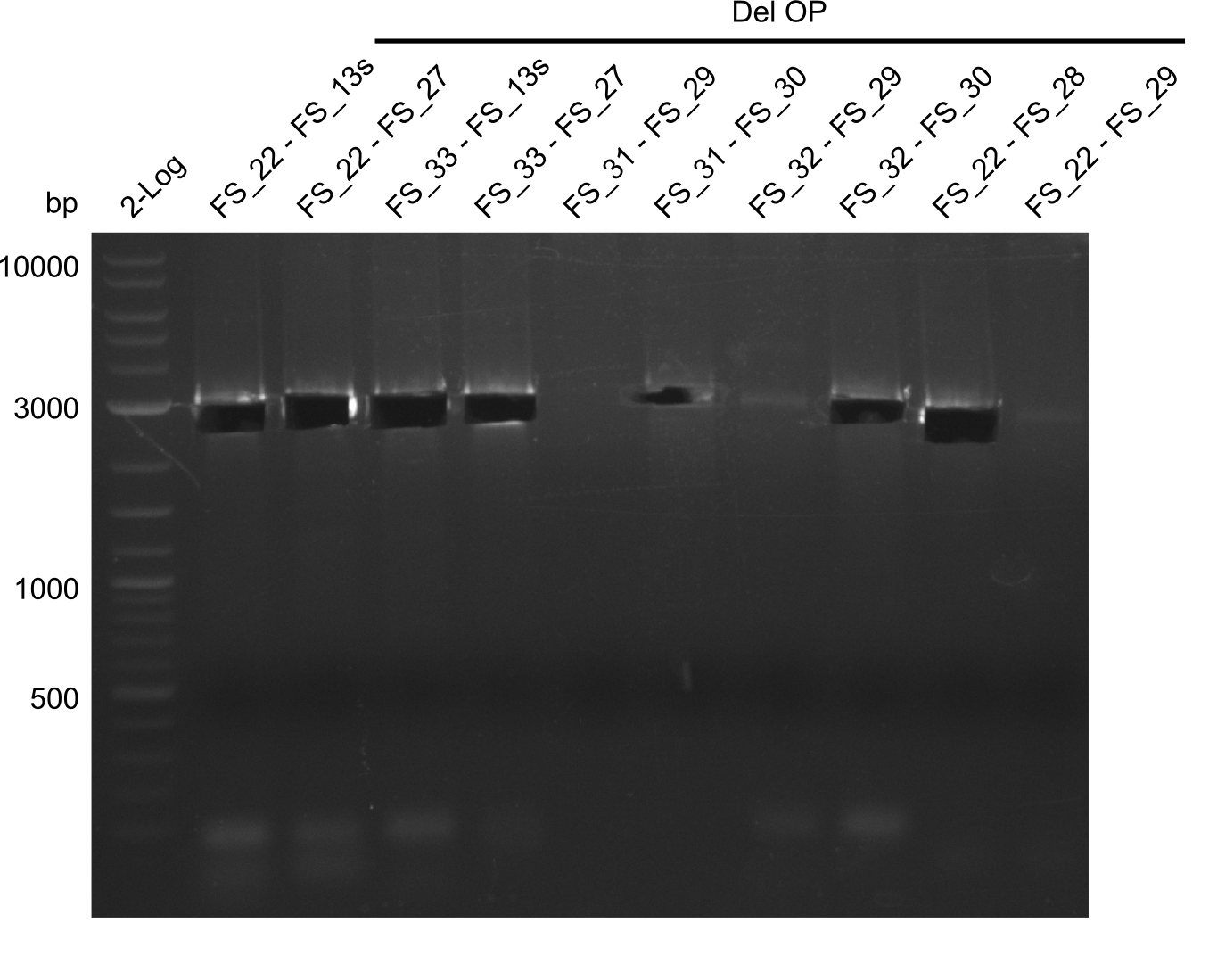
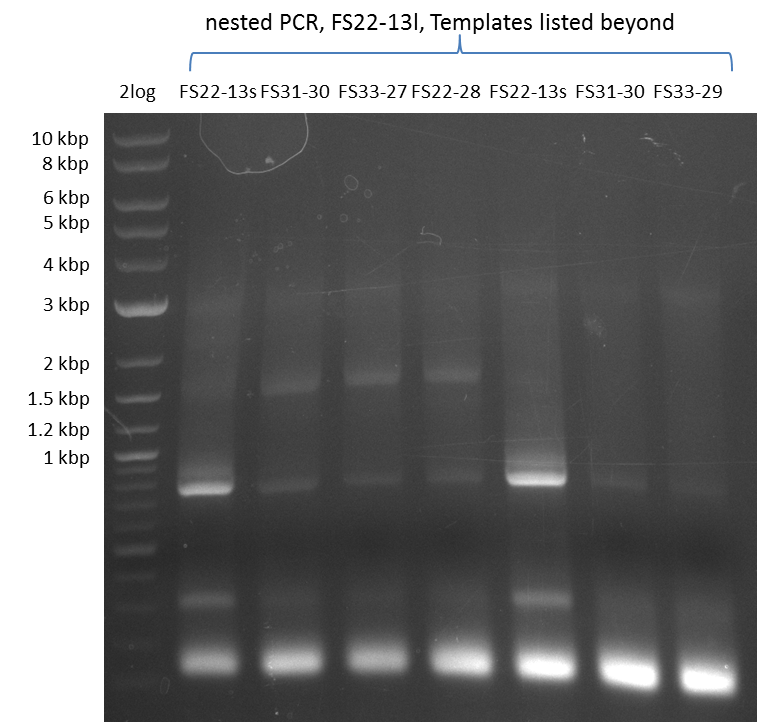
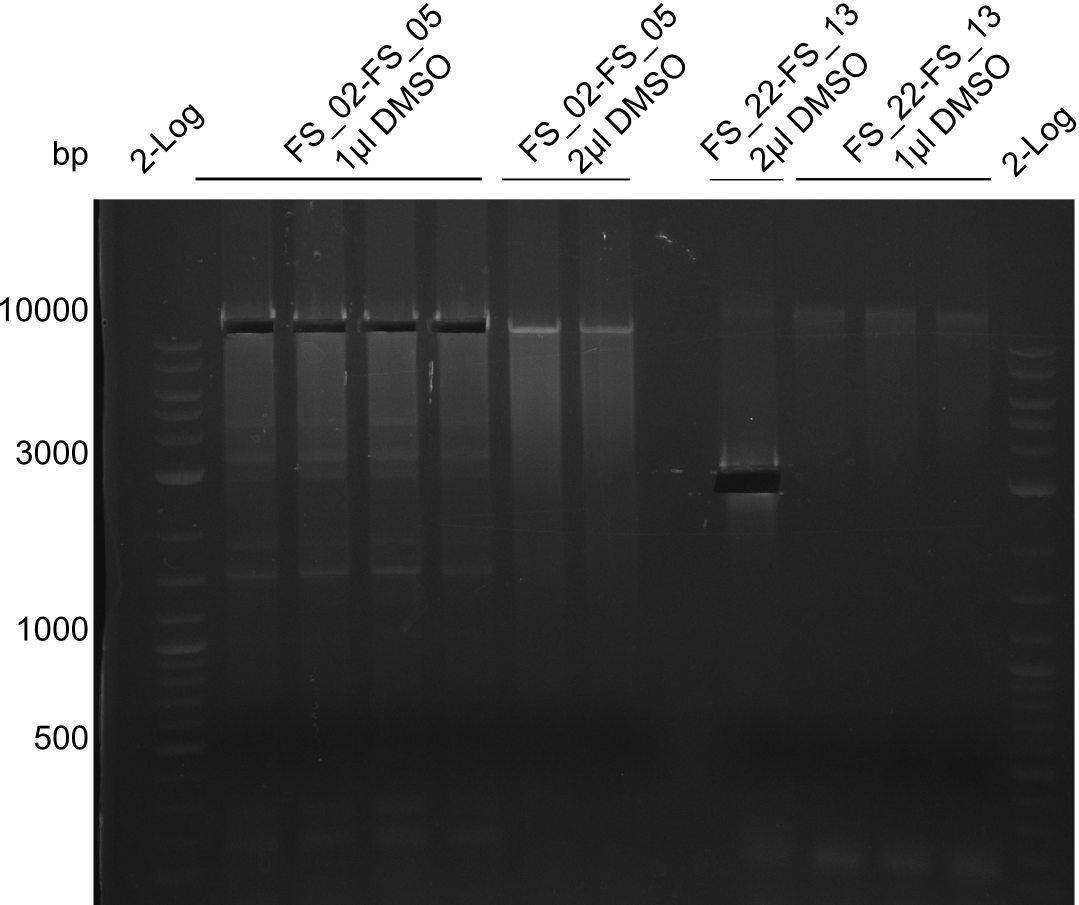
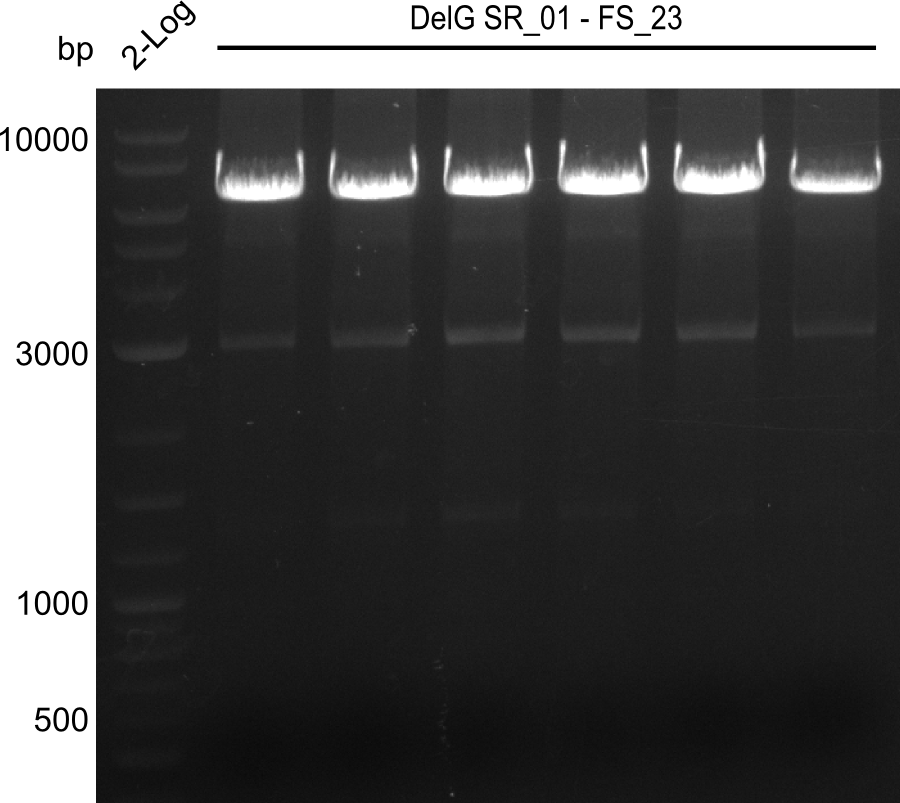
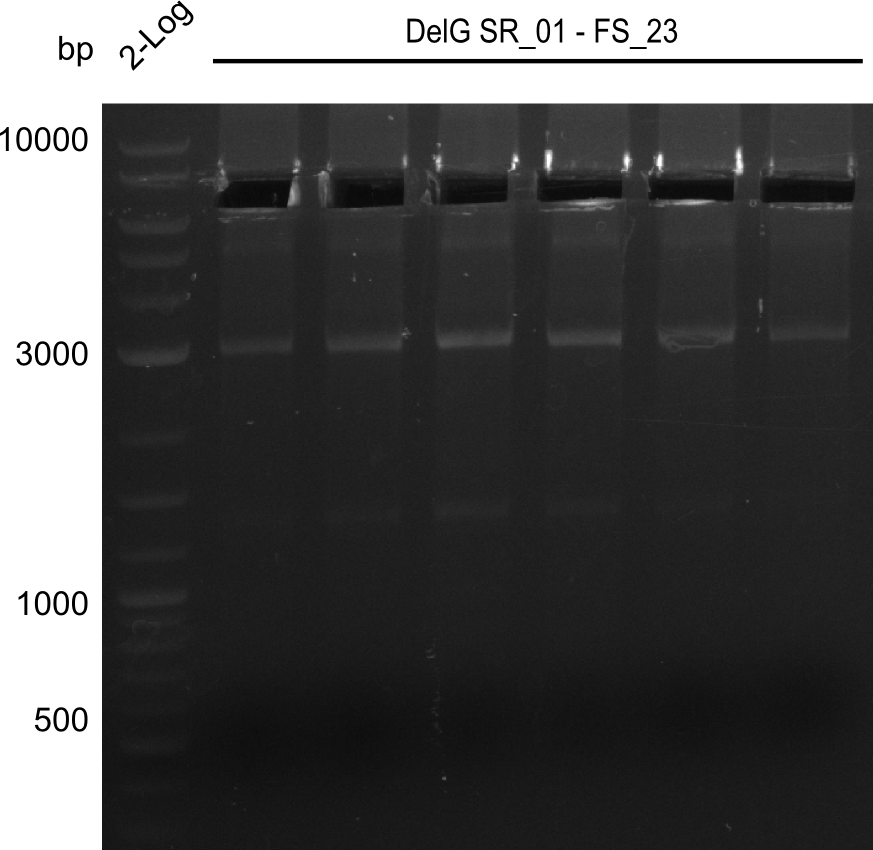
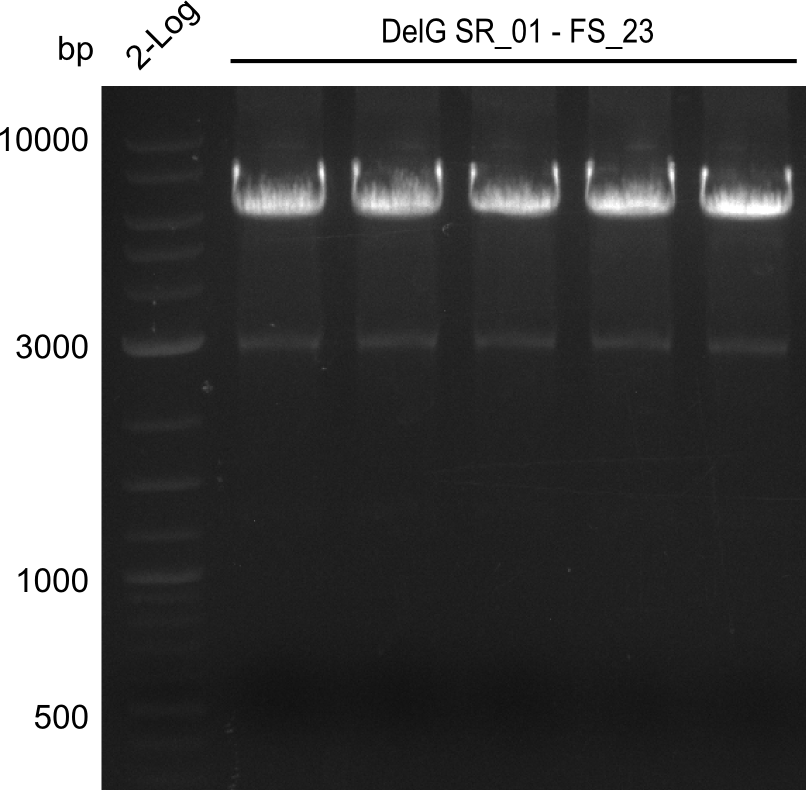
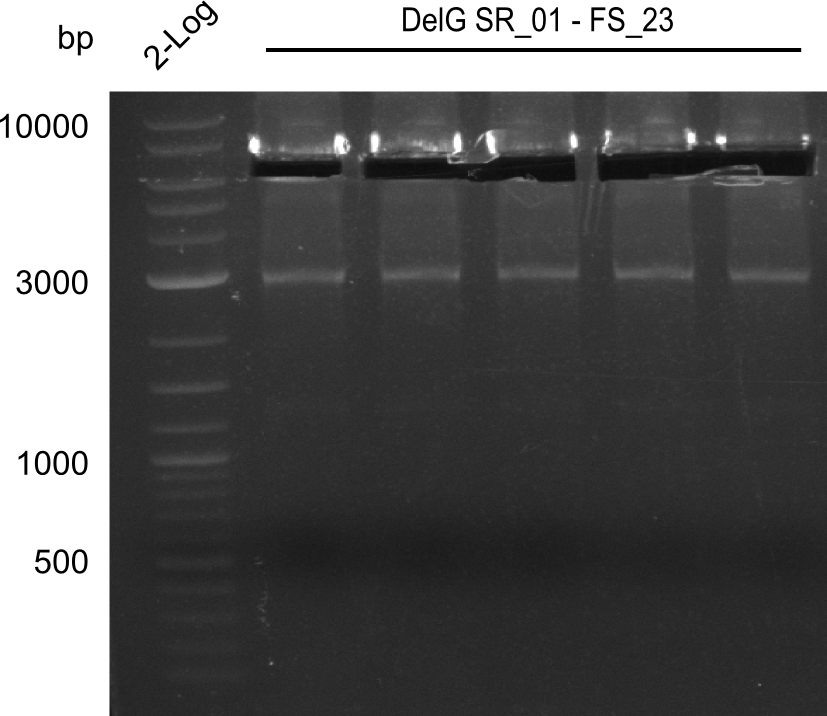
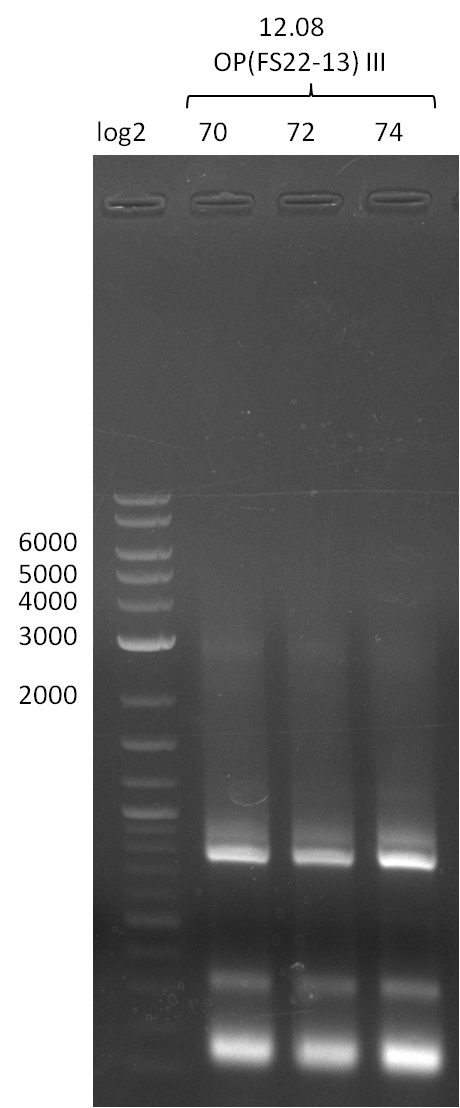
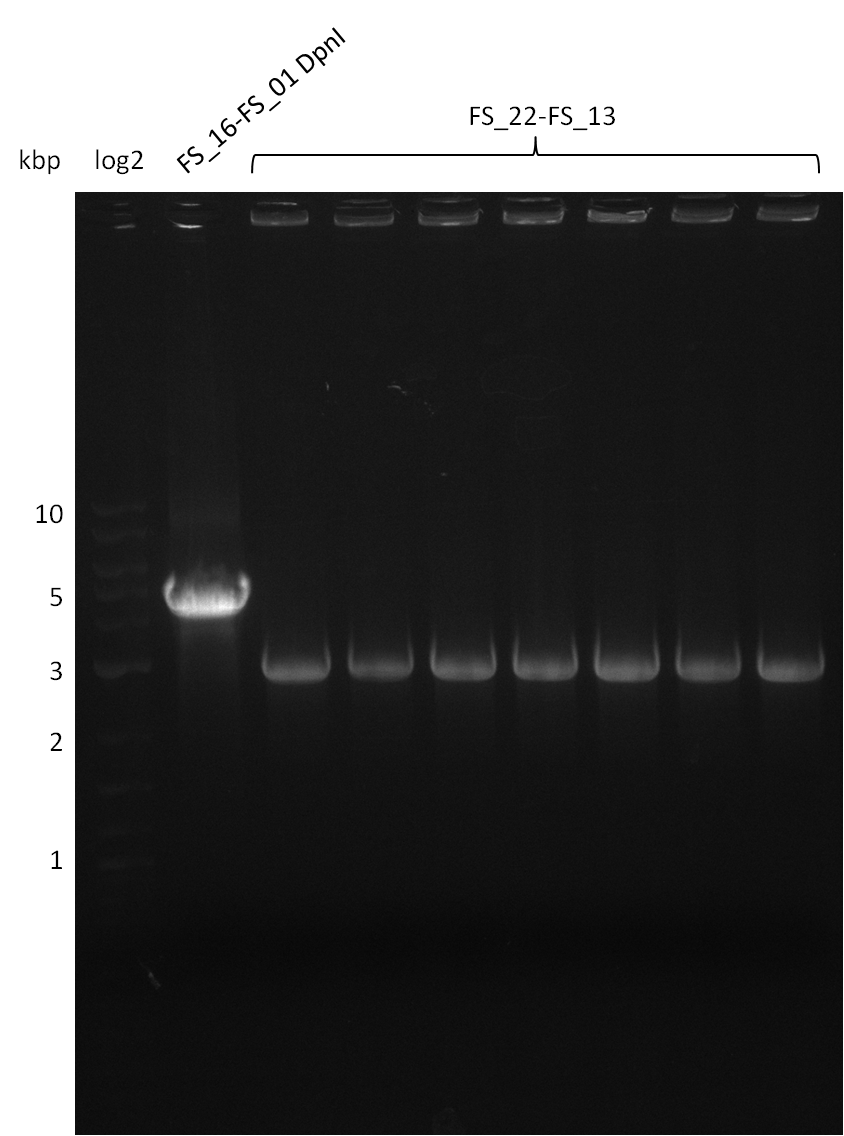
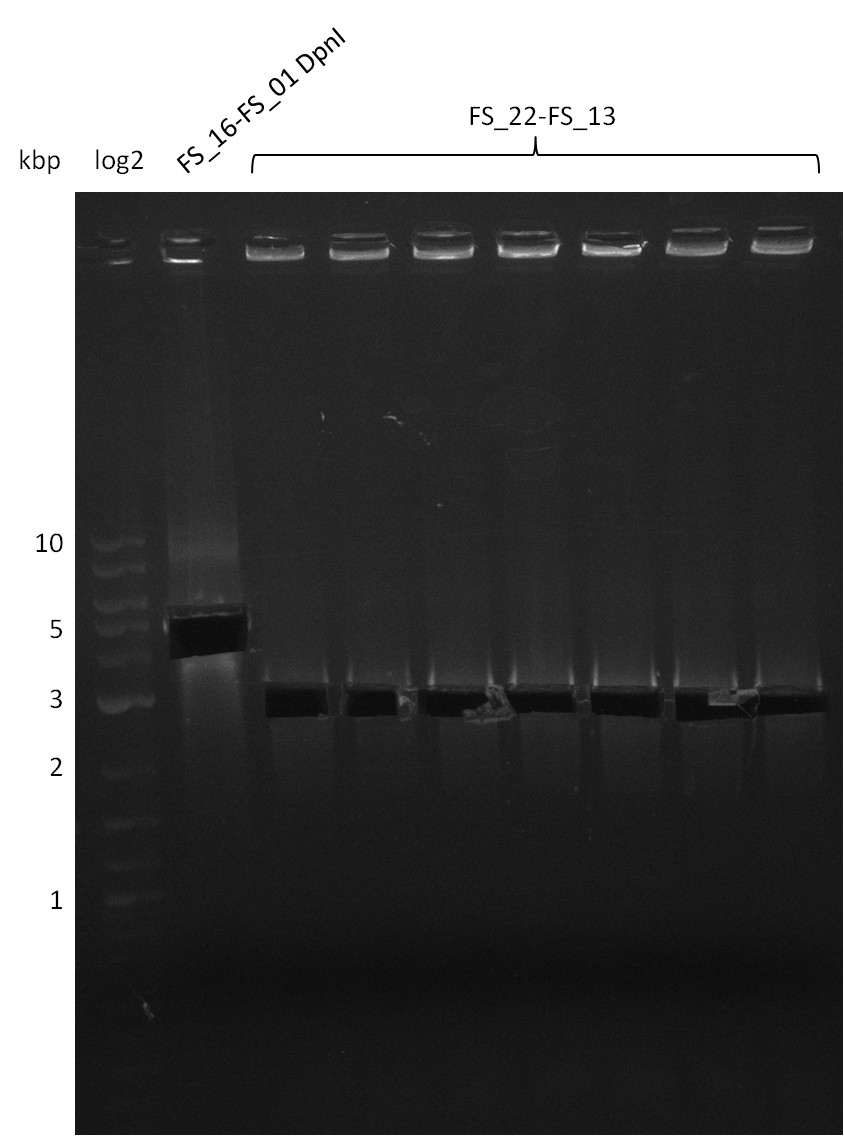
| - | In the previous week several of the | + | In the previous week, several of the newly ordered primer pairs improved our PCRs for DelO-P and DelF-G. However, results were not convincing enough to use these amplicons for Gibson assembly. Therefore, we spend this week optimizing the PCRs for the abovementioned fragments. In addition we validated the amplicon of DelG with restriction digest. Validation of the other PCR products did not succeed, mostly due to very low amount of DNA. |
</p> | </p> | ||
</div> | </div> | ||
| Line 113: | Line 116: | ||
<h1>Week 14</h1> | <h1>Week 14</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | + | We were still struggeling with getting the correct amplicons for the fragments encoding DelF-G as well as DelO-P. Furthermore, the restriction digests of the already successfully amplified fragments needed to be repeated using higher amounts of DNA, as results of the previous test digests were rather inconclusive. Furthermore, samples of these fragments were send for sequencing. | |
</p> | </p> | ||
</div> | </div> | ||
| Line 124: | Line 127: | ||
<h1>Week 15</h1> | <h1>Week 15</h1> | ||
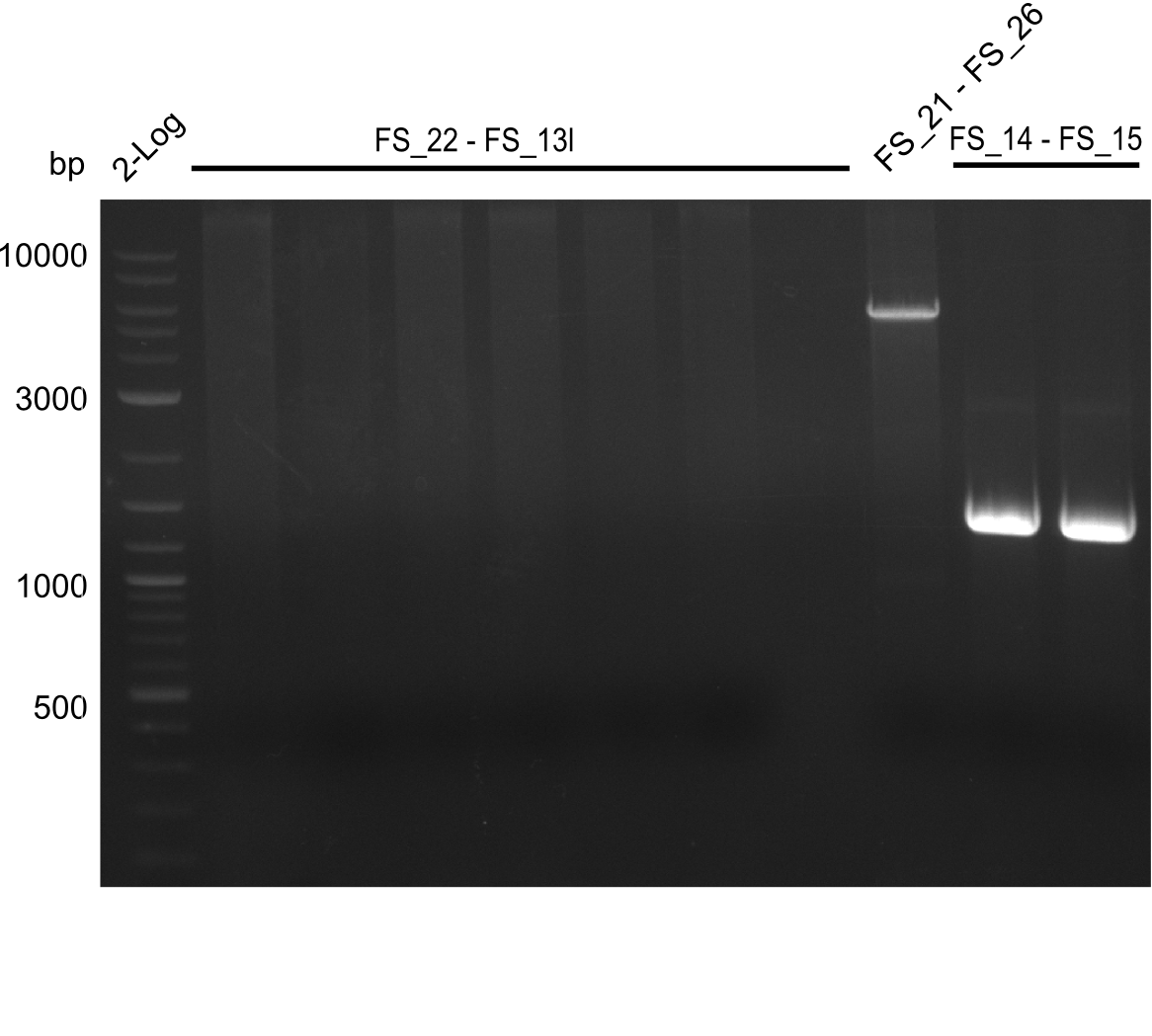
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
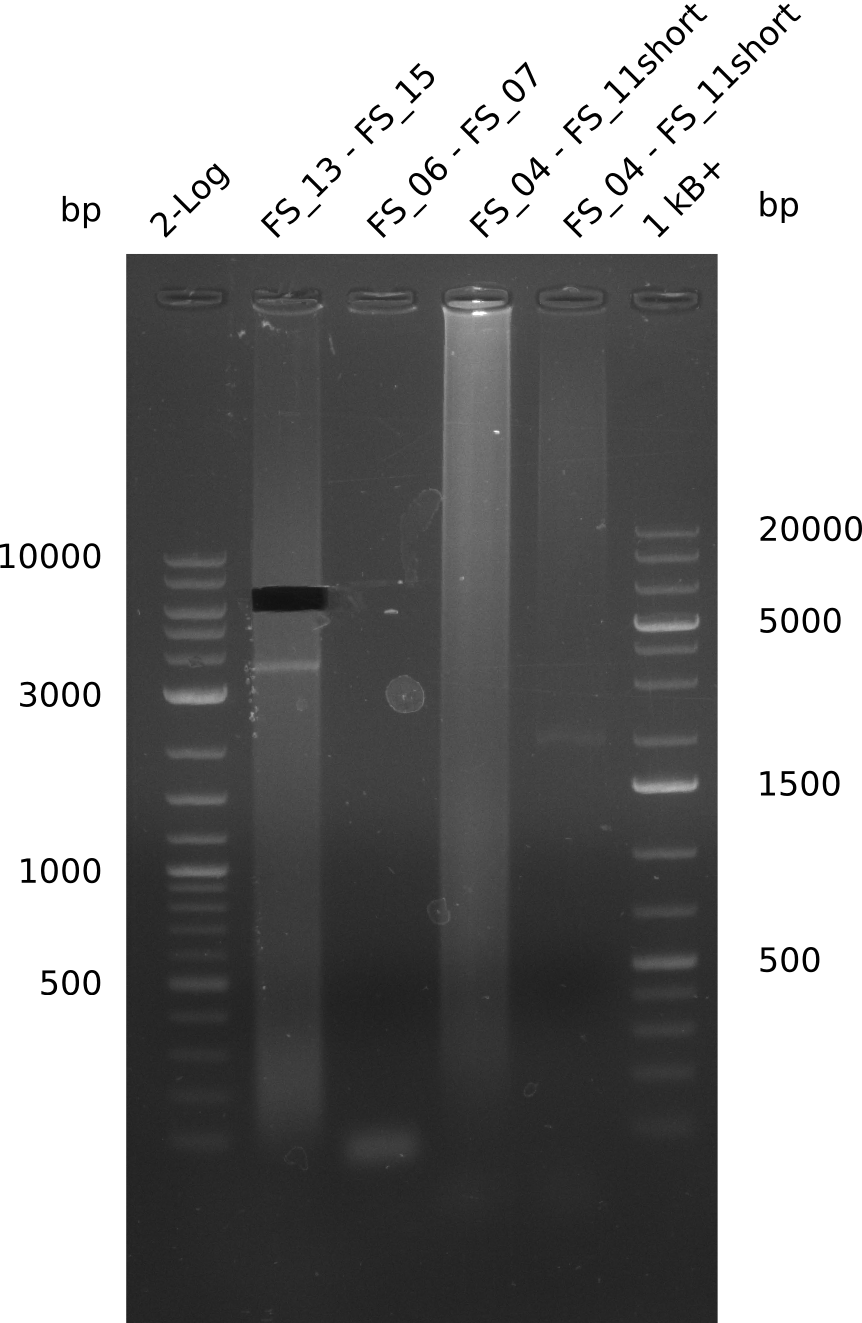
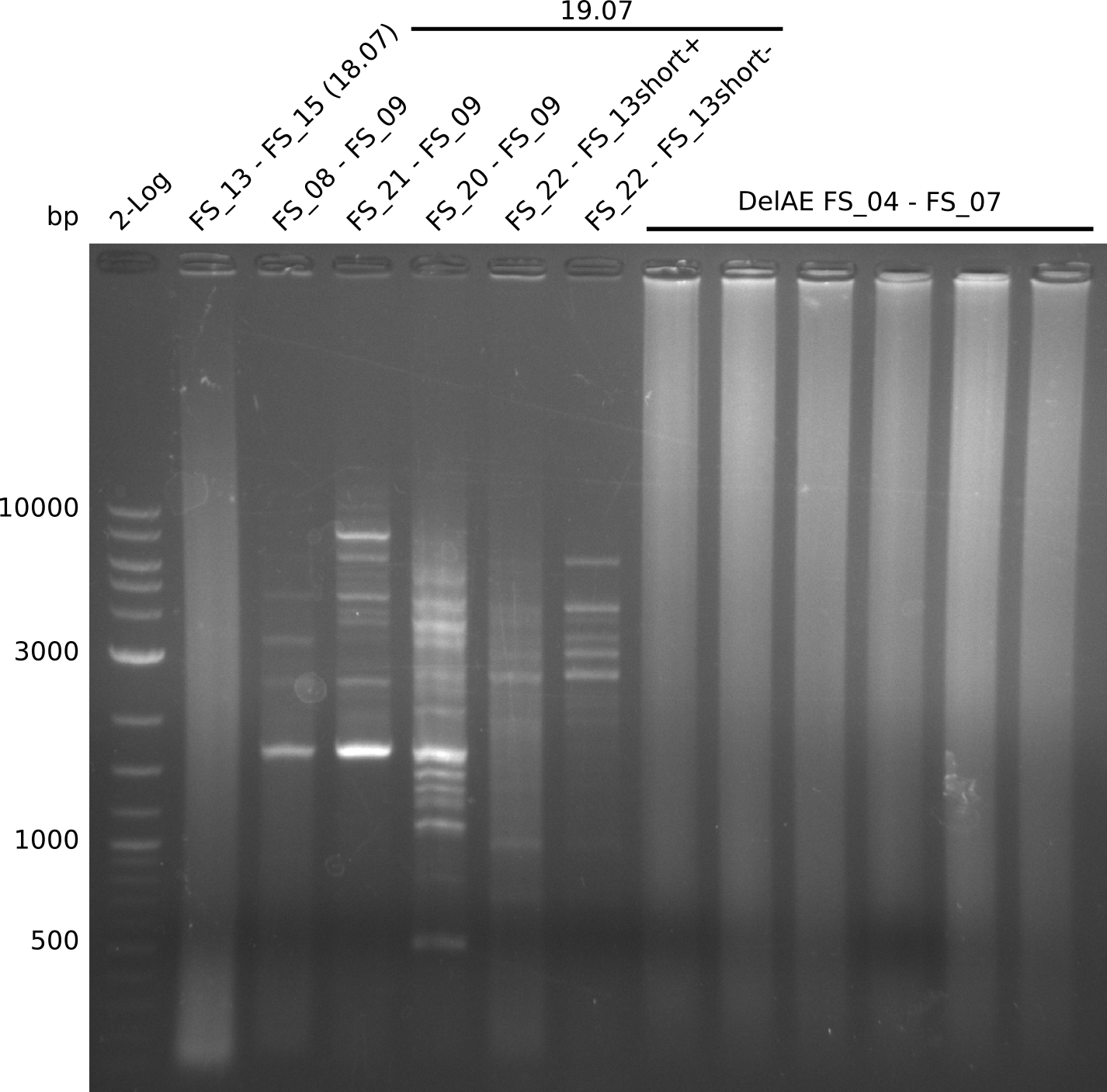
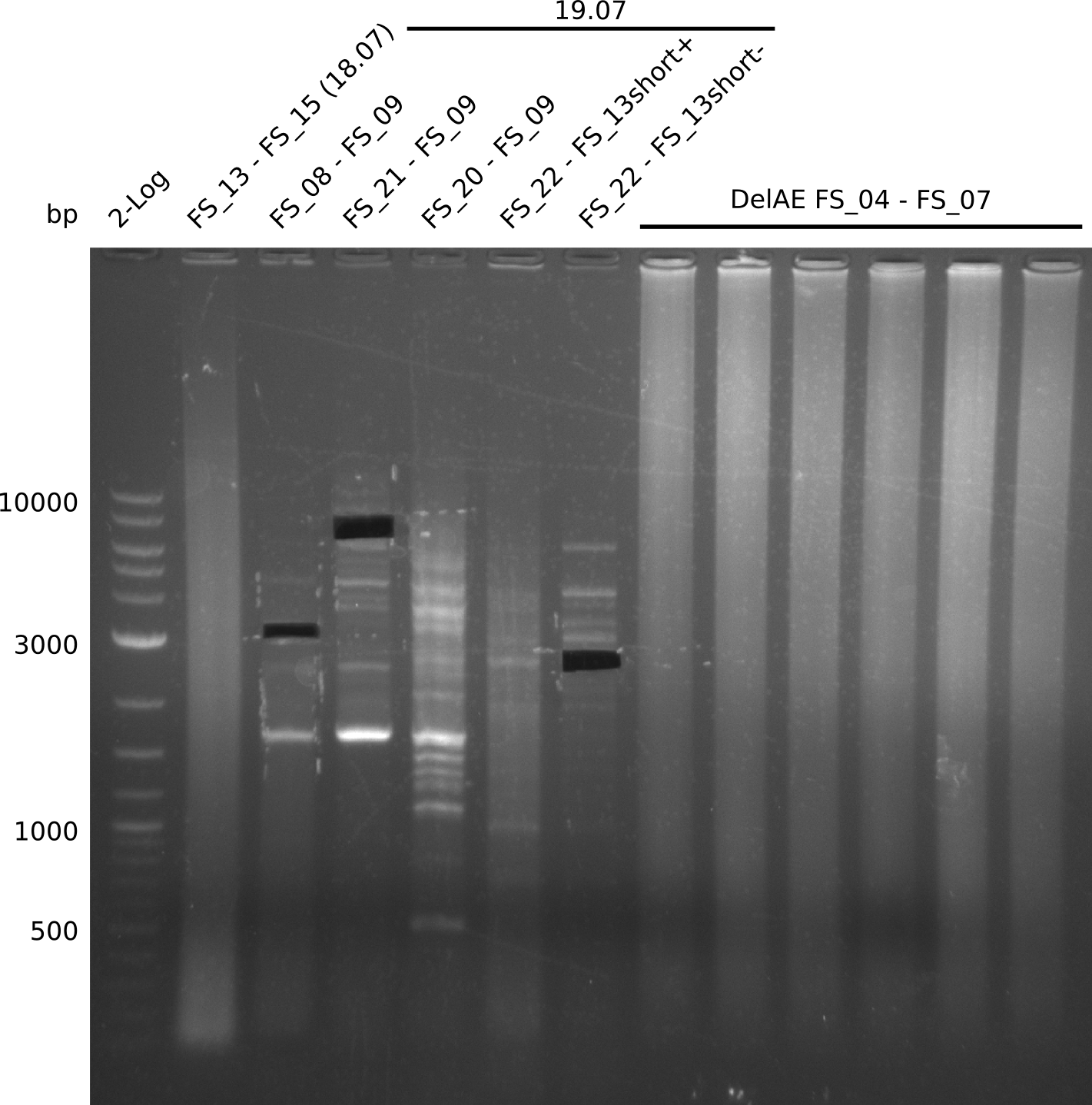
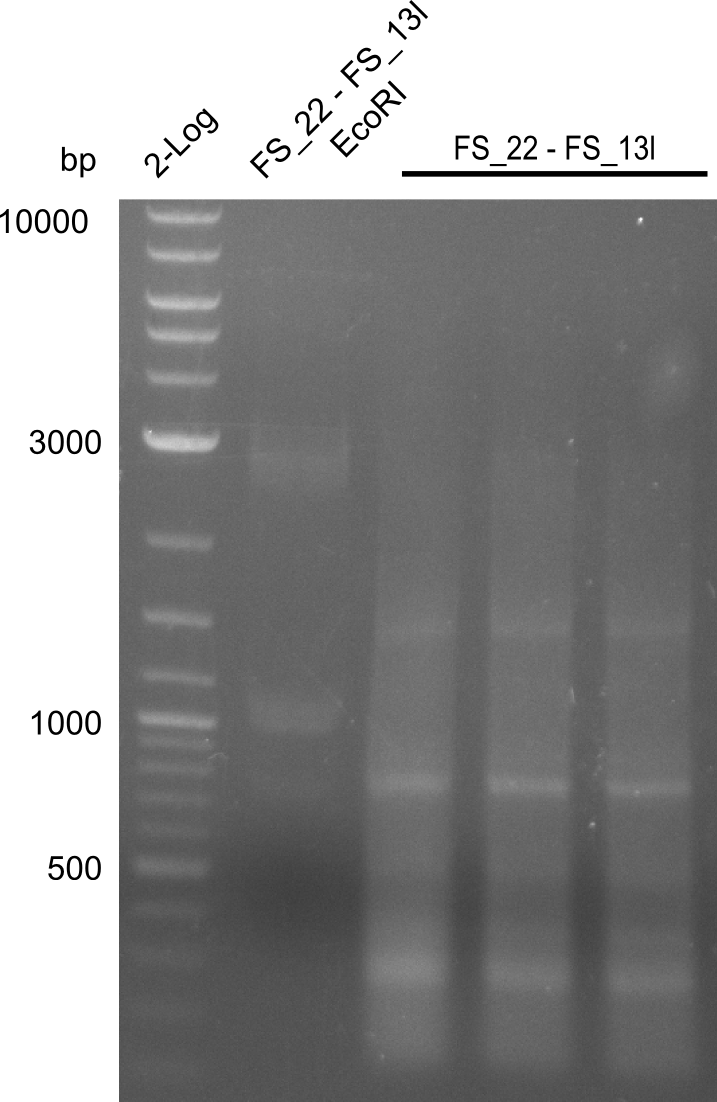
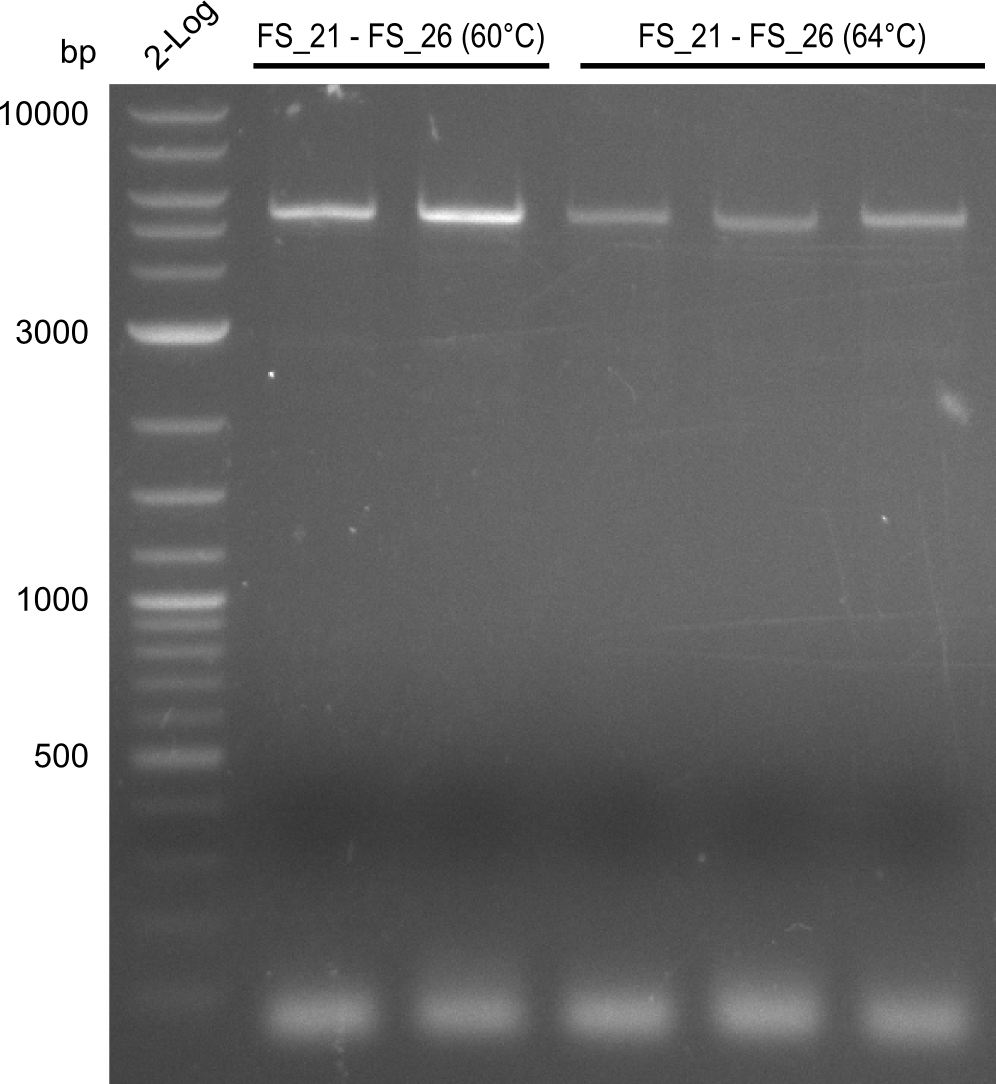
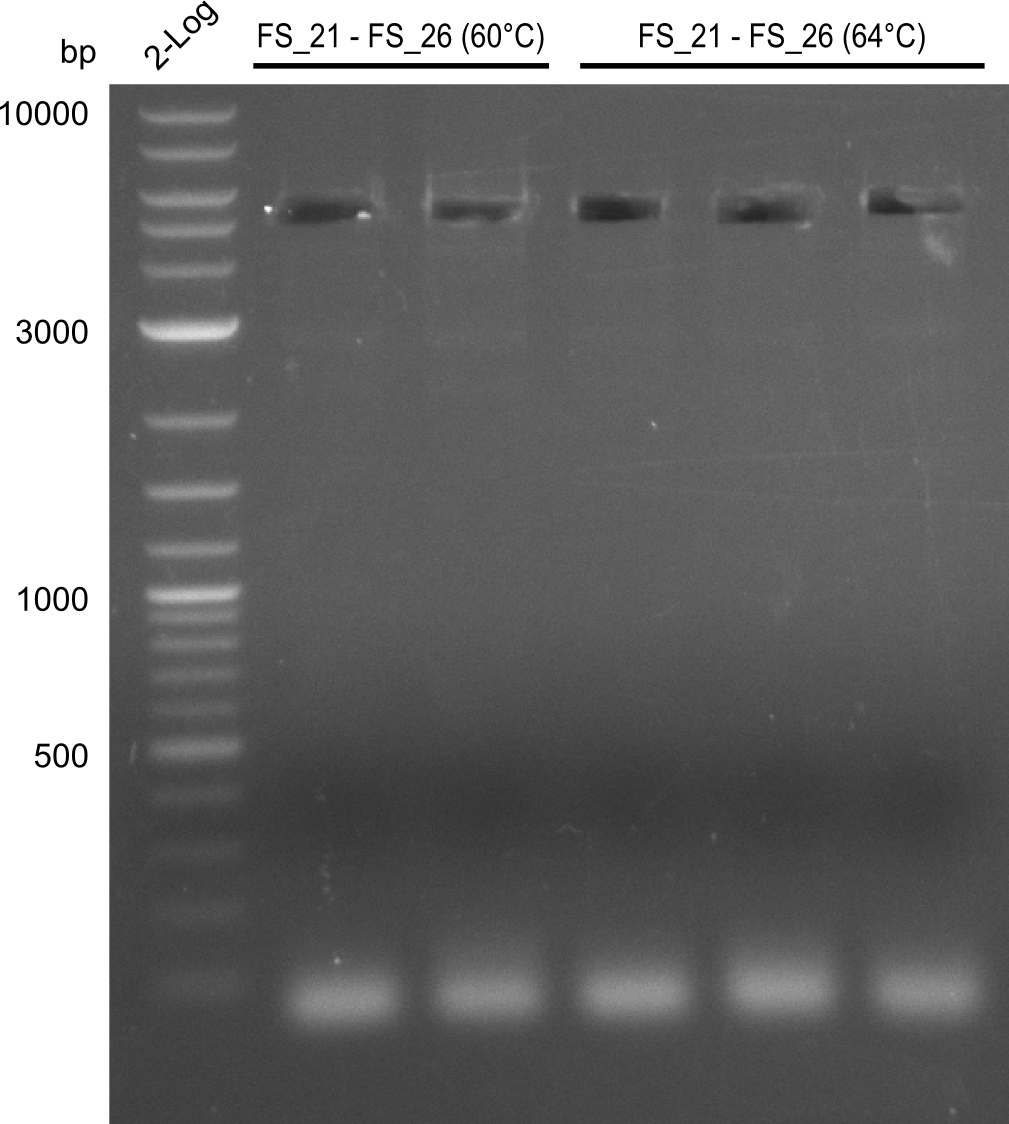
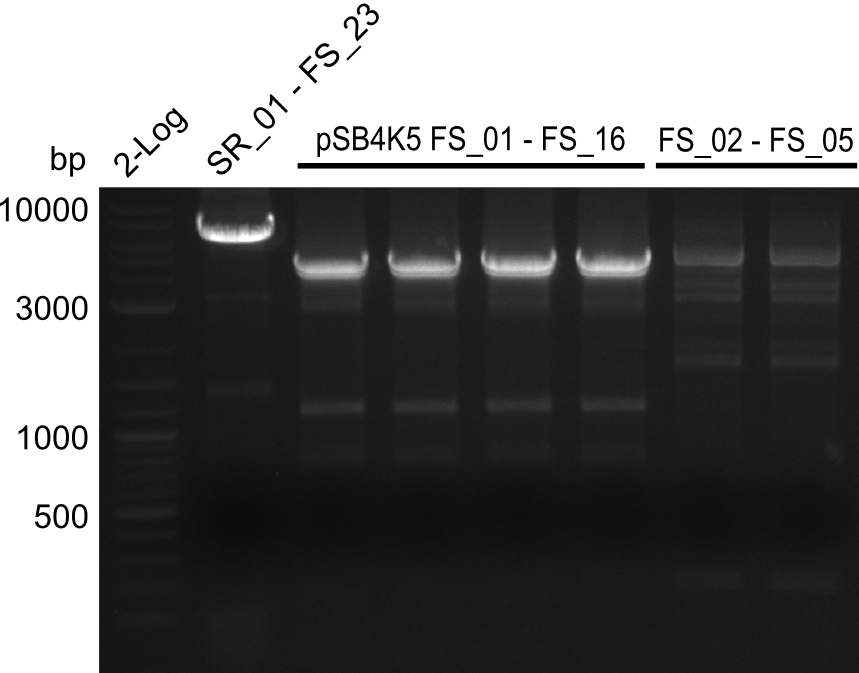
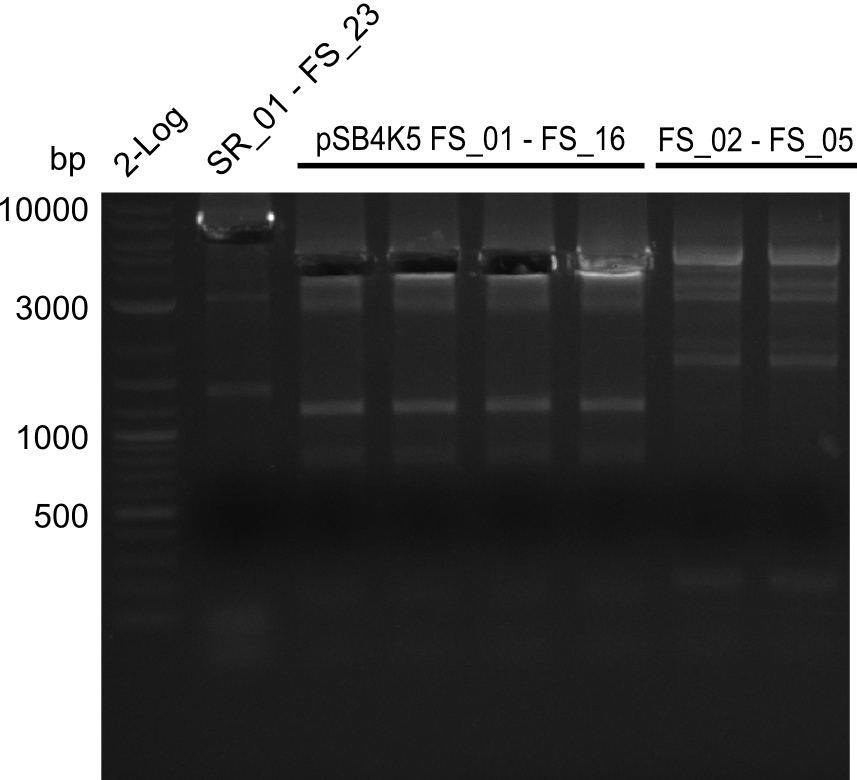
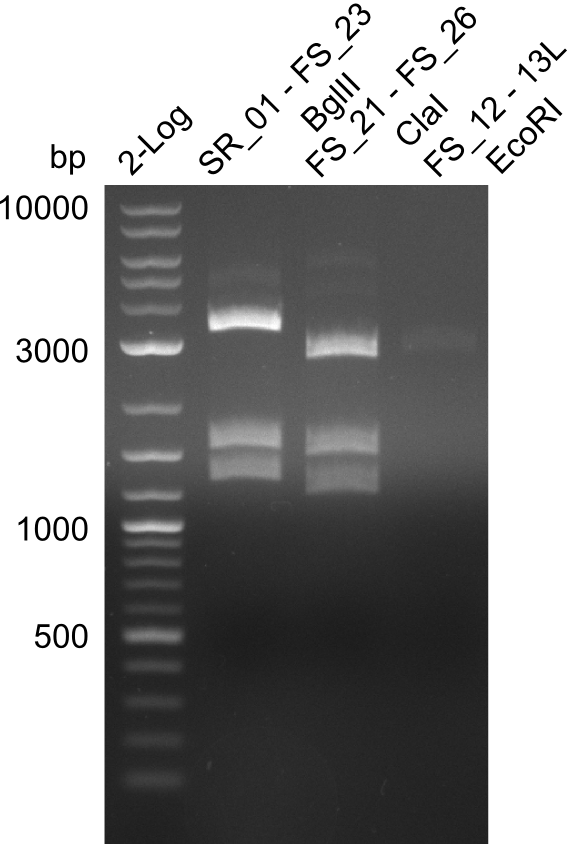
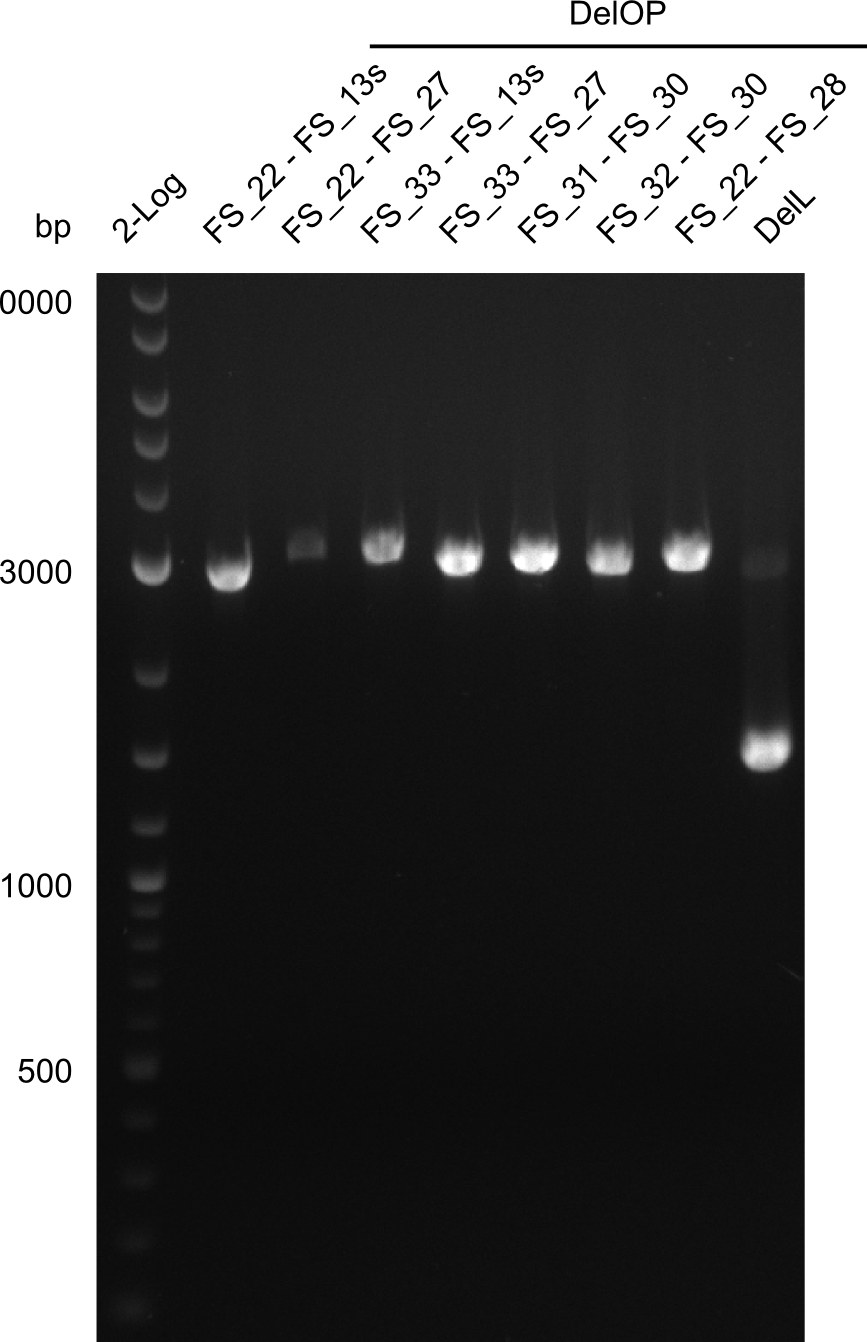
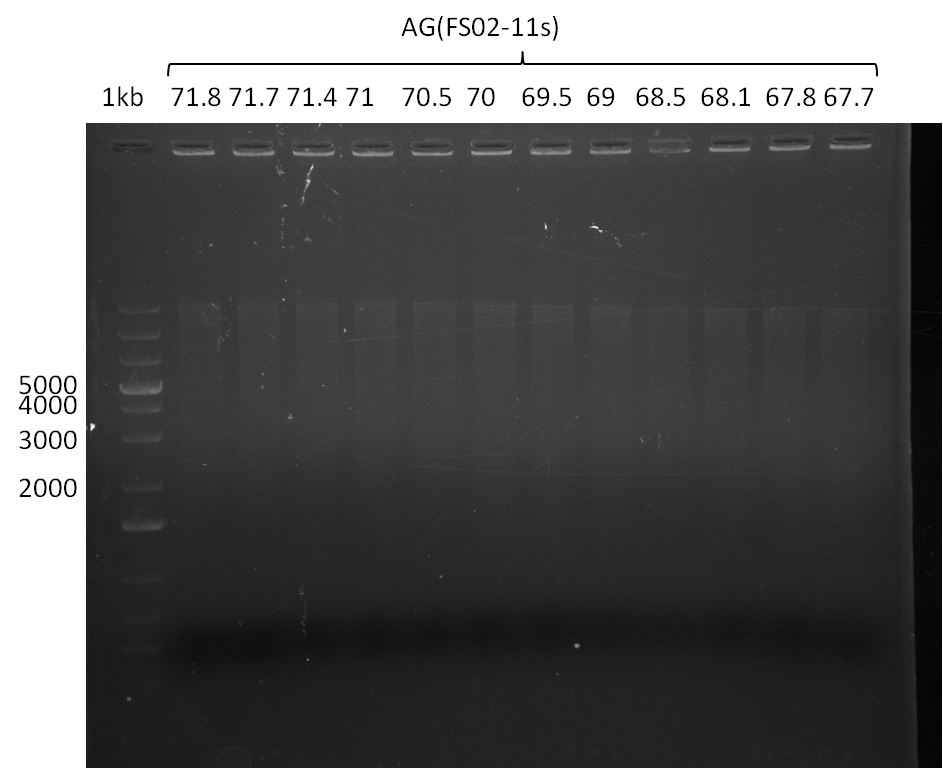
| - | Single read sequencing carried out by GATC | + | Single read sequencing of the PCR amplified fragments DelA-E, DelL as well as the backbone pSB4K5 |
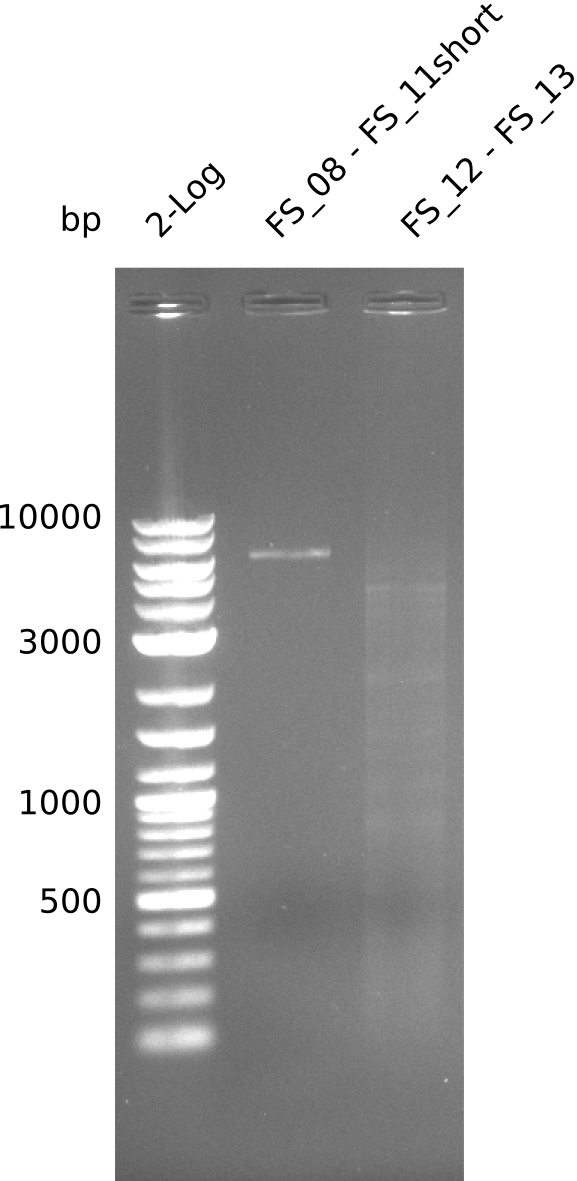
| - | + | was carried out by GATC. We blasted the obtained seqeuences against the reference sequence based on the <i>D. acidovorans SPH-1</i> strain available on NCBI. Although our sequence matched to the reference sequence, we found a significant number of missmatches, which was too diverse and high to be simply explained by random mutations introduced by our polymerase during PCR (note: we used a high-fidelity proofreading polymerase suitable for GC-rich templates and long amplicons). We thus hypothesized that the SPH-1 strain based on which our cloning strategy was designed might have a significant number of single-nucleotide polymorphisms in the Del cluster compared to the <i>D. acidovorans</i> DSM-39 strain, which we used as template for all PCRs (note: there is no complete genomic sequence available for the DSM-39 strain on NCBI). We further hypothesized, that this difference in sequence between the DSM-39 strain used as PCR template and the SPH-1 strain based on which our primers were designed could explain our troubles with the PCR amplifications of DelF-G and DelO-P. In consequence, we ordered the SPH-1 strain from the DSMZ in order to obtain a suitable template for our PCRs. This solved all our PCR problems right away and we were able to get all amplicons required for cloning the DelRest construct within this week. Furthermore, we successfully validated our amplicons by restriction digest and sequencing. | |
</p> | </p> | ||
</div> | </div> | ||
| Line 135: | Line 138: | ||
<h1>Week 16</h1> | <h1>Week 16</h1> | ||
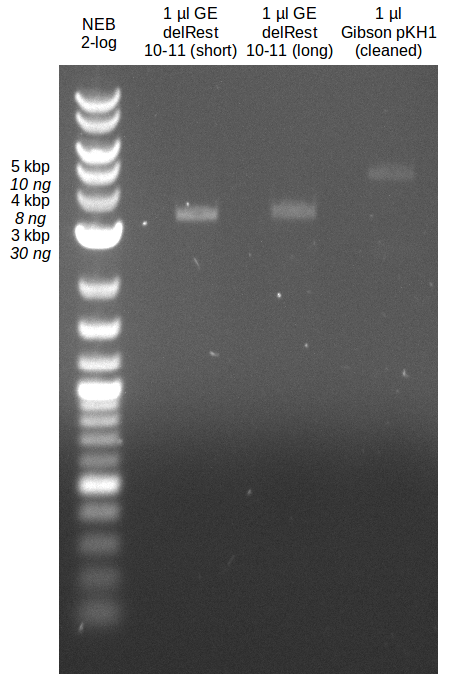
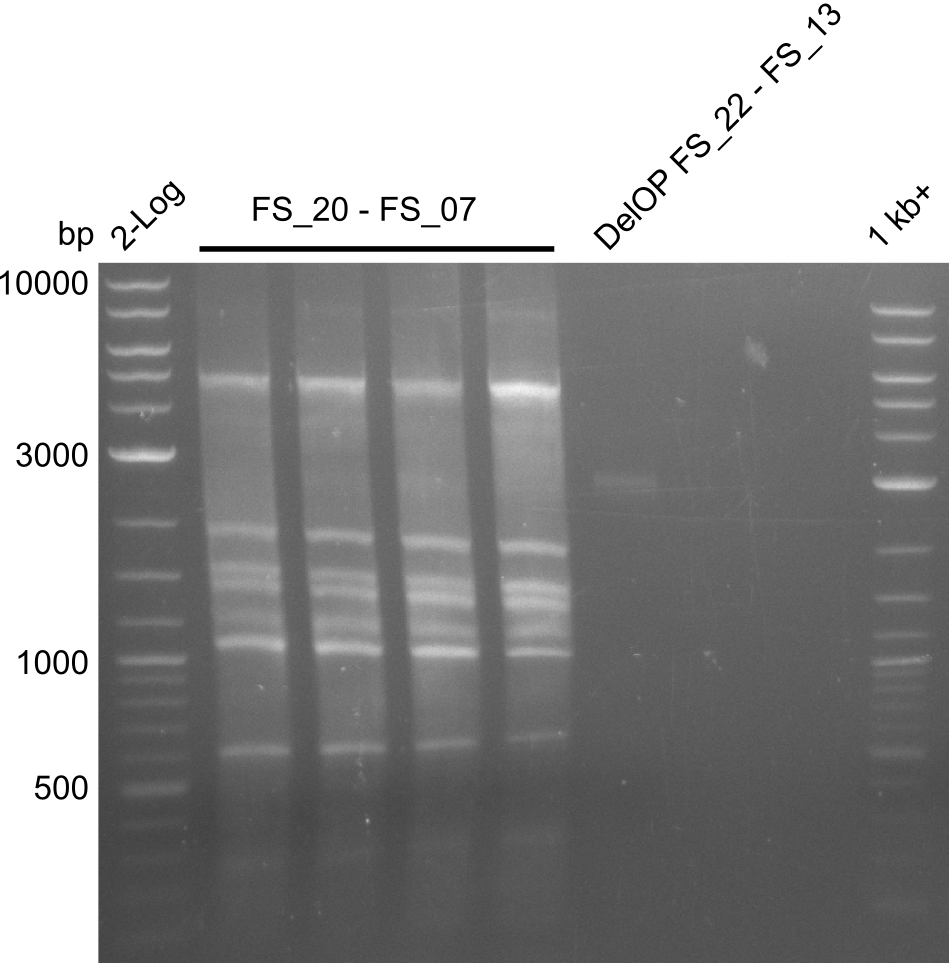
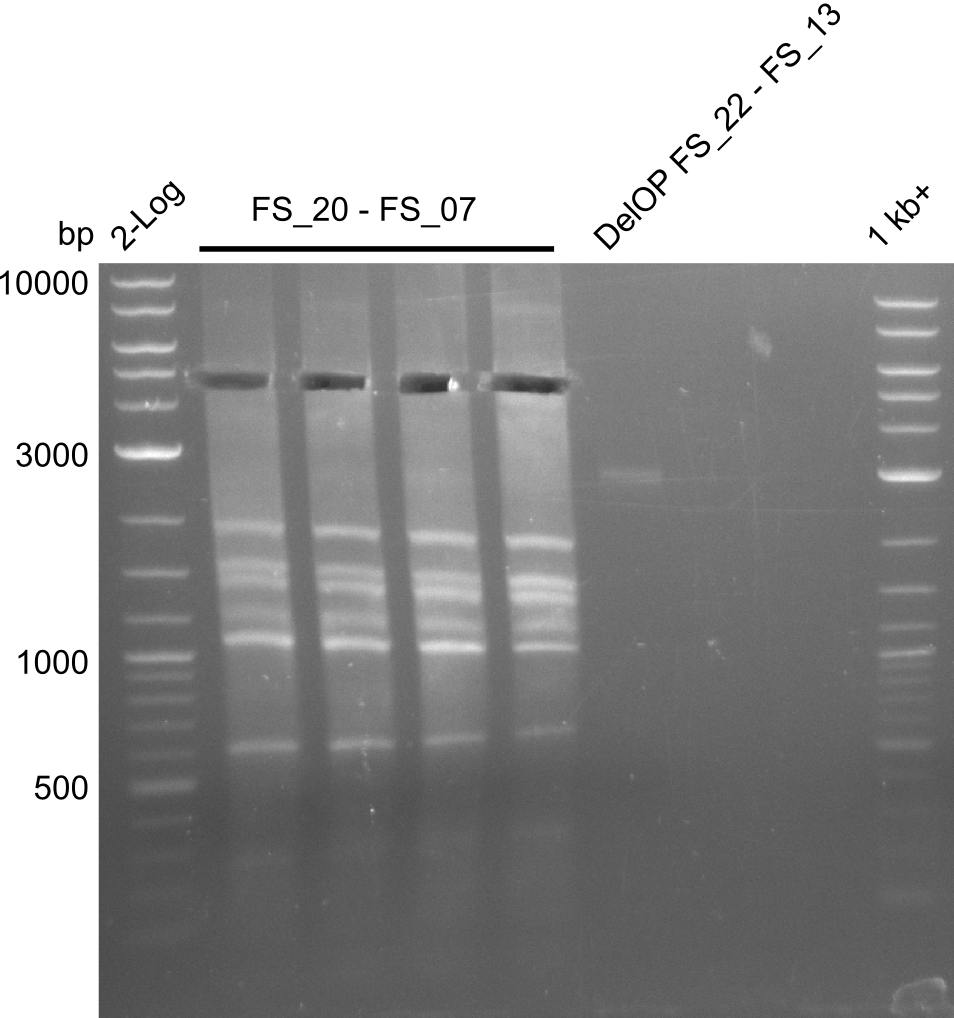
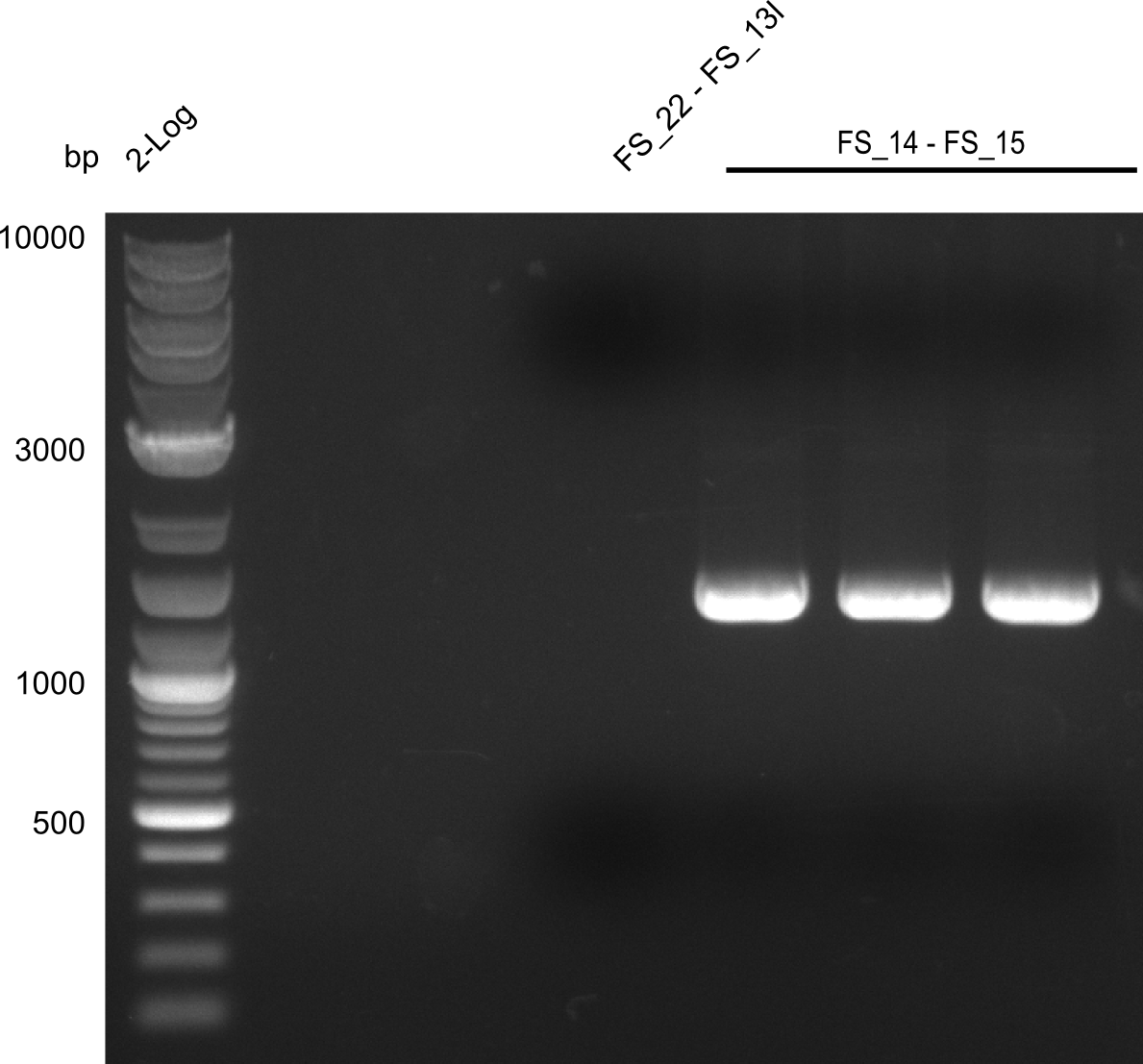
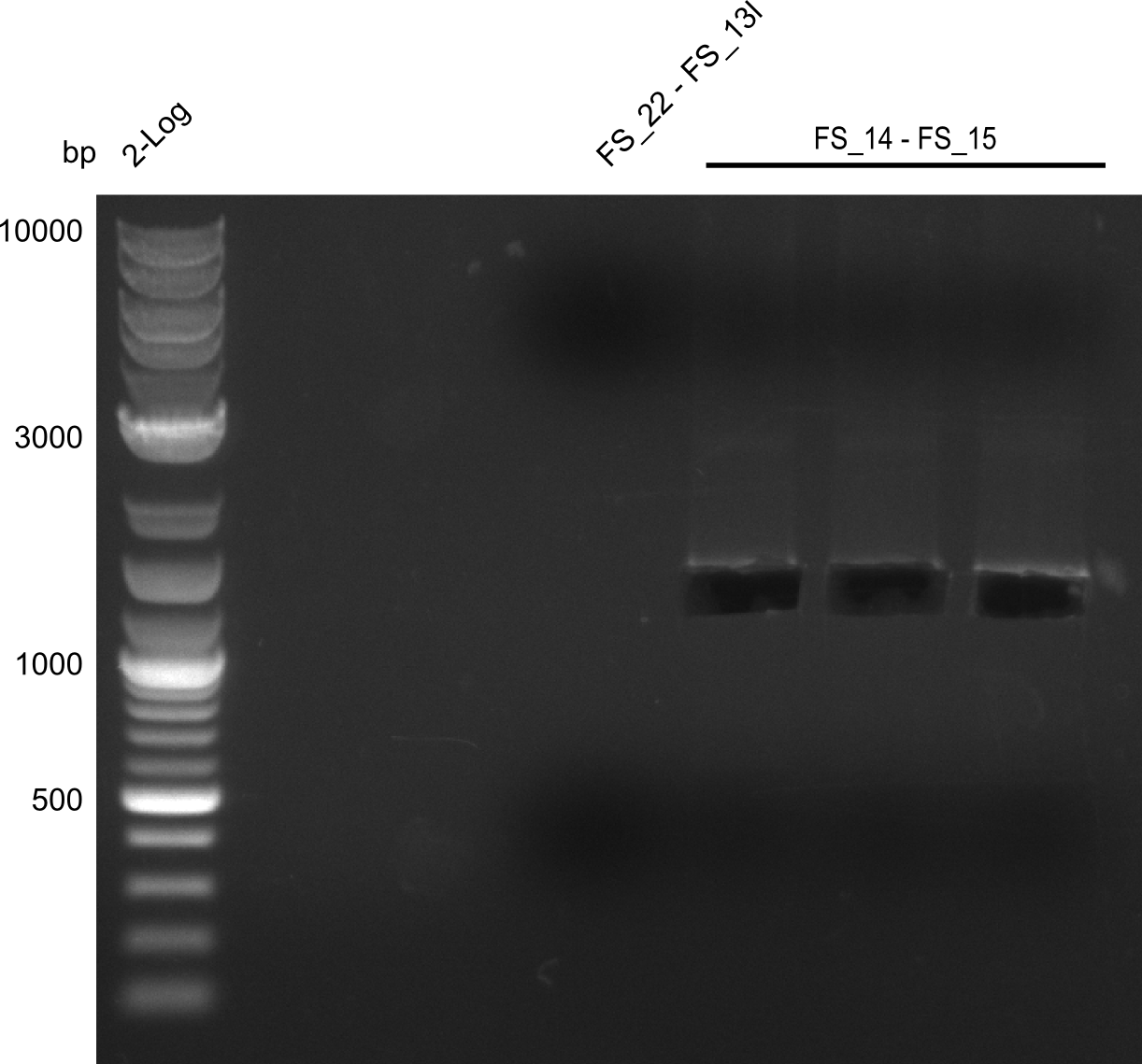
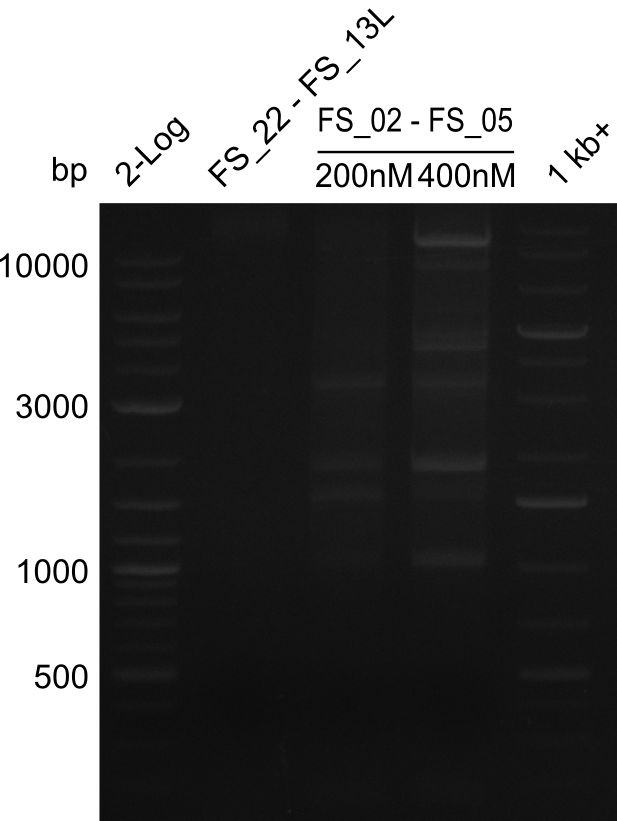
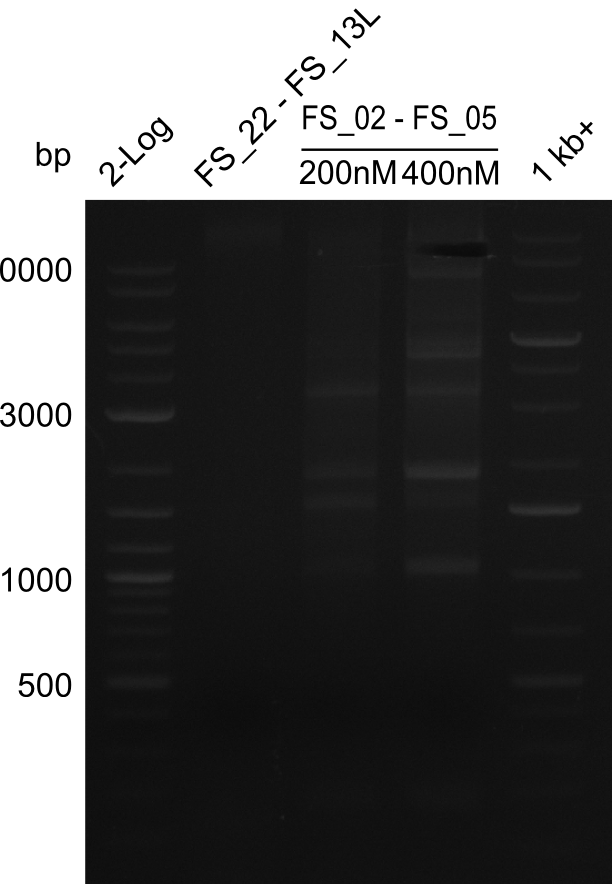
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
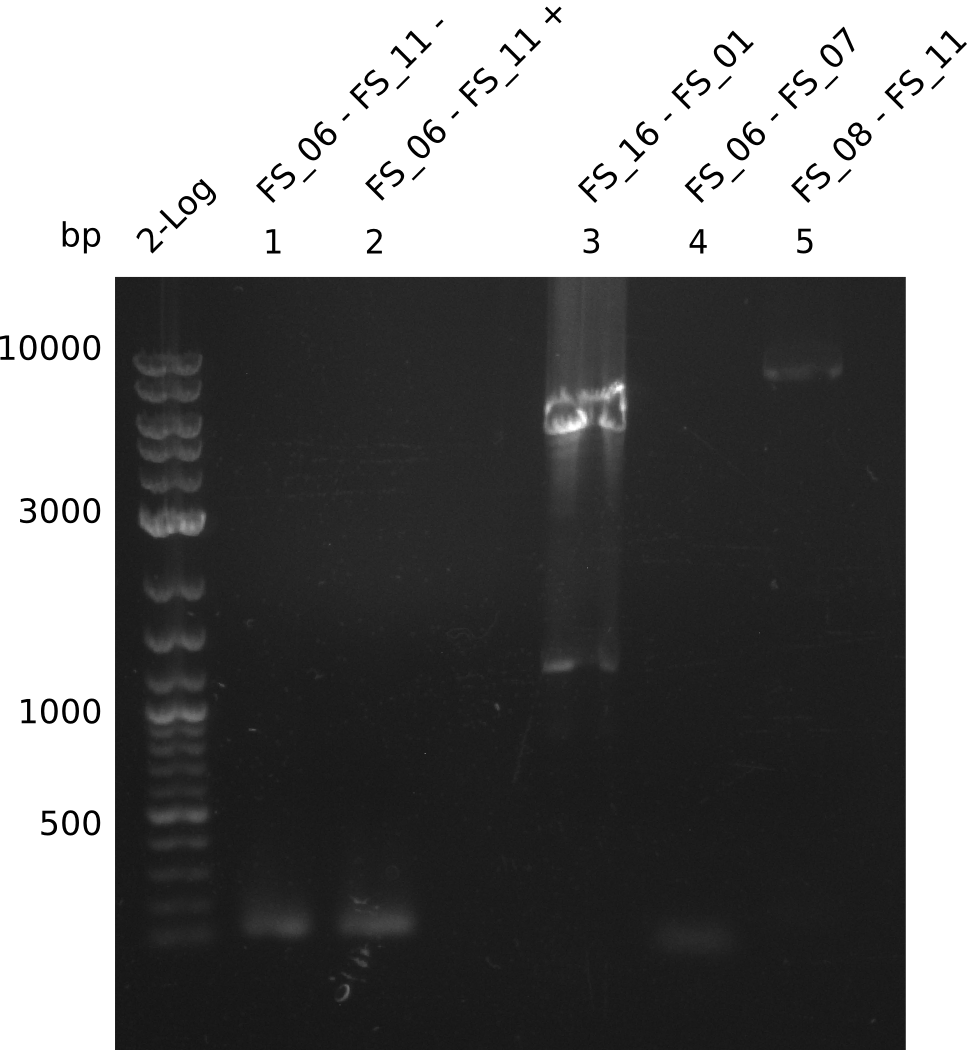
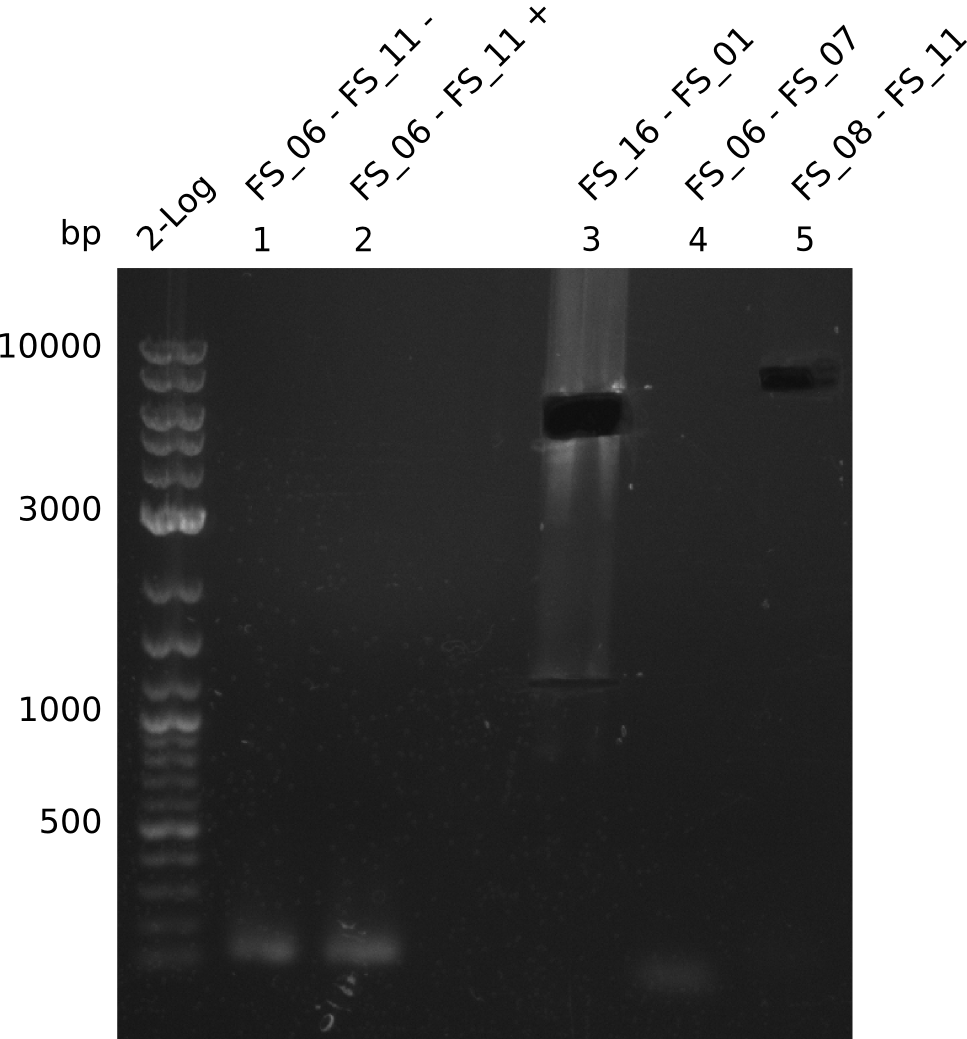
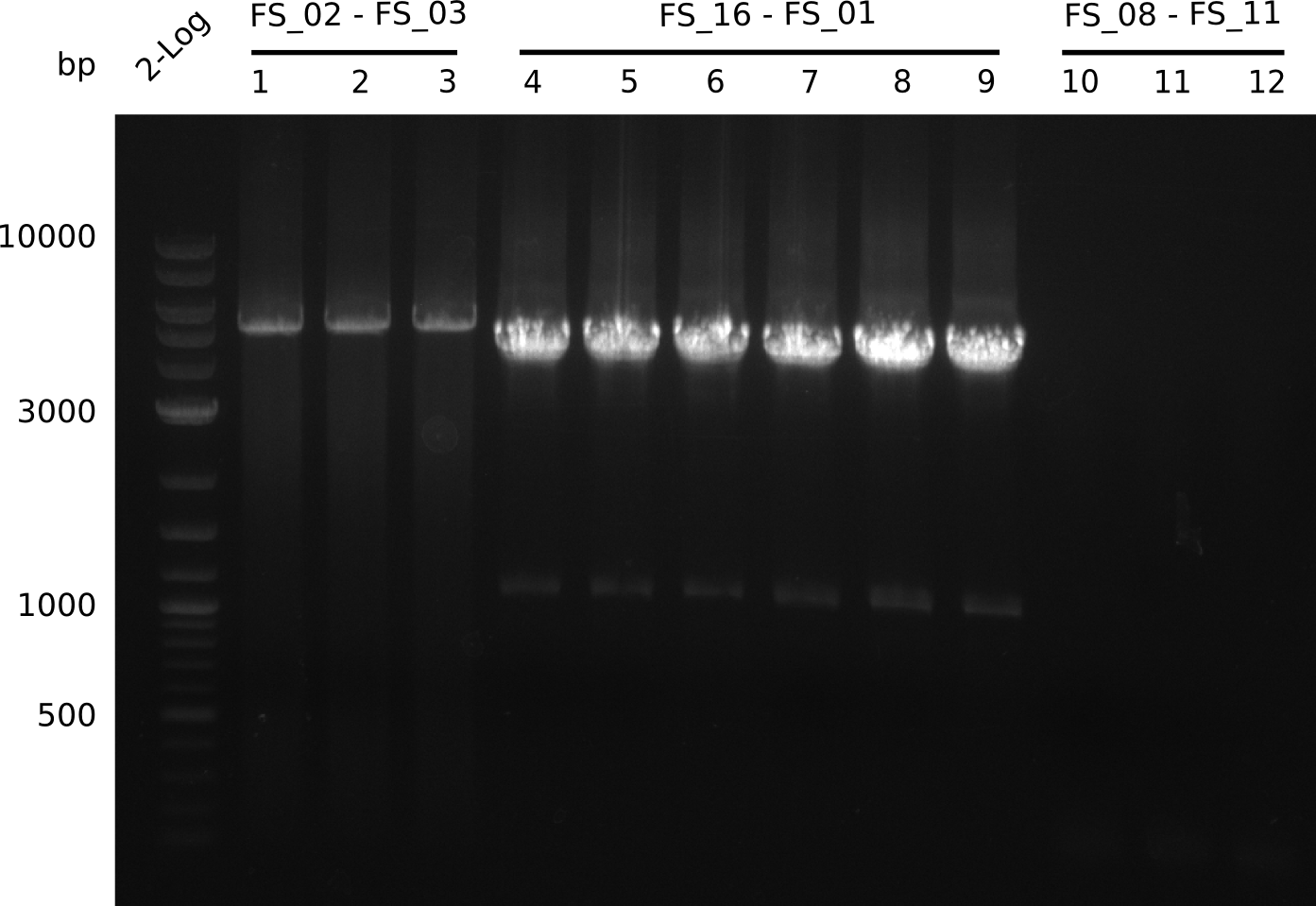
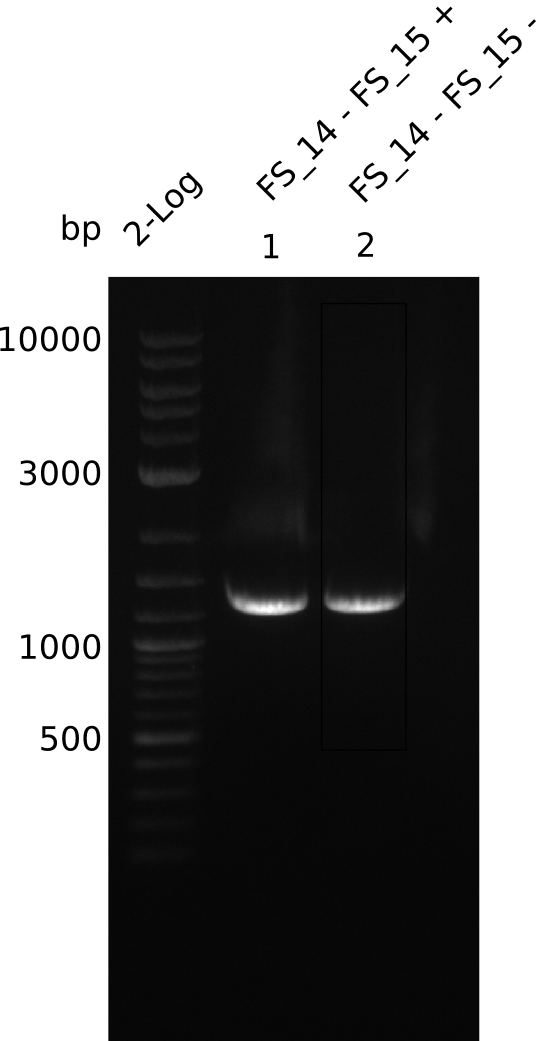
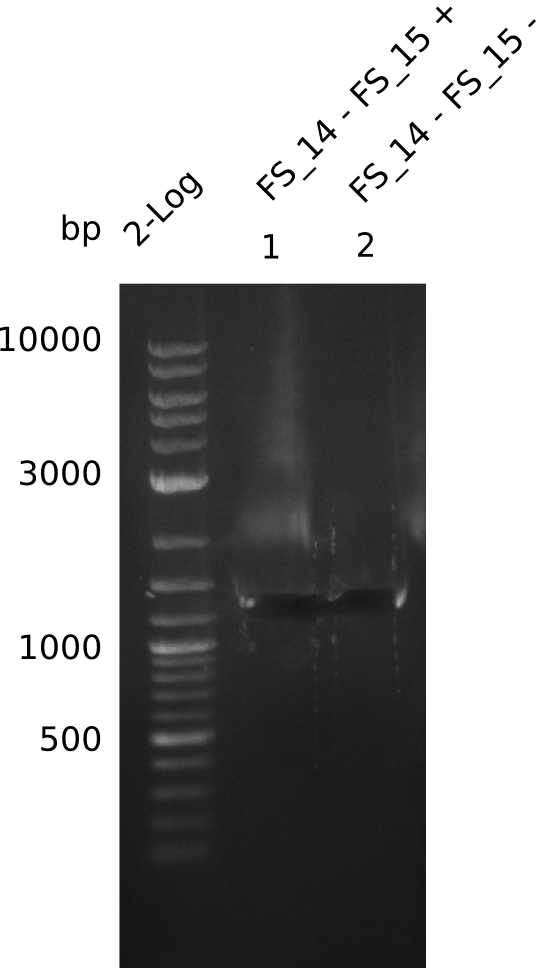
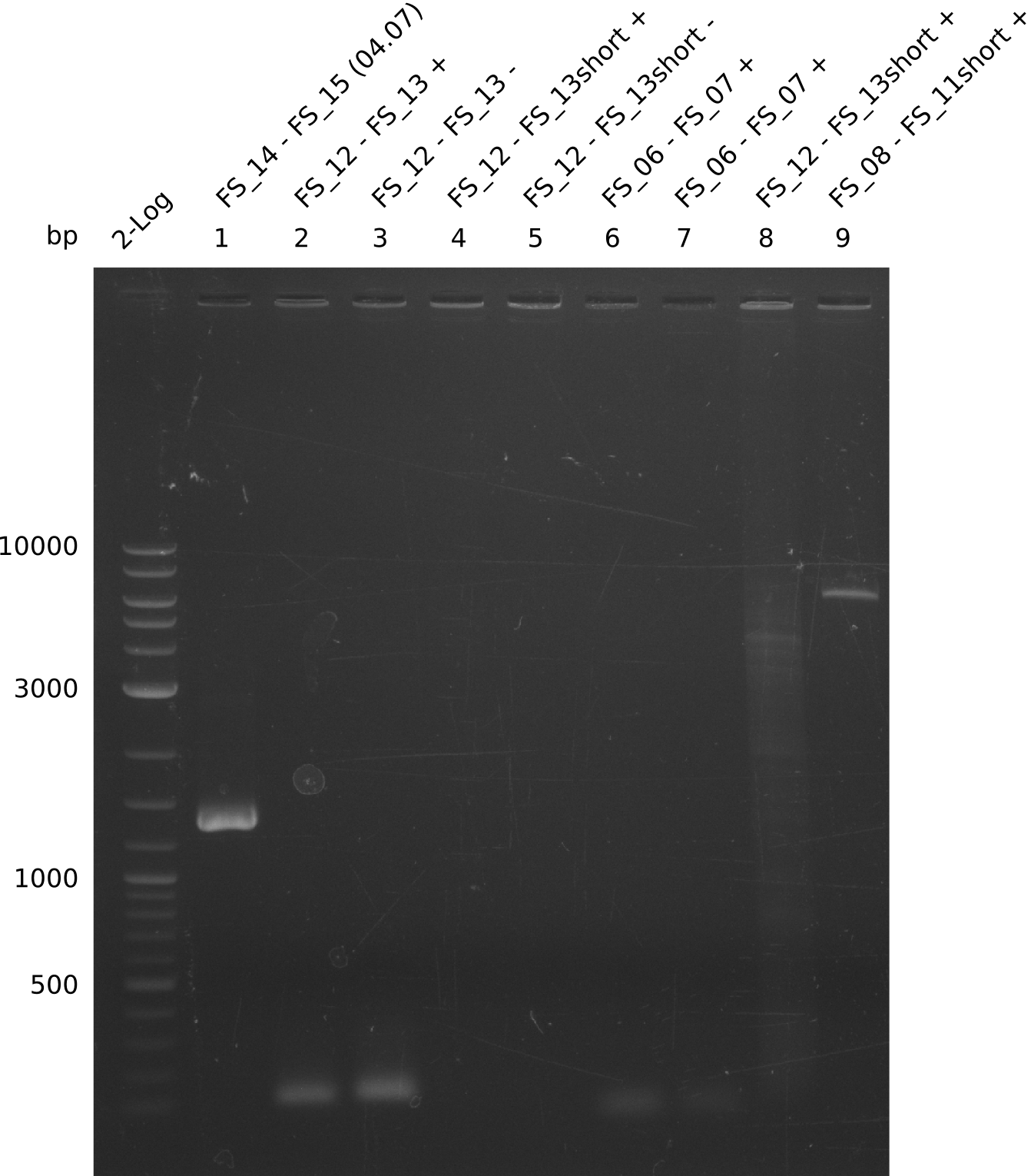
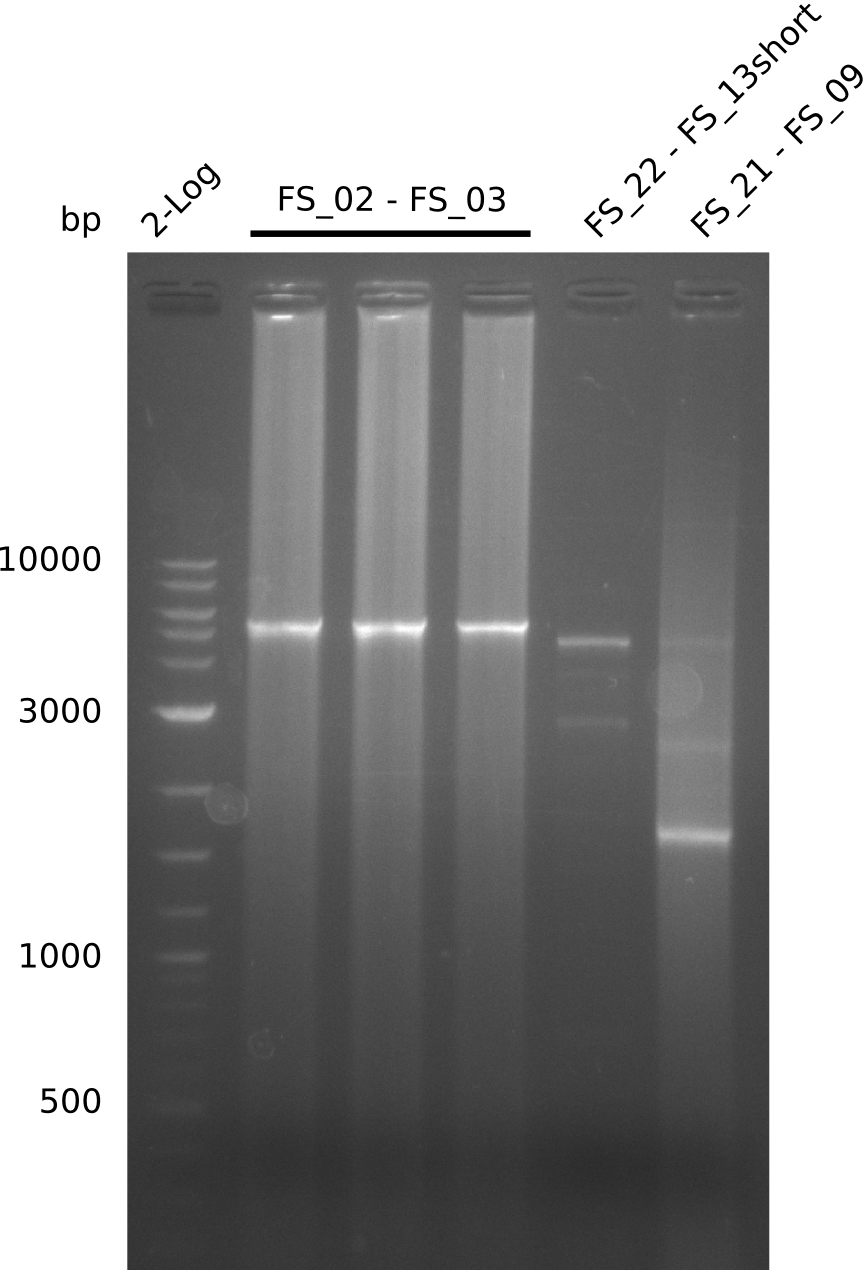
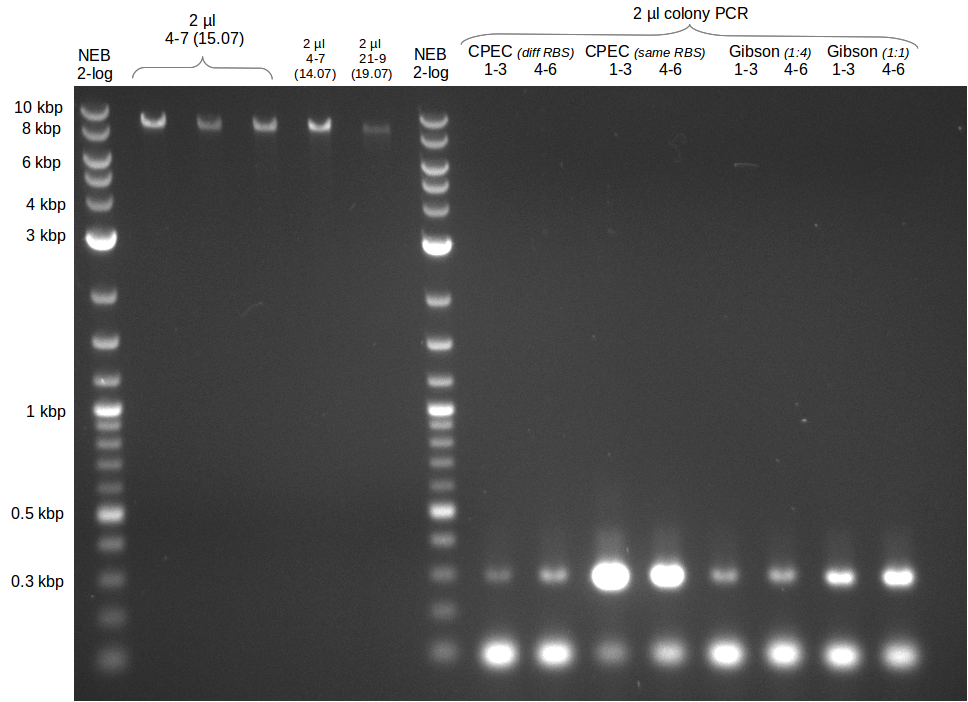
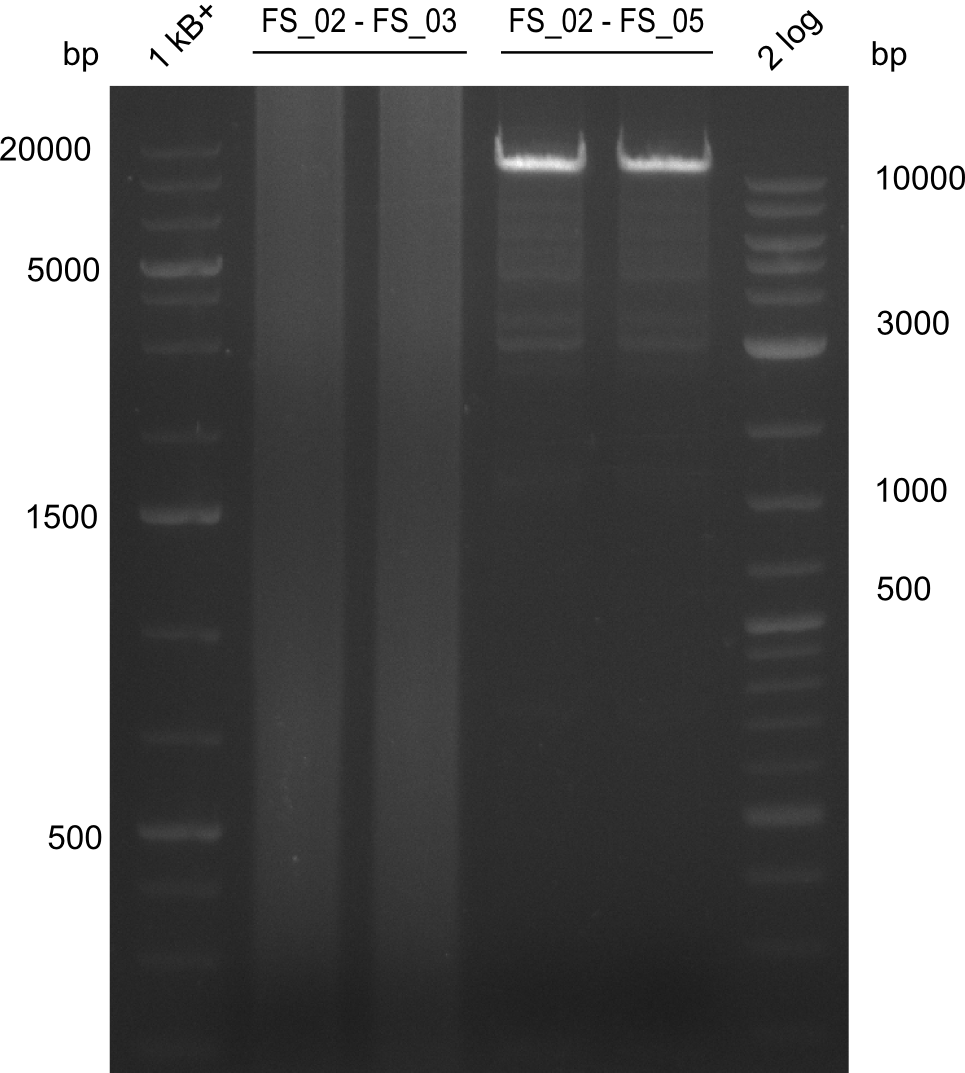
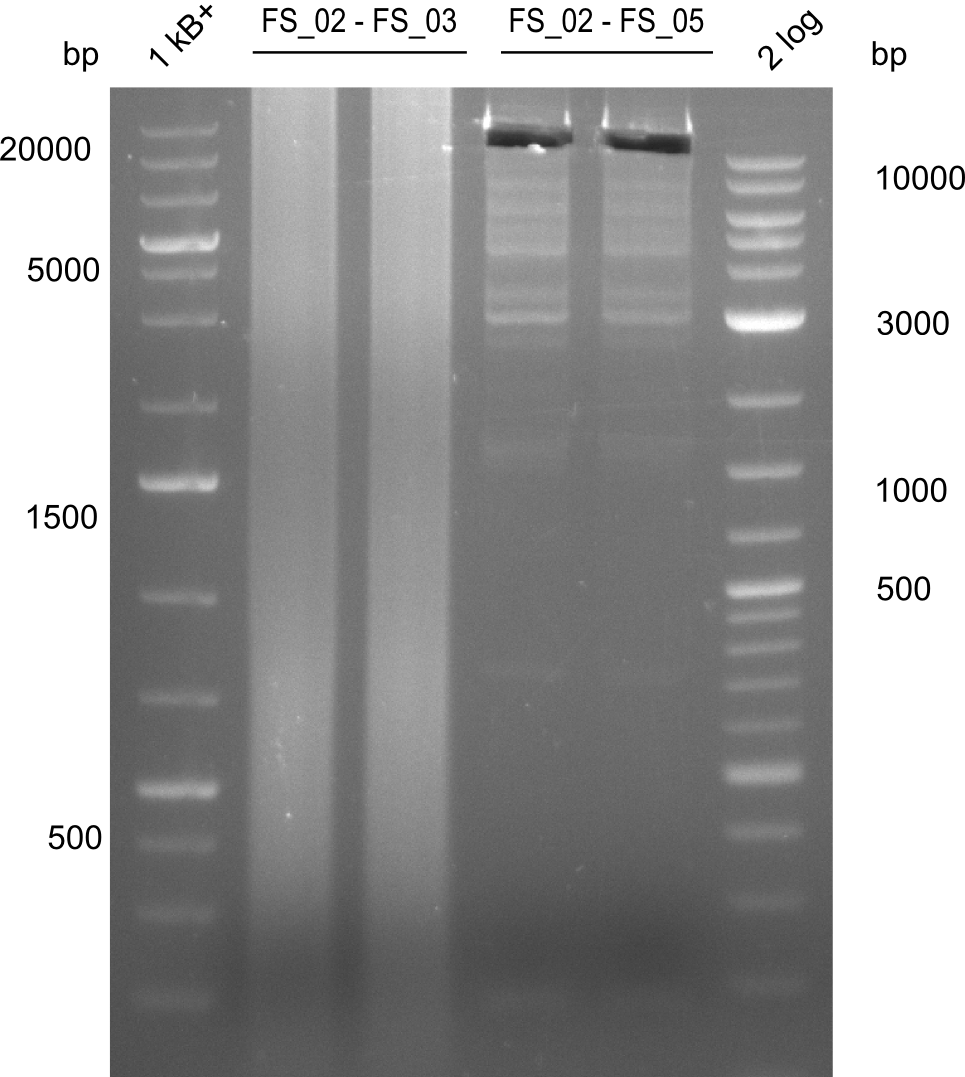
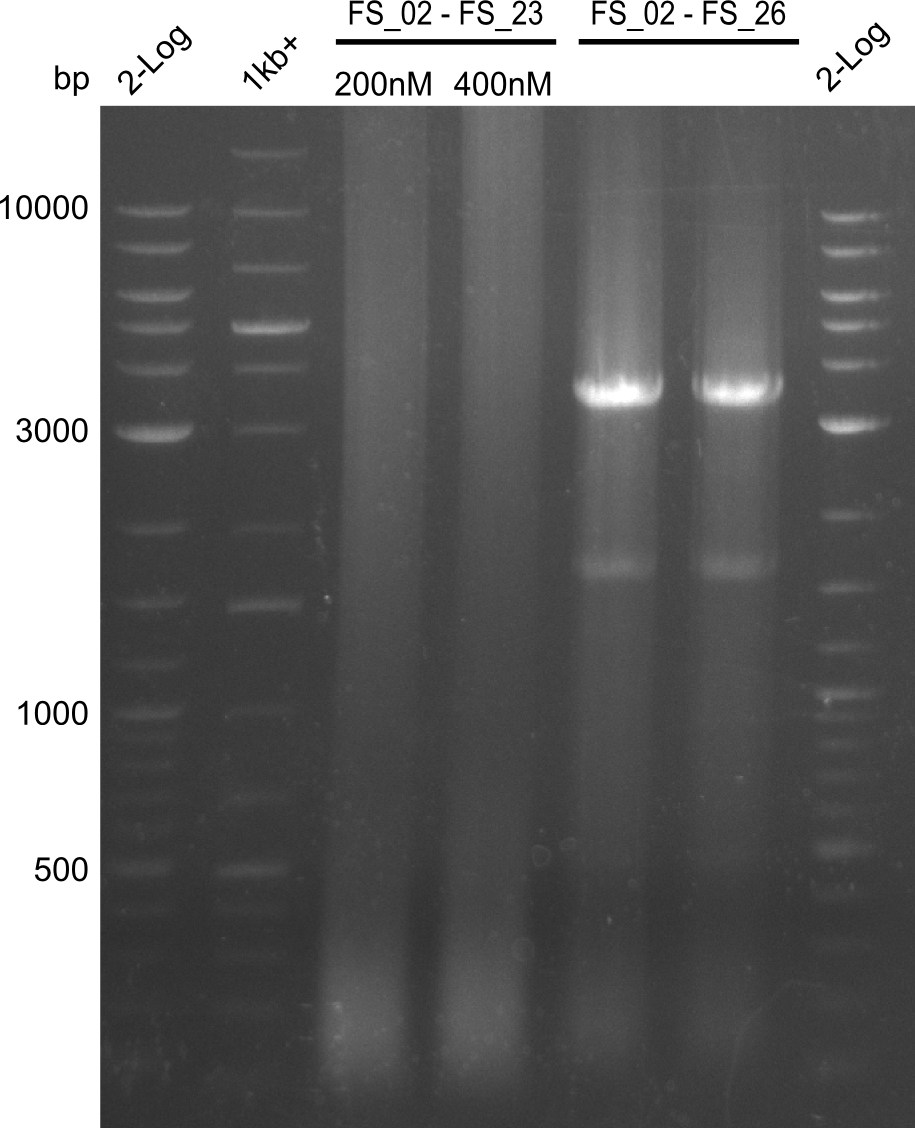
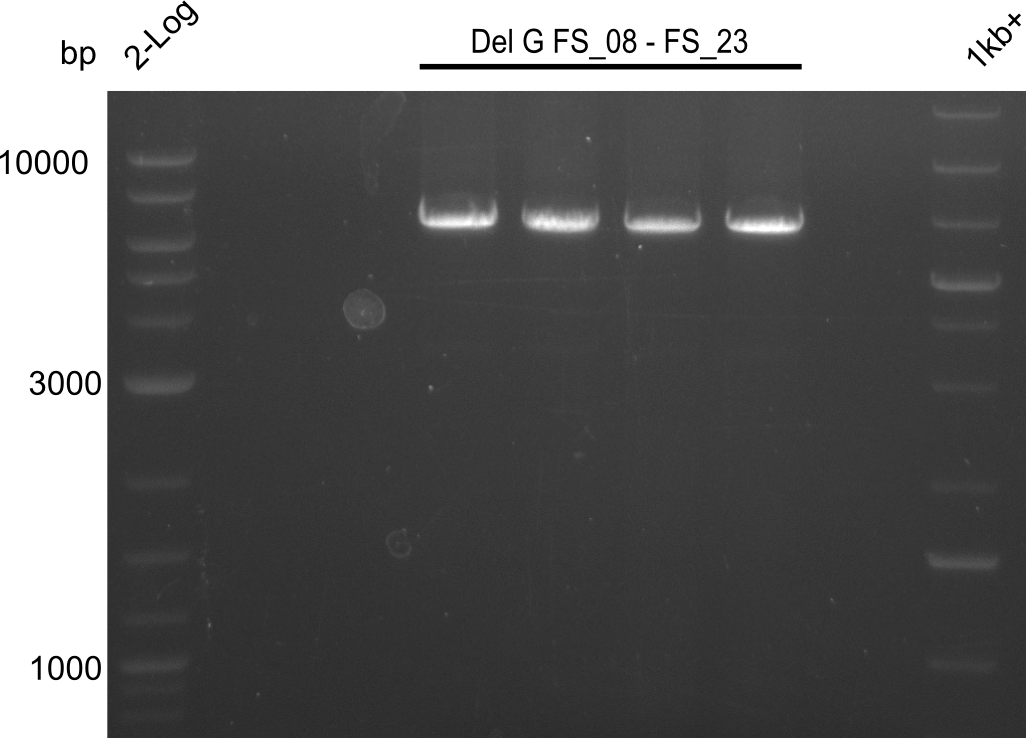
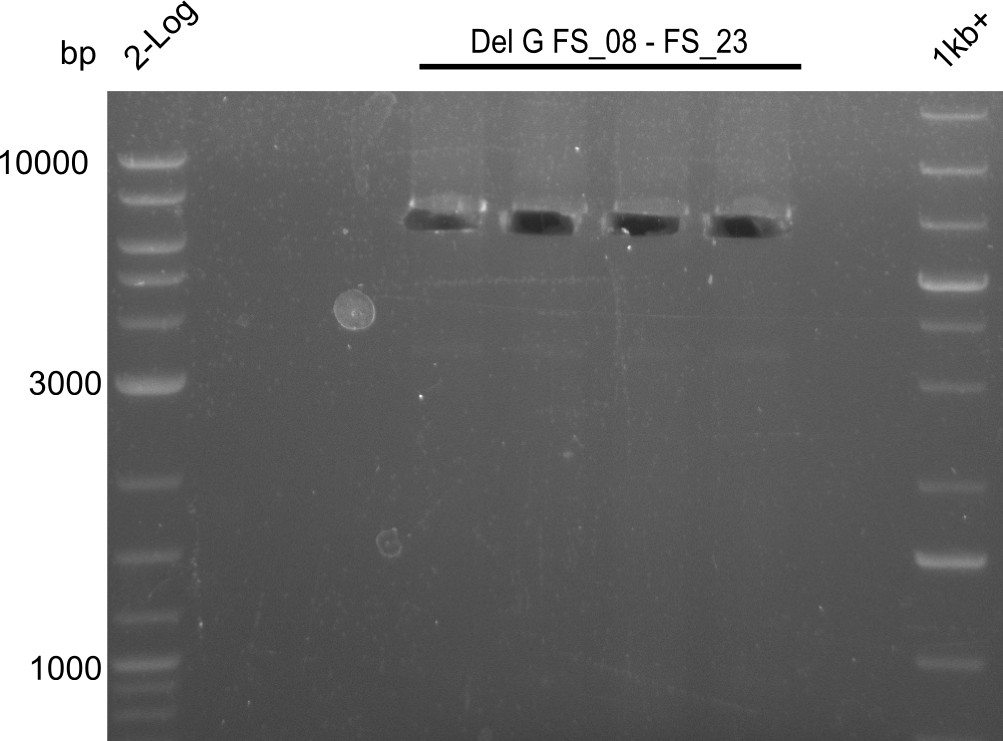
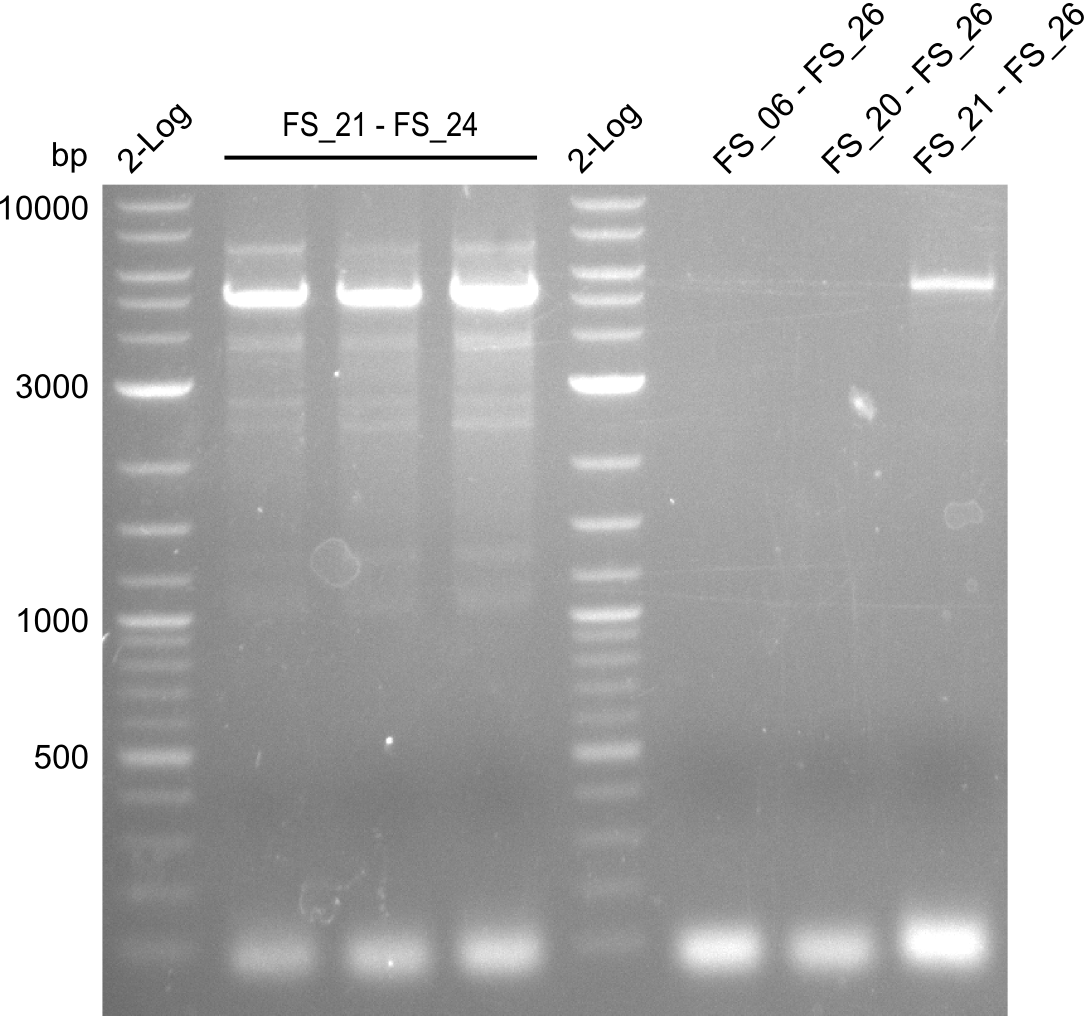
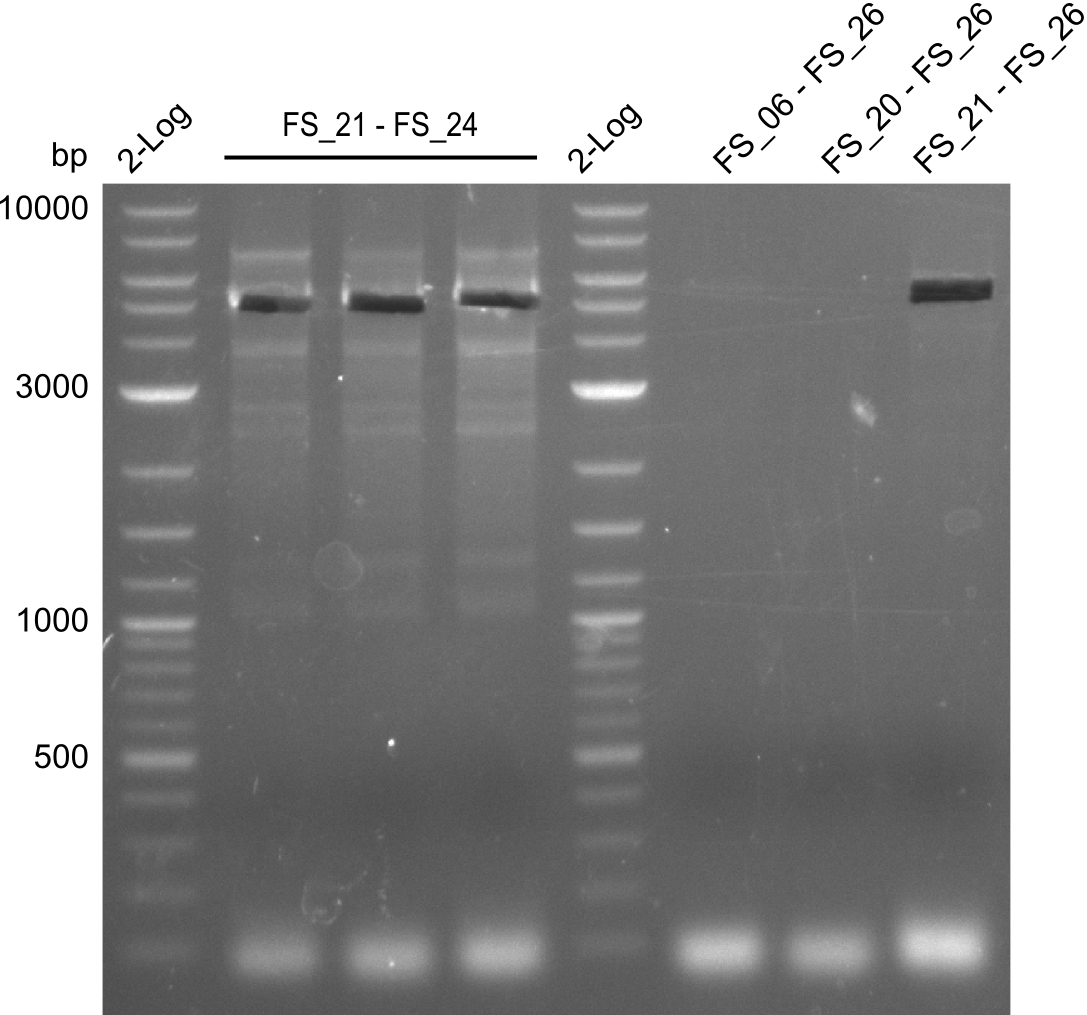
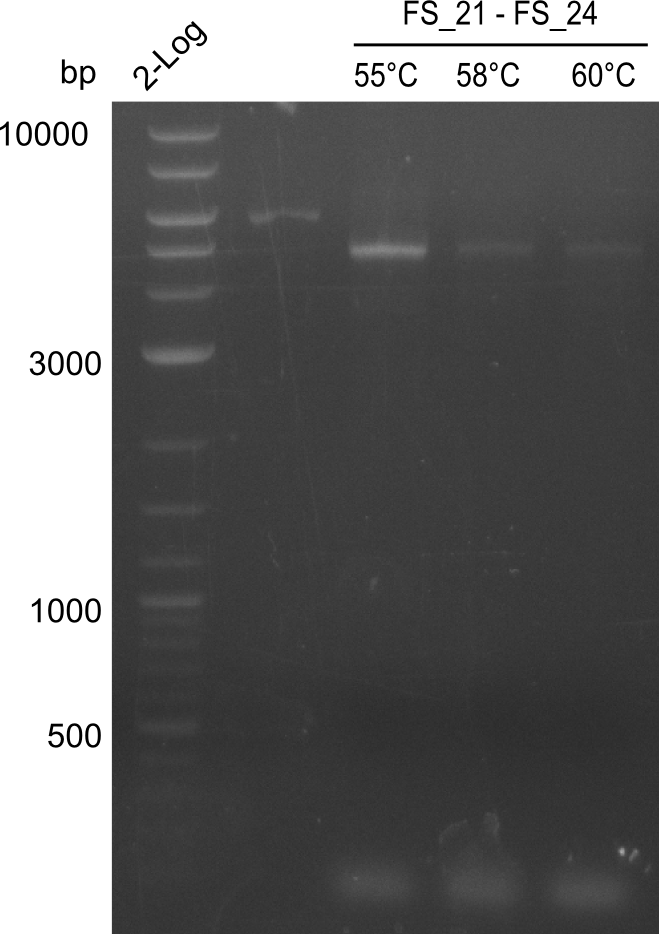
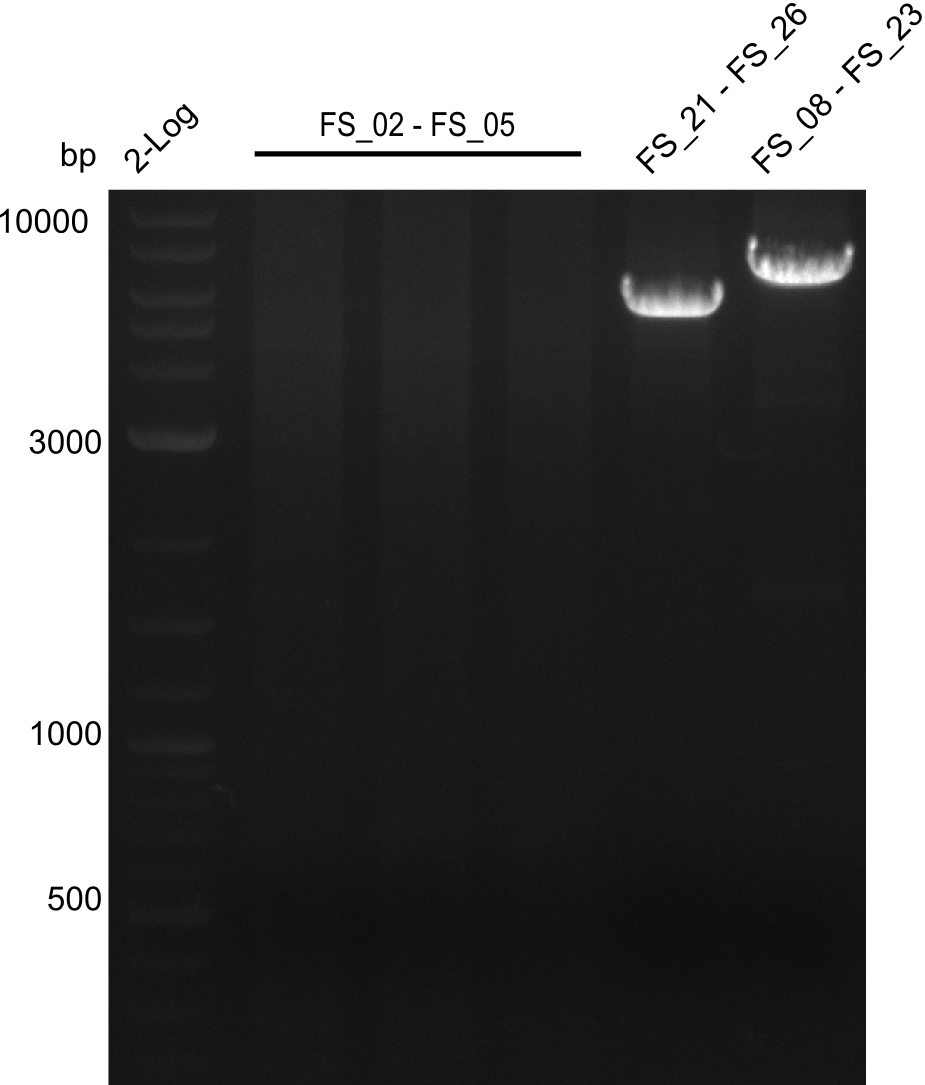
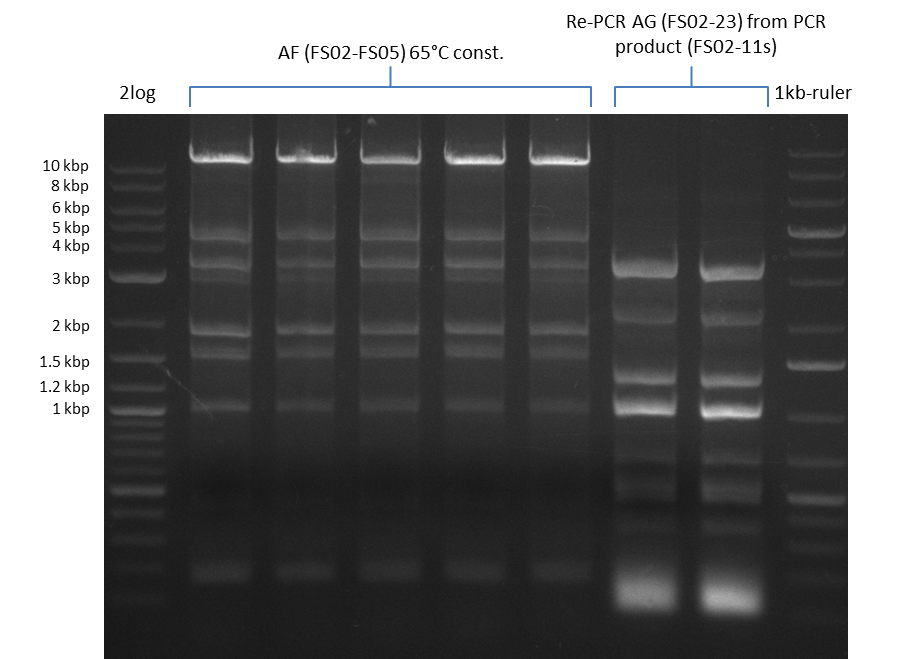
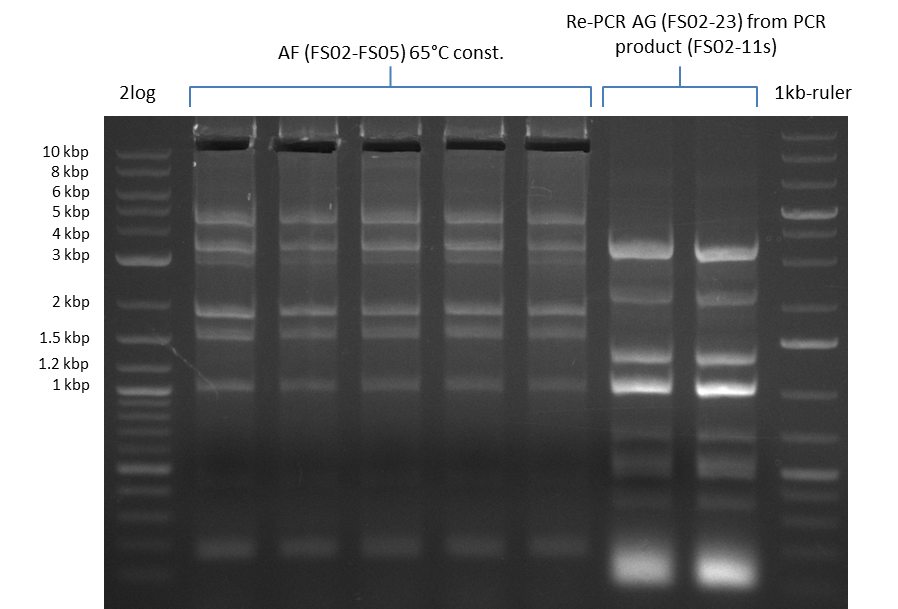
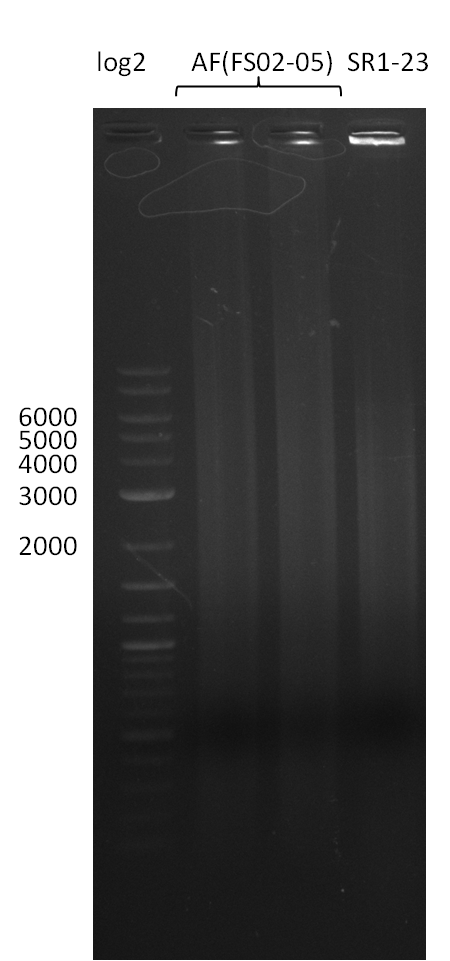
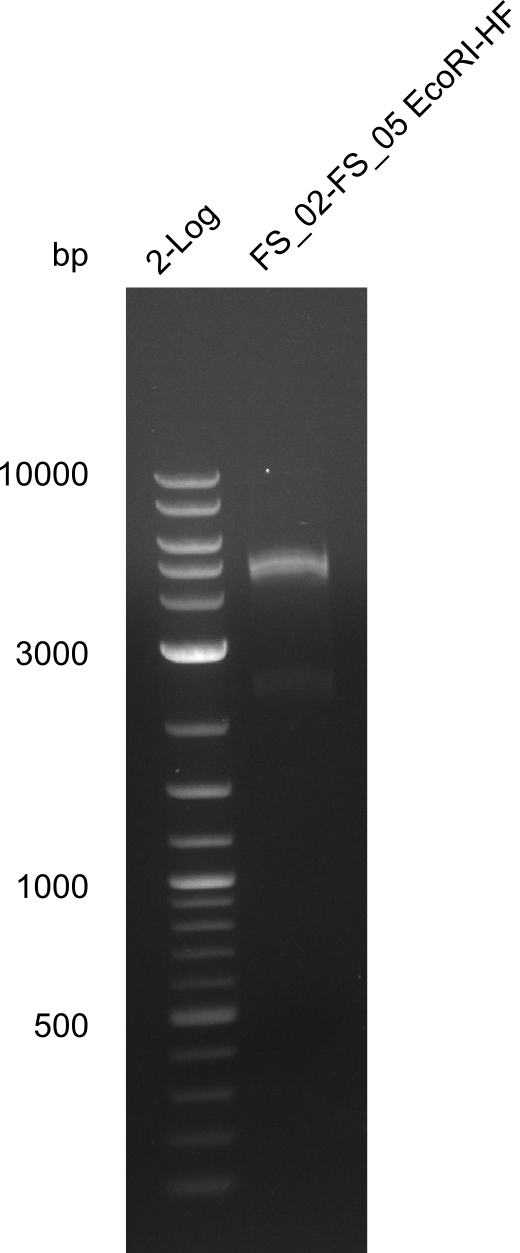
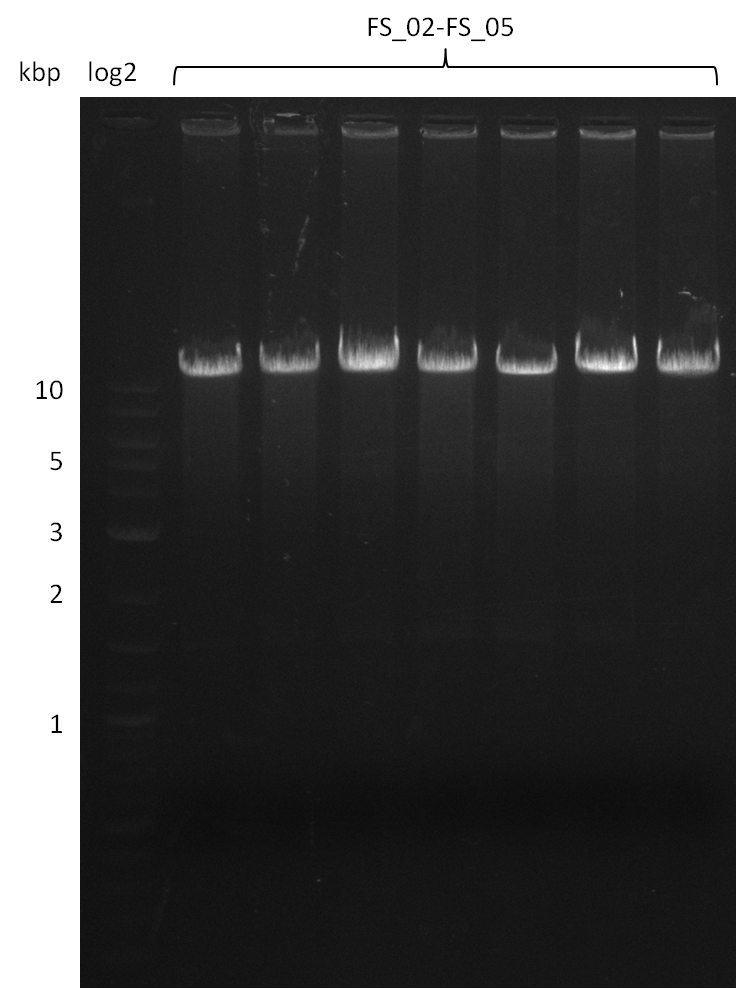
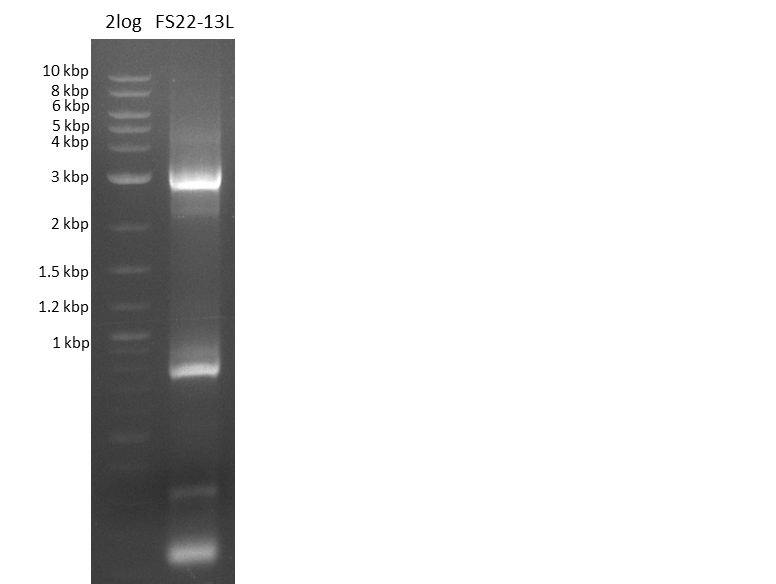
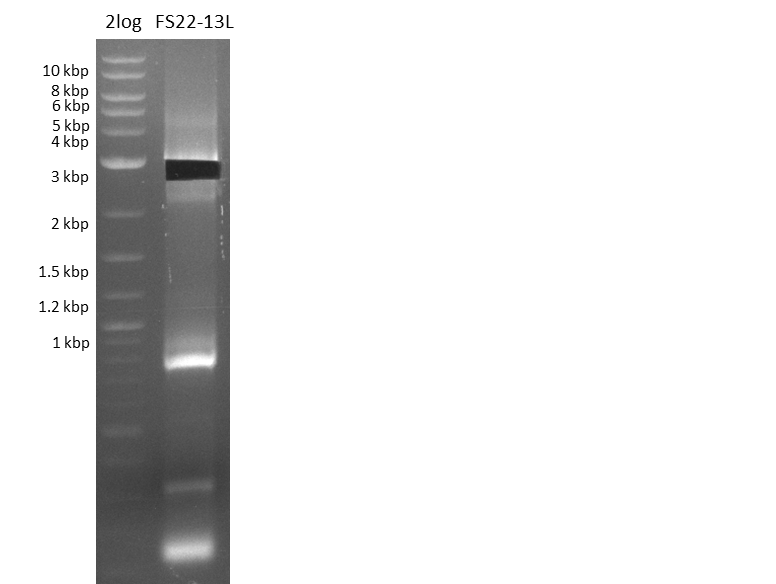
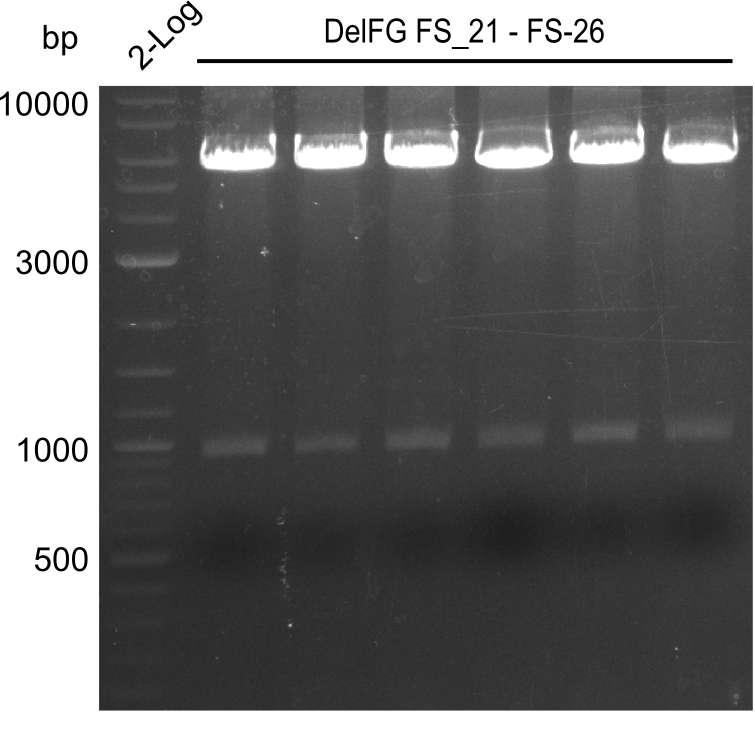
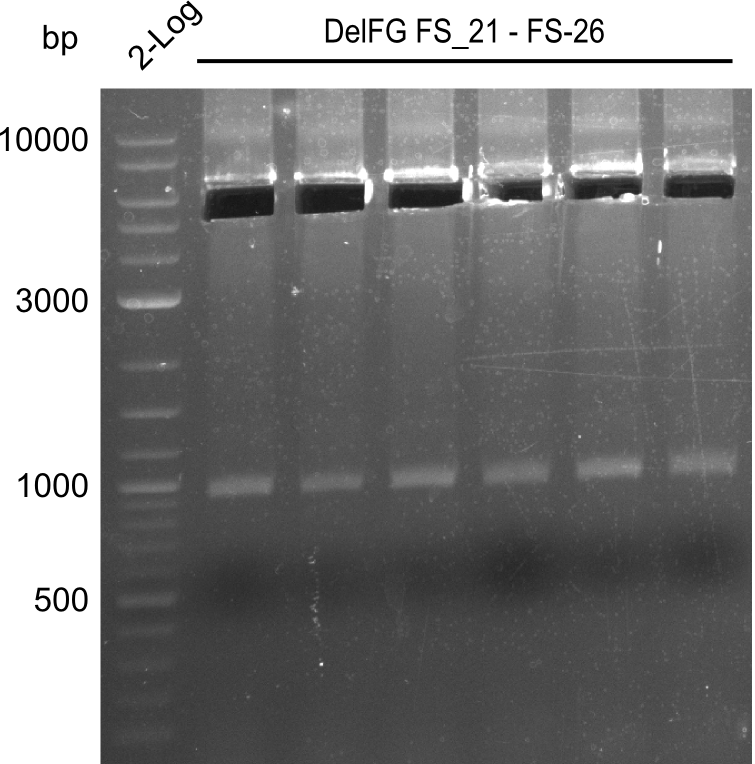
| - | Last week we successfully amplified all the | + | Last week we successfully amplified all the fragments needed to complete the DelRest cloning. Therefore, we went ahaed and performed Gibson assembly in order to assembel the final pFSN plasmid carrying bearing the DelRest genes. The assembly mix was transformed into <i>E. coli DH10beta</i> electrocompetent cells. Screening PCRs showed that the assembly was successful, calling for futher validation of the clones. |
</p> | </p> | ||
</div> | </div> | ||
| Line 146: | Line 149: | ||
<h1>Week 17</h1> | <h1>Week 17</h1> | ||
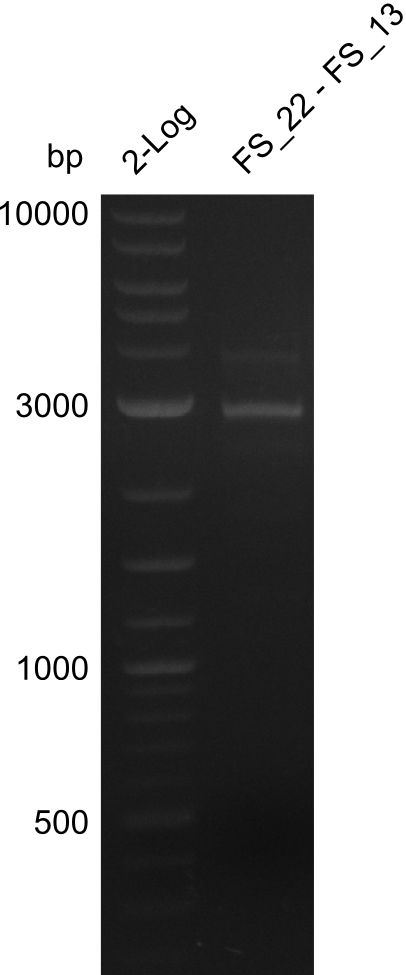
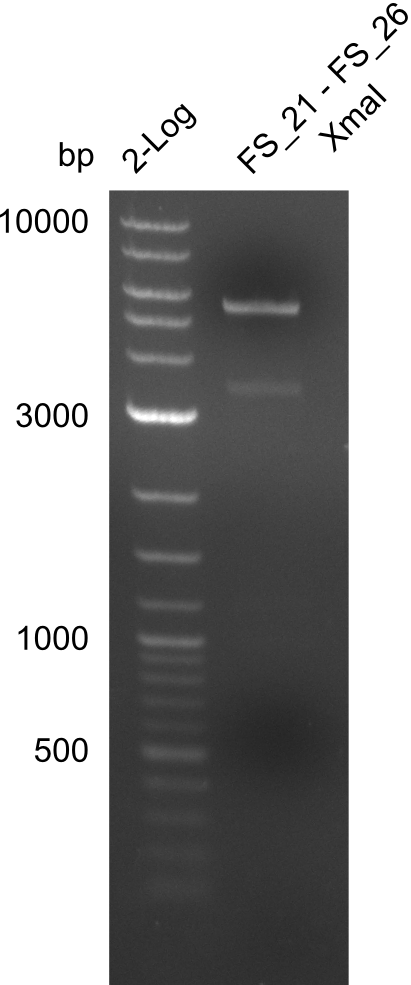
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
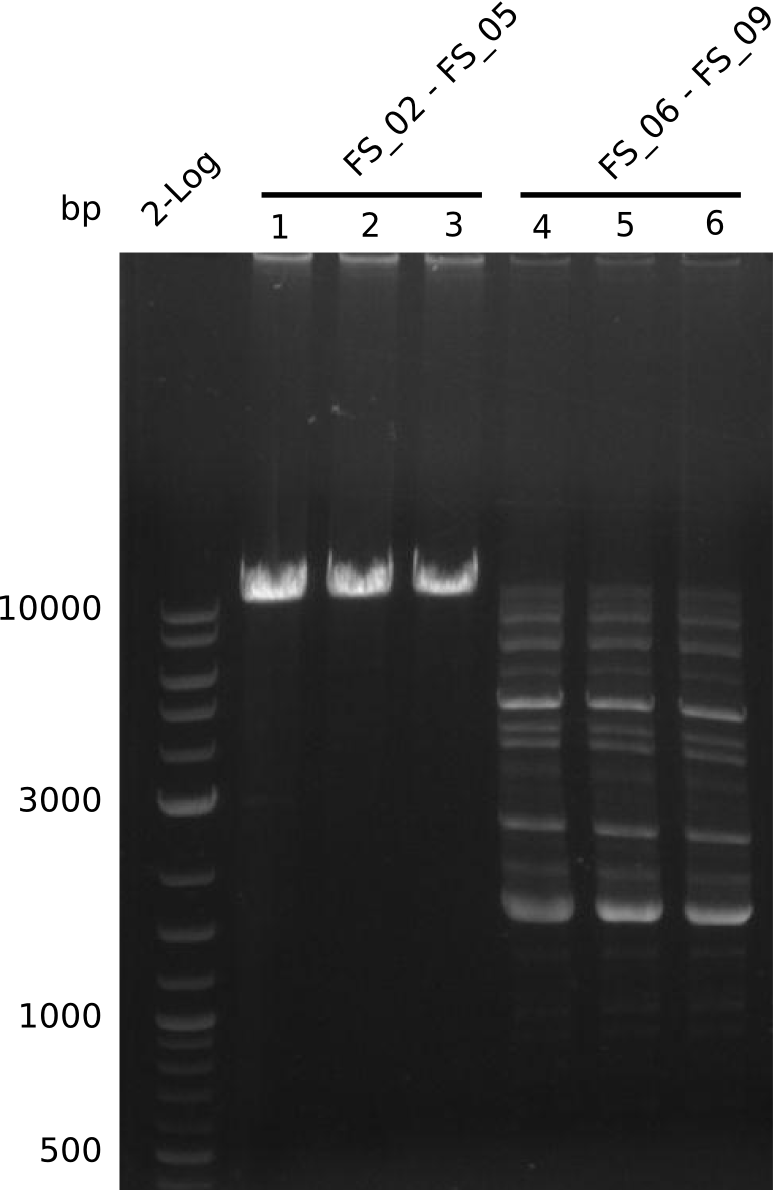
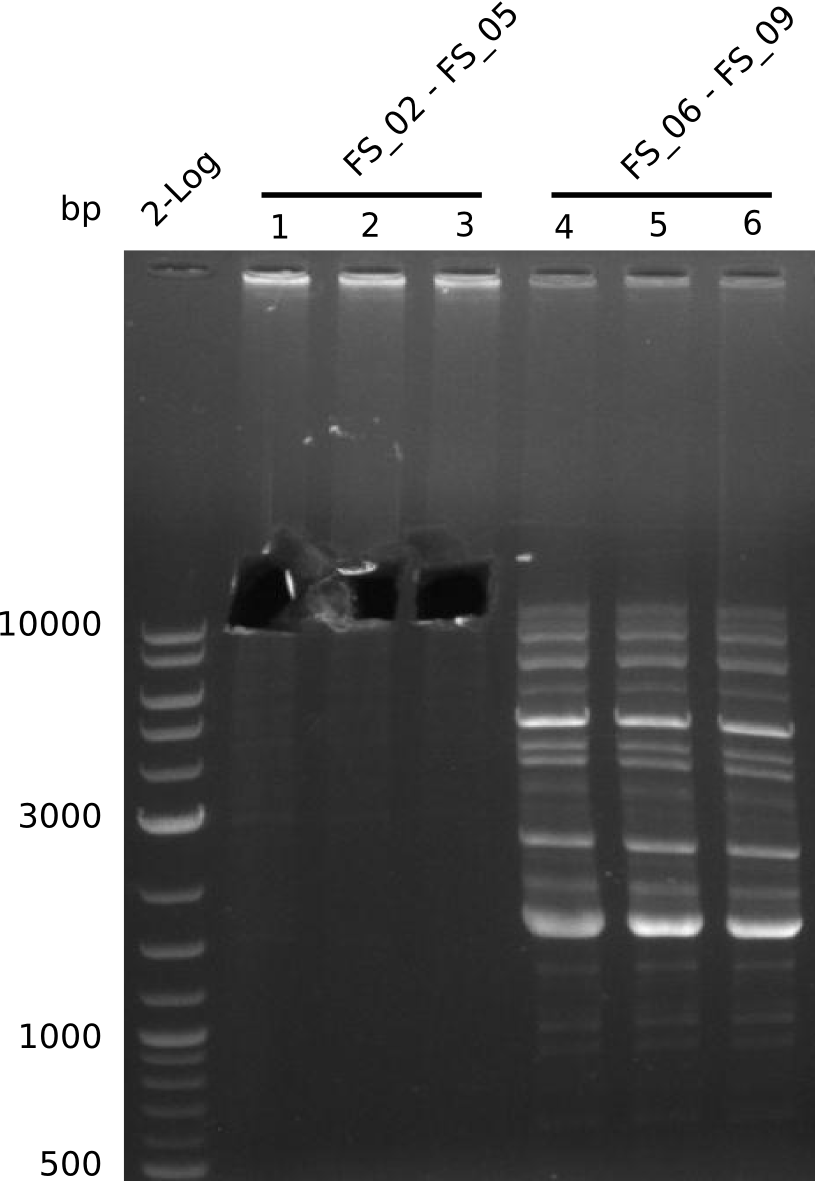
| - | + | From colonies which were positive for screening last week, we rescued 3 plasmids. Test restriction digest were conducted and showed our clones to be correct. | |
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 160: | Line 160: | ||
<h1>Week 18</h1> | <h1>Week 18</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
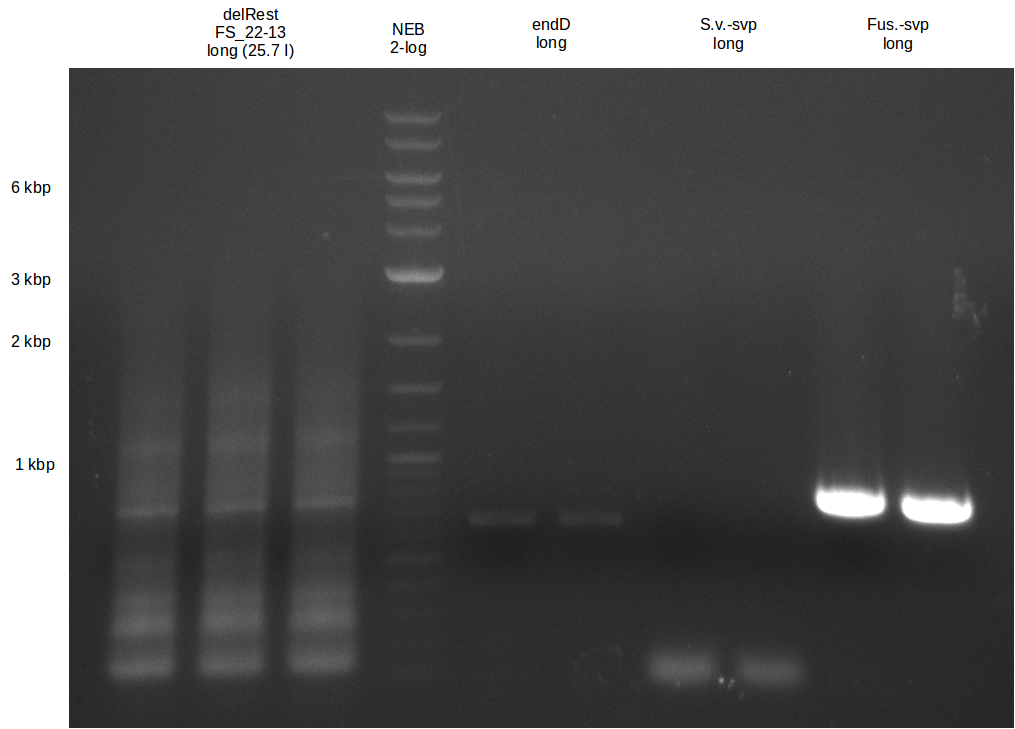
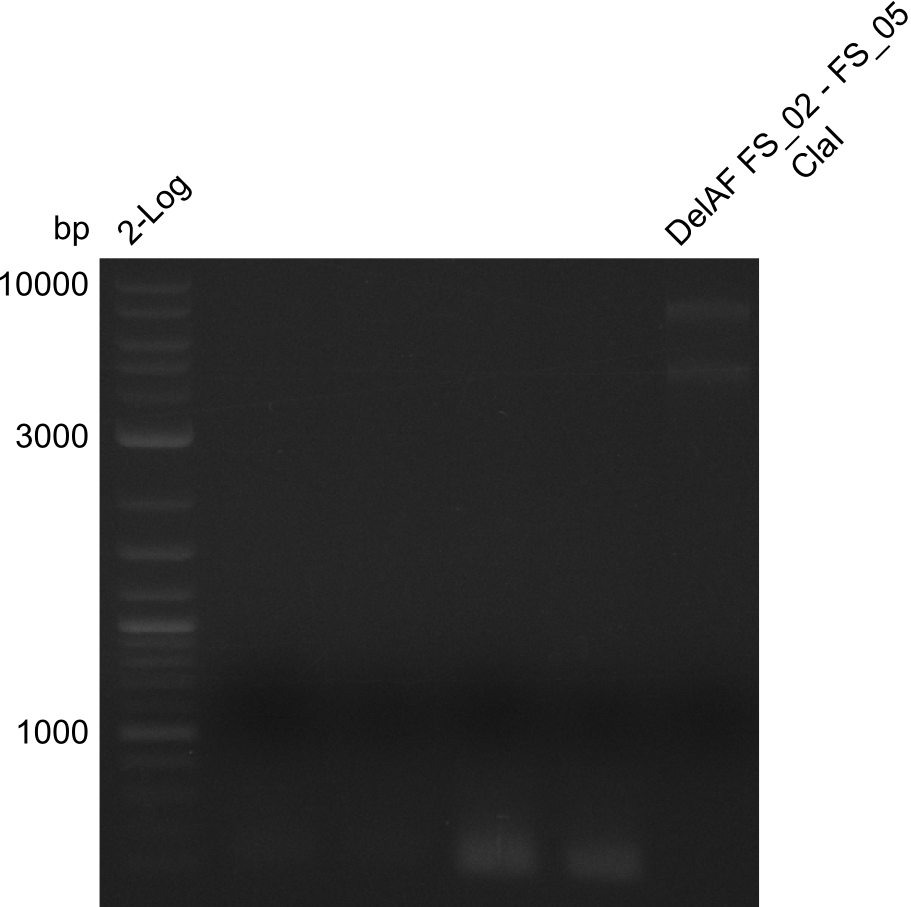
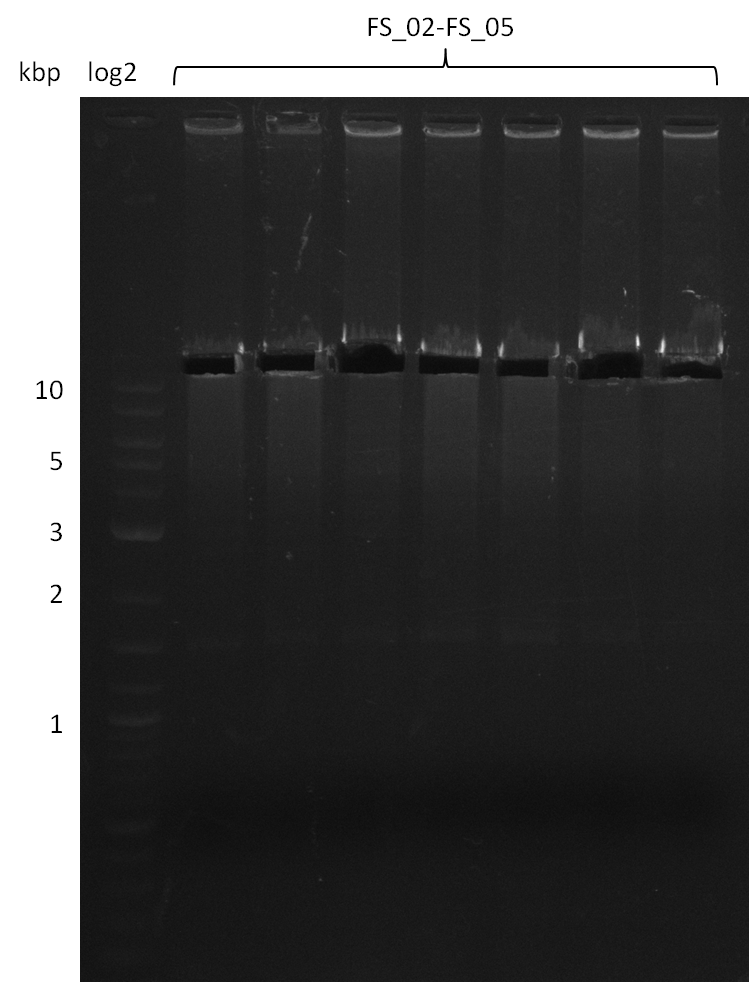
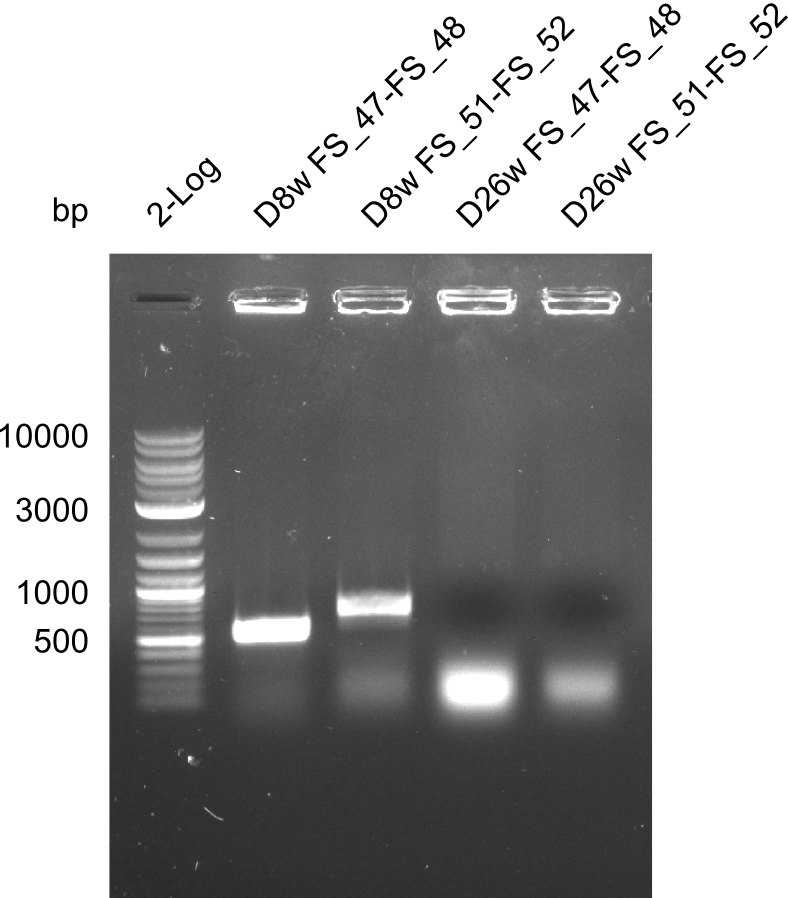
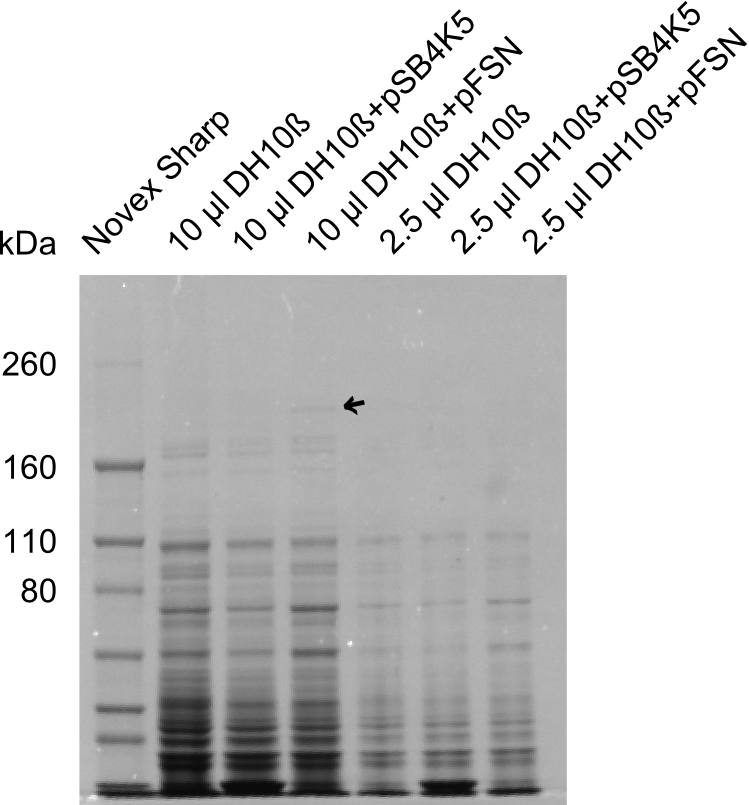
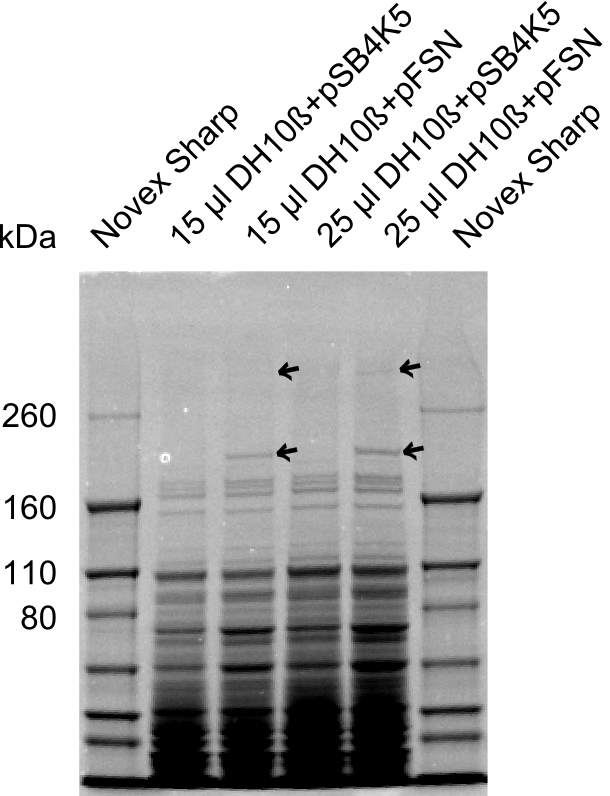
| - | + | We send one of our DelRest constructs that showed the right restriction pattern in the test digest for sequencing. As it would be quite costly to sequence the whole 32 kb plasmid, we focused on the ligation sites between the different assembly fragments, which are most prone to insertion of errors. Although all insert fragments sequenced, including the ligation sites between DelA and DelF, DelO-P and DelL, were 100 % correct, we detected a mutation within the mRFP cds (note: we wanted to use mRFP in order to confirm expression from our DelRest plasmid). FACS analysis of <i> E. coli </i> bearing the DelRest construnct confirmed that mRFP expression was impaired, likely due to the corresponding mutation. However, as mRFP was only meant to be a general expression control on our construct, we did not start correcting out construct by mutagenesis. Instead, we started preparing samples for an SDS page in order to directly proof the expression of the Del genes by Coomassie staining. | |
</p> | </p> | ||
</div> | </div> | ||
| Line 170: | Line 170: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 19</h1> | <h1>Week 19</h1> | ||
| - | + | The Coomassie staining we carried out last week did not display all expected bands clearly. This occured most likely due to the low amount of protein loaded onto the corresponding SDS page. Therefore the SDS page was repeated. | |
| + | This expression analysis confirmed the results indicated by the previous SDS-page. We were able to show that DelE and DelG are expressed at a detectable level. | ||
| + | As we could confirm the successful cloning and functioning of the DelRest expression plasmid named pFSN (for Florian-Sophie-Nils :)) the DelRest subproject was succssfully completed and the personal resources of the DelRest group were shifted to the DelH group as well as the wiki. | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
</p> | </p> | ||
| Line 182: | Line 184: | ||
</div> | </div> | ||
</div><!-- /.carousel --> | </div><!-- /.carousel --> | ||
| + | </div> | ||
| + | <div class="col-sm-12 col-md-6" style="margin-top:7%"> | ||
| + | <div class="jumbotron"> | ||
| + | |||
| + | <div style="width:100%;"> | ||
| + | <a class="fancybox fancyGraphical" rel="group" href="https://static.igem.org/mediawiki/2013/e/e3/Heidelberg_ga_delf.png"> | ||
| - | + | <img style="width:100%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 4px; border-color: grey;" src="https://static.igem.org/mediawiki/2013/e/e3/Heidelberg_ga_delf.png"></img> | |
| - | + | ||
| - | + | </a> | |
| - | + | ||
</div> | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| - | <div class="col-sm- | + | <div class="container"> |
| + | <div class="row"> | ||
| + | <div class="col-sm-12"> | ||
<!--Start Weekly Labjournal--> | <!--Start Weekly Labjournal--> | ||
| Line 209: | Line 223: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 215: | Line 229: | ||
<div class="tab-pane active" id="a10"> | <div class="tab-pane active" id="a10"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week10_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 296: | Line 310: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a11"> | <div class="tab-pane active" id="a11"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Templates/Del_week11_overview}}<html> | ||
</p> | </p> | ||
| Line 381: | Line 396: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 387: | Line 402: | ||
<div class="tab-pane active" id="a12"> | <div class="tab-pane active" id="a12"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week12_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 449: | Line 464: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a13"> | <div class="tab-pane active" id="a13"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week13_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 527: | Line 542: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 533: | Line 548: | ||
<div class="tab-pane active" id="a14"> | <div class="tab-pane active" id="a14"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week14_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 596: | Line 611: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a15"> | <div class="tab-pane active" id="a15"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week15_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 683: | Line 698: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a16"> | <div class="tab-pane active" id="a16"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week16_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 754: | Line 769: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a17"> | <div class="tab-pane active" id="a17"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week17_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 791: | Line 806: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a18"> | <div class="tab-pane active" id="a18"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week18_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 820: | Line 835: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a19"> | <div class="tab-pane active" id="a19"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week19_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 841: | Line 856: | ||
</div> | </div> | ||
</div> | </div> | ||
| - | + | </div> | |
| - | + | </div> | |
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
| + | {{:Team:Heidelberg/Templates/Footer-Nav}} | ||
{{:Team:Heidelberg/Templates/Footer-DelRest}} | {{:Team:Heidelberg/Templates/Footer-DelRest}} | ||
Latest revision as of 01:37, 29 October 2013


 "
"