Team:Heidelberg/Delftibactin/DelH
From 2013.igem.org
(Difference between revisions)
| (43 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:Heidelberg/Templates/Navigation}} | {{:Team:Heidelberg/Templates/Navigation}} | ||
| + | {{:Team:Heidelberg/Templates/scrollbar-css}} | ||
| + | |||
<html> | <html> | ||
<style type="text/css"> | <style type="text/css"> | ||
| Line 11: | Line 9: | ||
p { | p { | ||
font-size:10px; | font-size:10px; | ||
| + | } | ||
| + | .carousel-inner { | ||
| + | margin-top:17%; | ||
} | } | ||
</style> | </style> | ||
<div class="container"> | <div class="container"> | ||
<!--Project Description--> | <!--Project Description--> | ||
| - | <div | + | <div> |
| - | <h1><span style="font-size: | + | <h1><span style="font-size:180%;color:#FFCC00;">Del H.</span><span class="text-muted" style="font-size:120%"> This nasty 18 kb fragment.</span></h1> |
| - | <p> | + | <p style="font-size:14px">Facing the challenge to clone 18 kbp of genomic DNA from <i>D. acidovorans</i>.</p> |
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col-sm-6"> | + | <div class="col-xs-12 col-sm-12 col-md-6"> |
<!--Months--> | <!--Months--> | ||
<ul class="pagination" style="margin-bottom:2%; margin-left:15%;"> | <ul class="pagination" style="margin-bottom:2%; margin-left:15%;"> | ||
| Line 31: | Line 32: | ||
<li class="month_tab" id="august"><a href="#" style="width:100px; text-align:center">August</a></li> | <li class="month_tab" id="august"><a href="#" style="width:100px; text-align:center">August</a></li> | ||
<li class="month_tab" id="september"><a href="#" style="width:100px; text-align:center">September</a></li> | <li class="month_tab" id="september"><a href="#" style="width:100px; text-align:center">September</a></li> | ||
| - | + | <li class="month_tab" id="october"><a href="#" style="width:100px; text-align:center">October</a></li> | |
<li><a href="#" id="forwards">»</a></li> | <li><a href="#" id="forwards">»</a></li> | ||
</ul> | </ul> | ||
| Line 58: | Line 59: | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
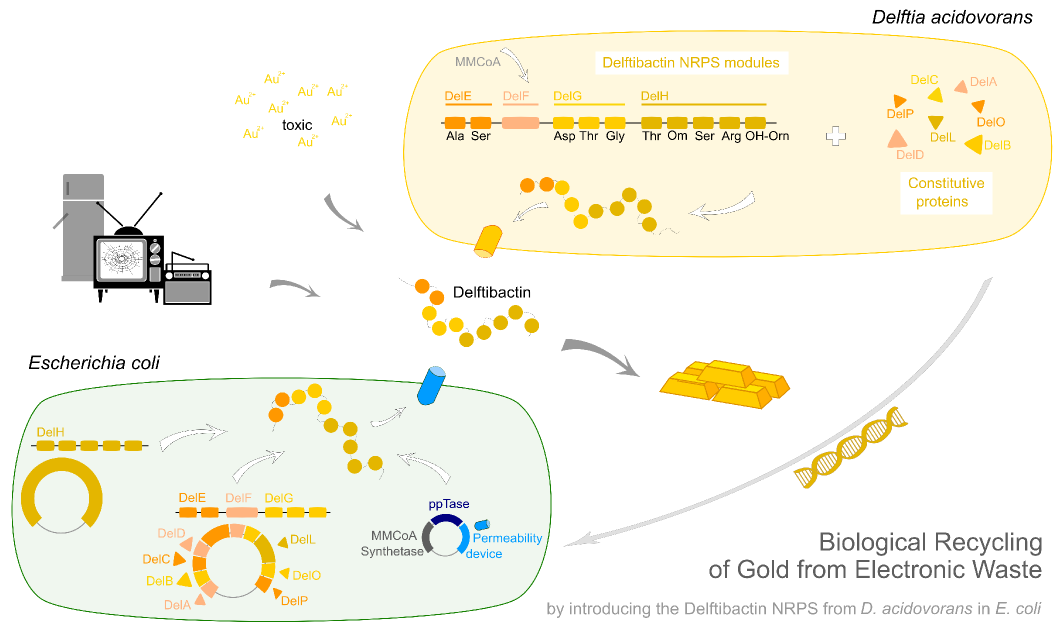
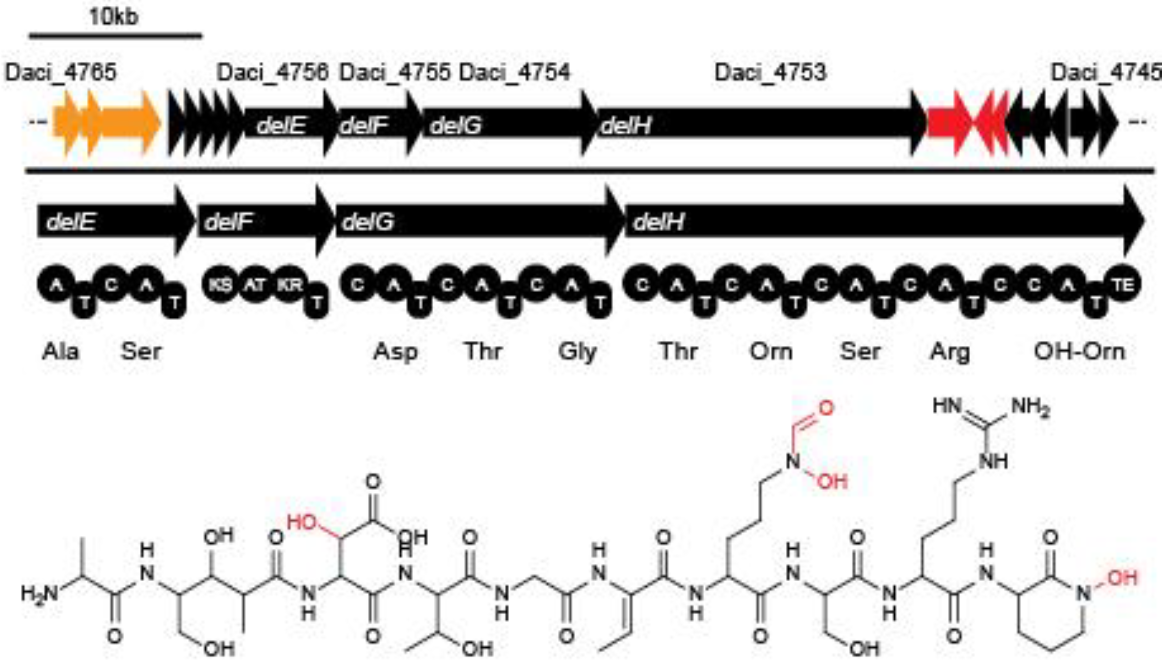
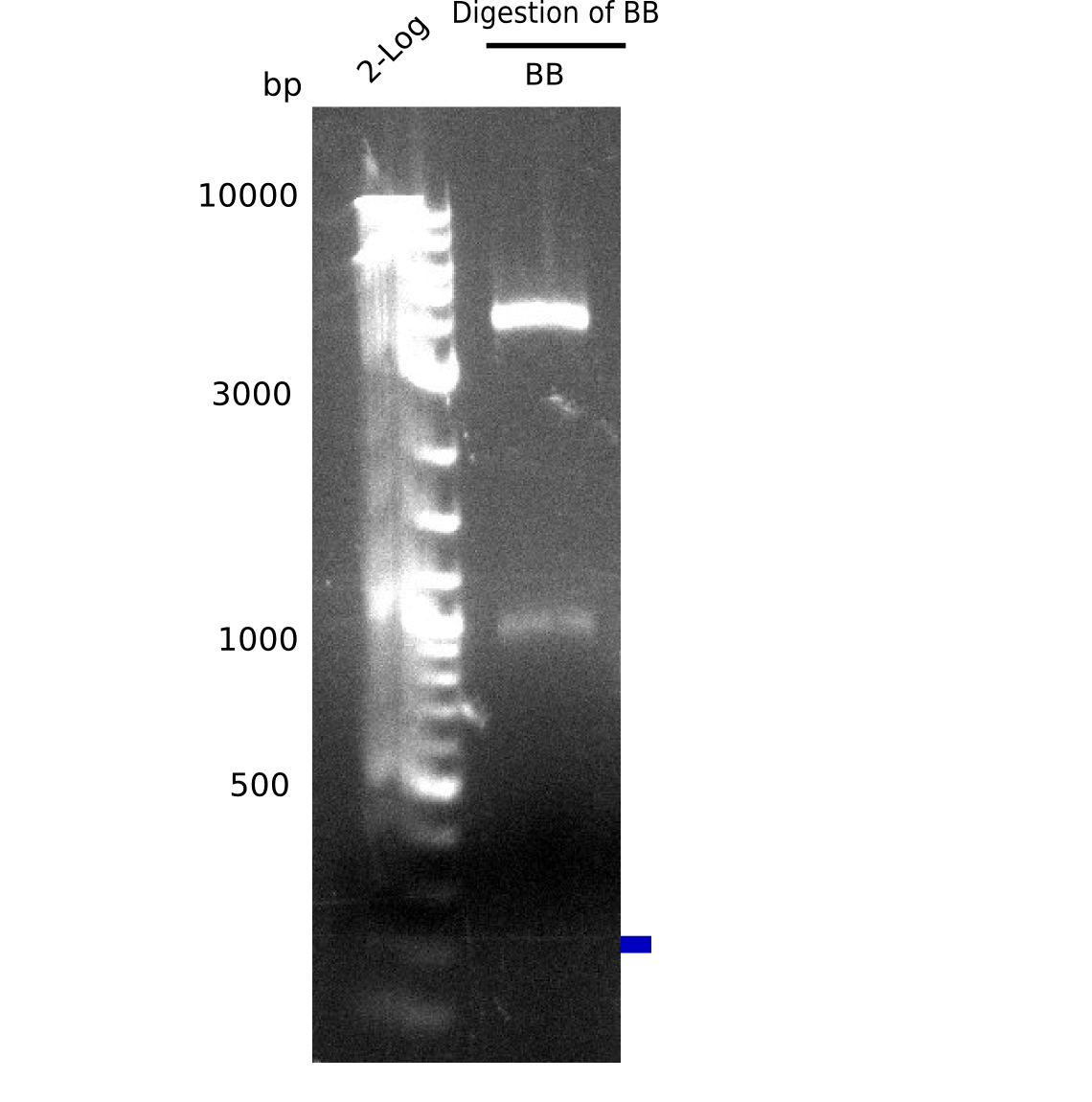
| - | In order to transfer the gold precipitating NRPS from <i>D. acidovorans</i> to <i>E.coli</i>, the necessary modules will be amplified from the D. acidovorans genome and assembled as plasmids. Due to its large size of 18 | + | In order to transfer the gold precipitating NRPS from <i>D. acidovorans</i> to <i>E.coli</i>, the necessary modules will be amplified from the <i>D. acidovorans</i> genome and assembled as plasmids. Due to its large size of 18 kb, the module DelH will be expressed on a separate plasmid. A strategy was developed, primers designed accordingly and necessary BioBricks retrieved from the distribution. The <i>D. acidovorans</i> was obtained from the DSMZ and cultured in Acidovorax complex medium. |
</p> | </p> | ||
| Line 68: | Line 69: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 2</h1> | <h1>Week 2</h1> | ||
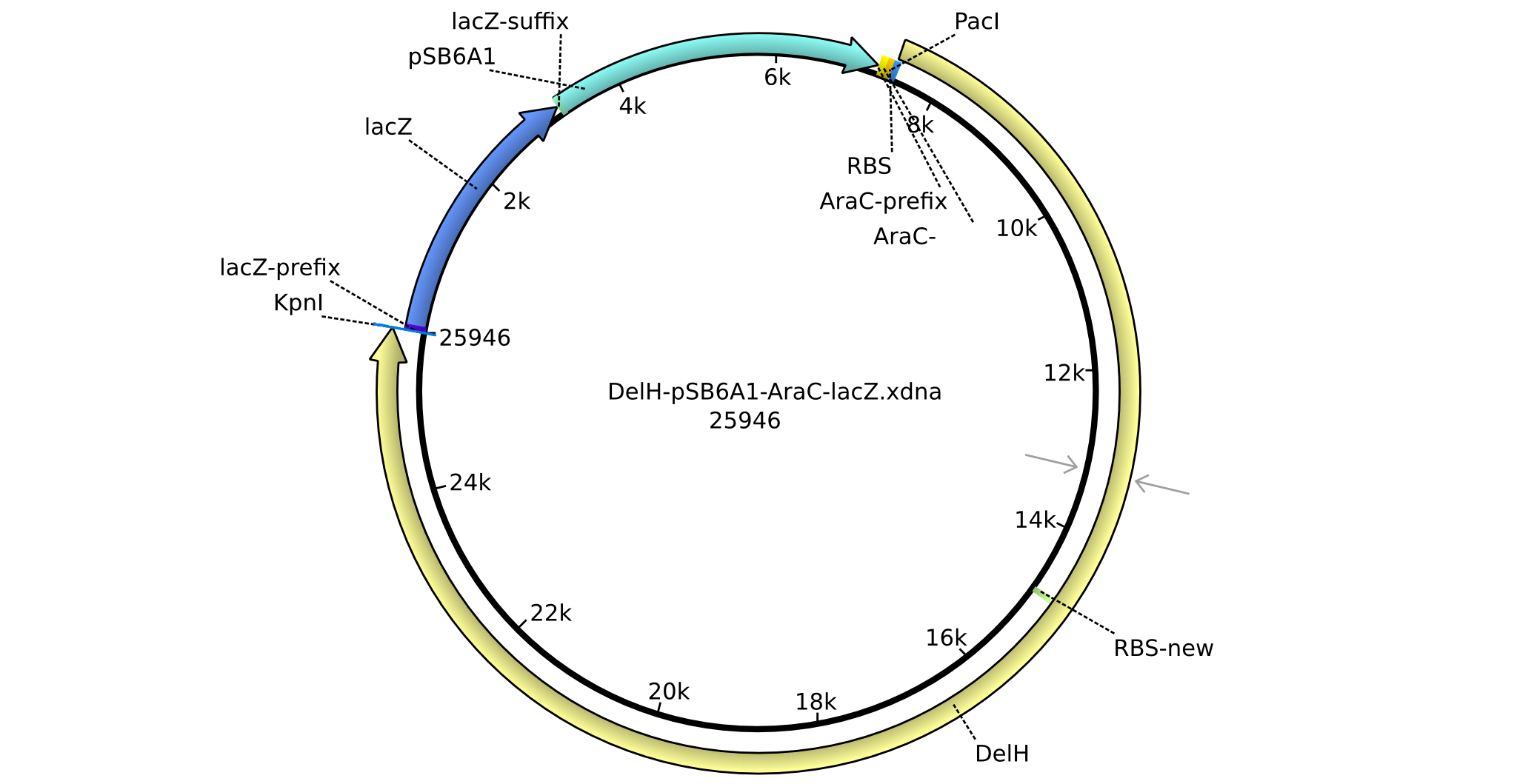
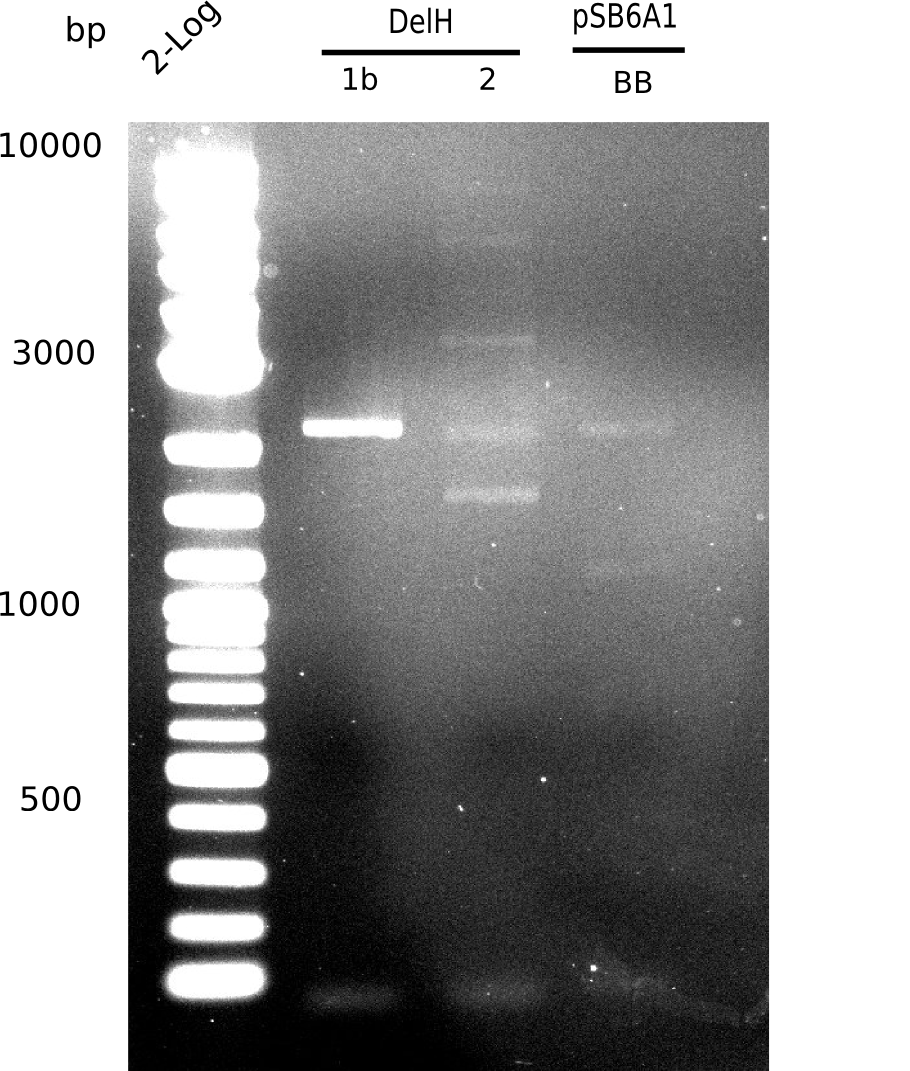
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Using the designed primers DelH_f1_PacI_fw and DelH_f1_SalI_rev, the first PCRs to amplify DelH F1 as well as DelH_f2_SalI_fw and DelH_f2_KpnI_rev to amplify DelH F2 were performed and conditions optimized. Additionally, necessary backbone fragments pSB6A1 and lacZ were restriction | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Using the designed primers DelH_f1_PacI_fw and DelH_f1_SalI_rev, the first PCRs to amplify DelH F1 as well as DelH_f2_SalI_fw and DelH_f2_KpnI_rev to amplify DelH F2 were performed and conditions optimized. Additionally, necessary backbone fragments pSB6A1 and lacZ were generated by restriction digest. New BioBricks to obtain the AraC promotor (I13453 and K206000) were chosen. |
</p> | </p> | ||
</div> | </div> | ||
| Line 77: | Line 78: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 3</h1> | <h1>Week 3</h1> | ||
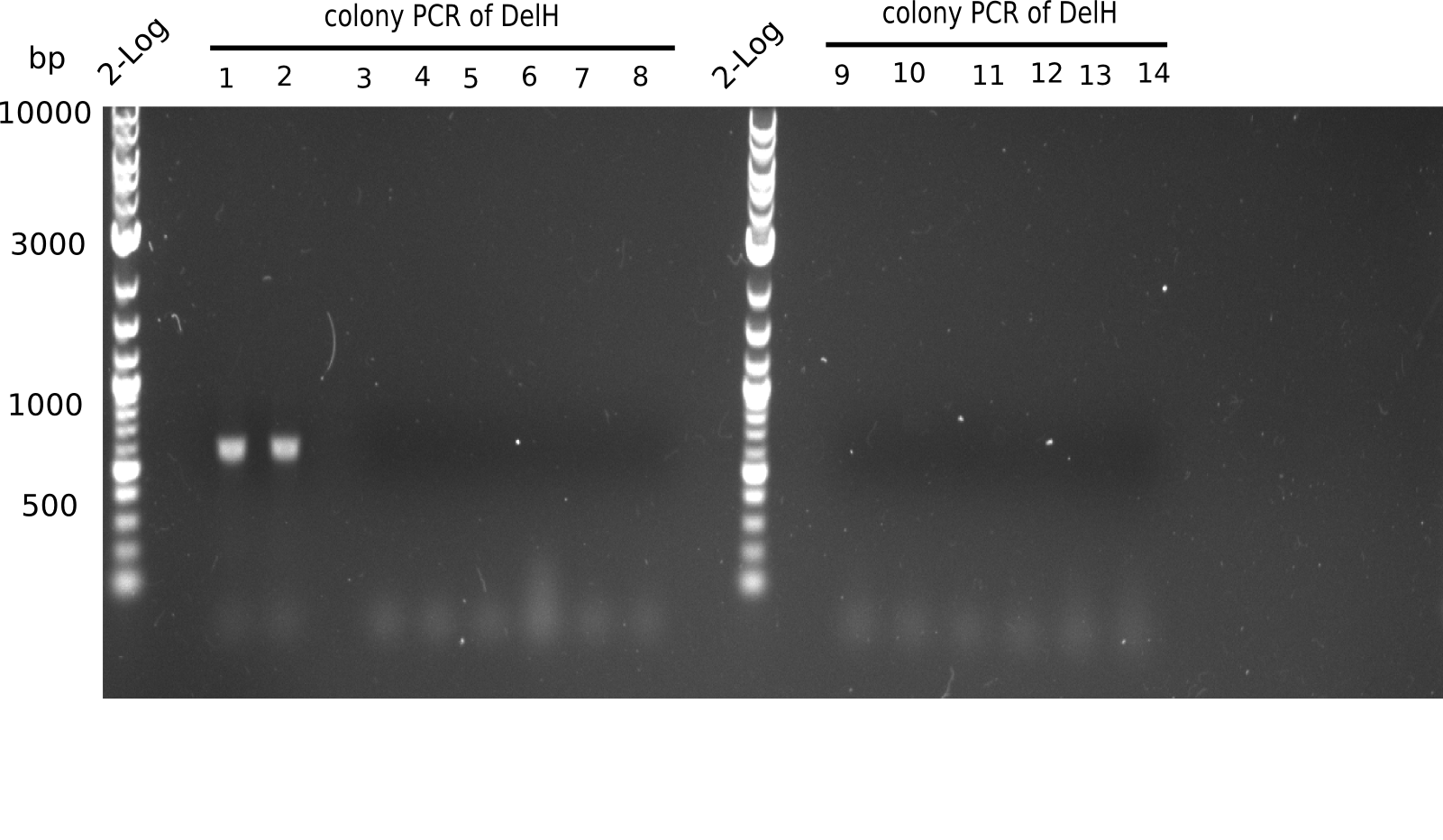
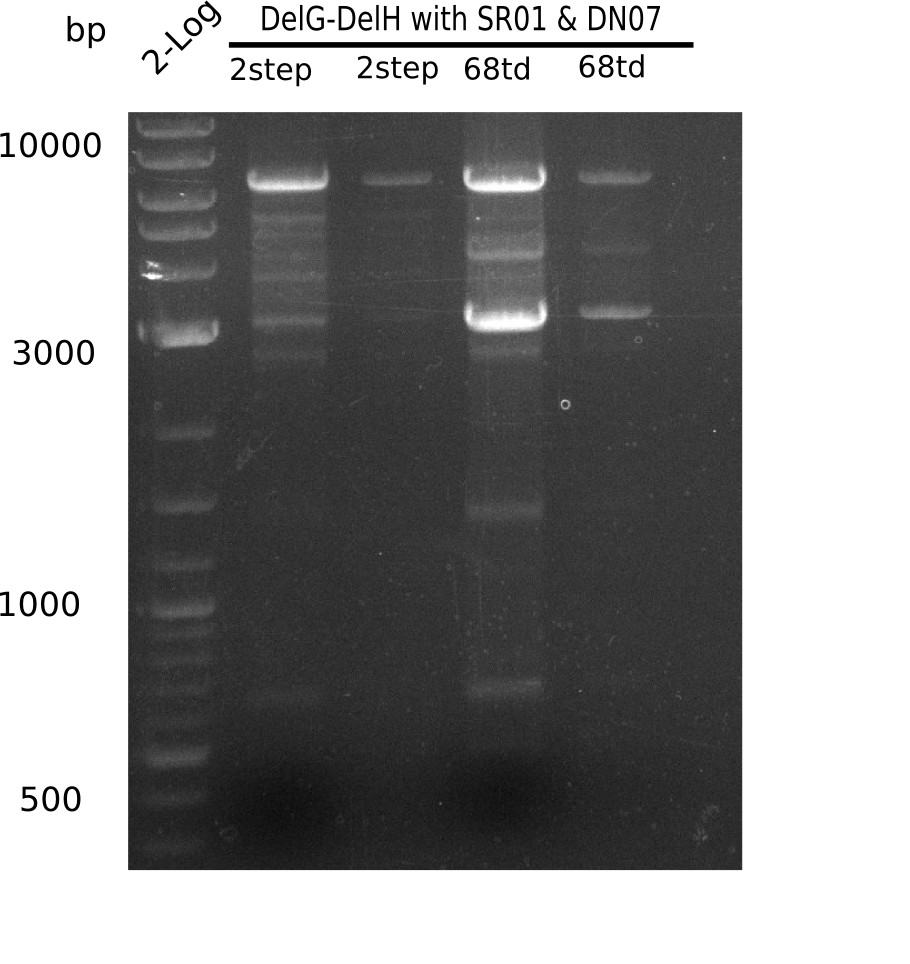
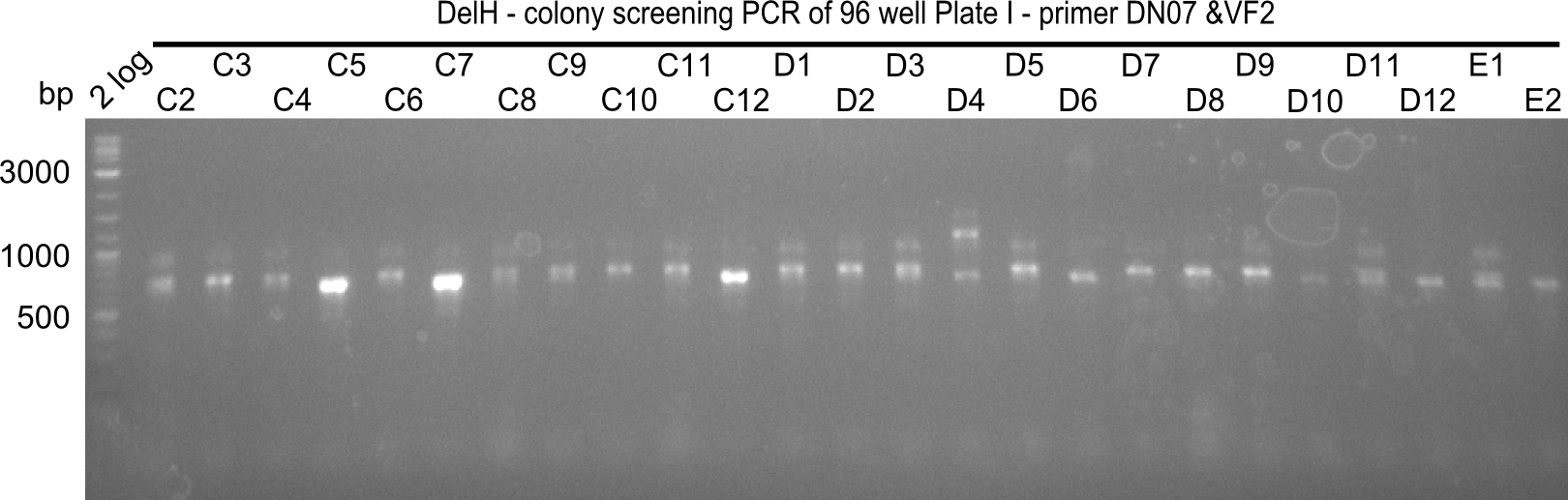
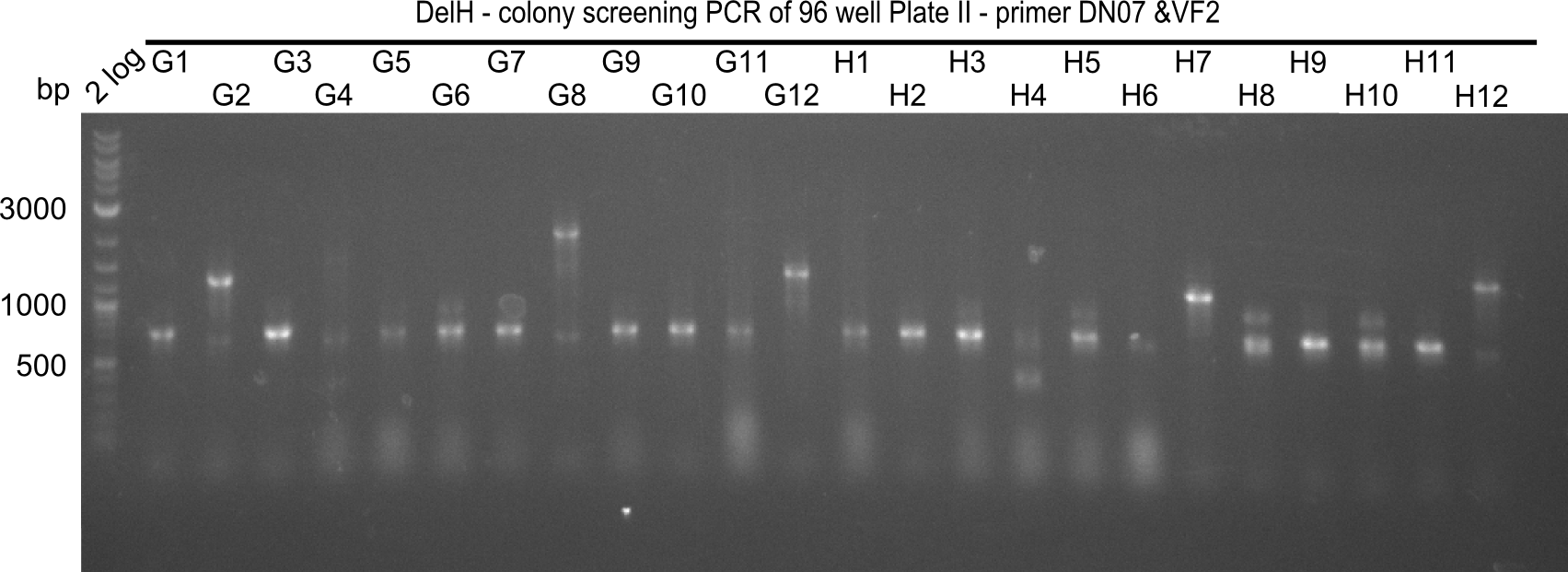
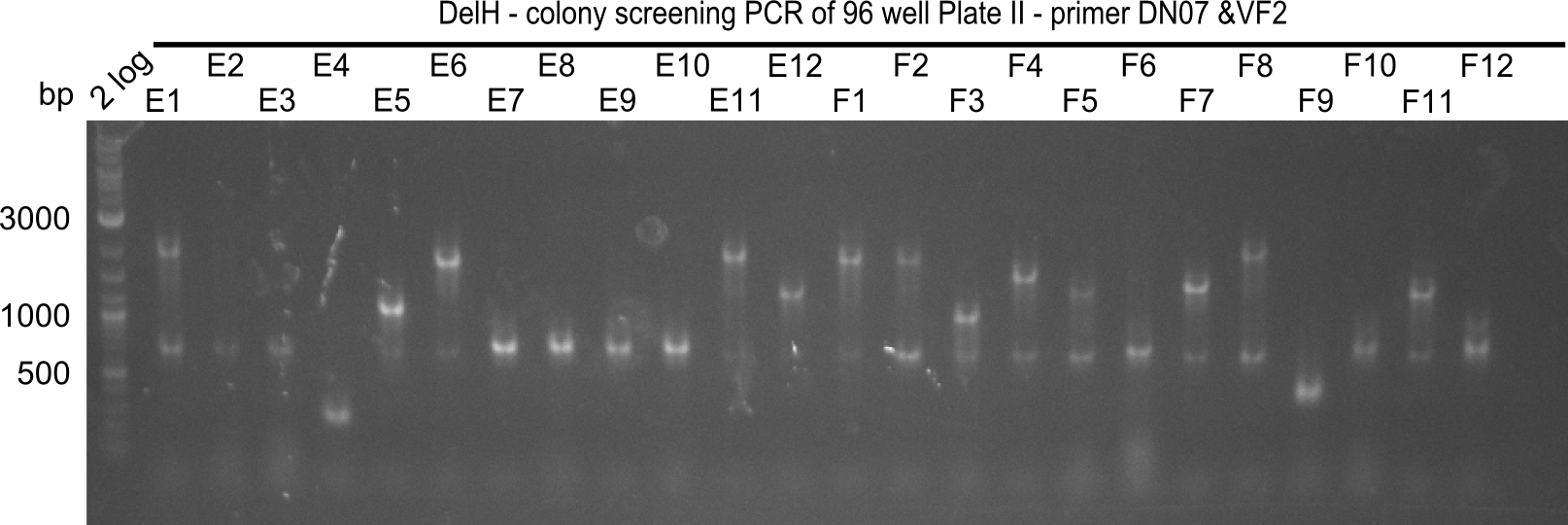
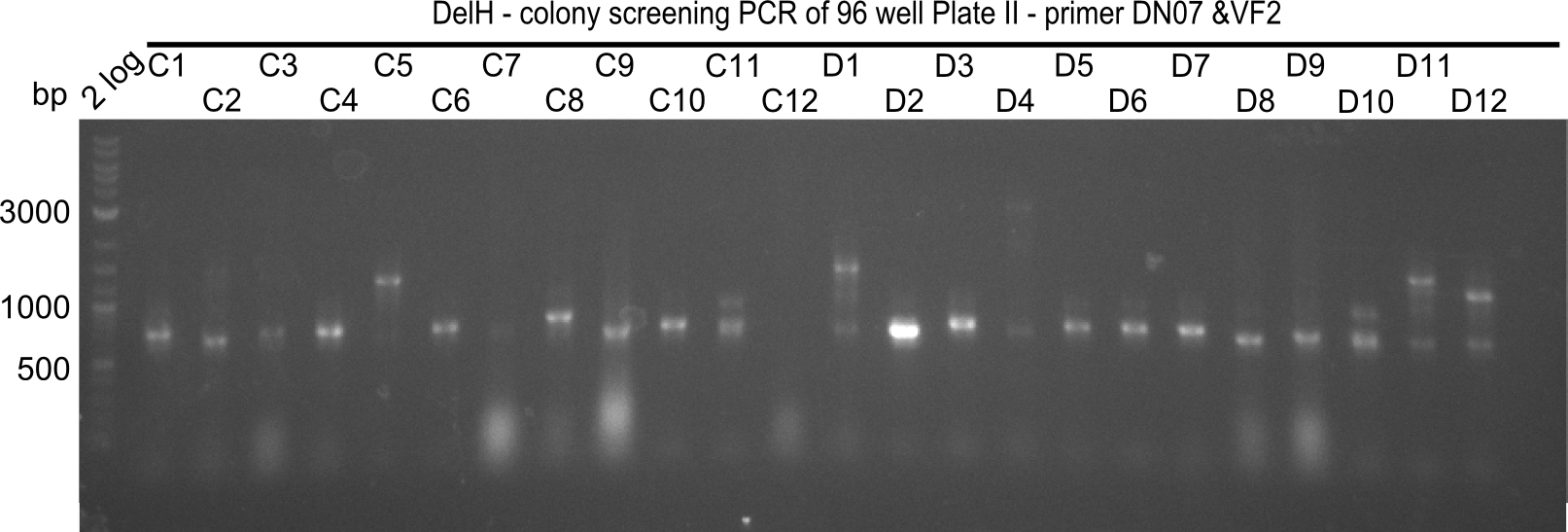
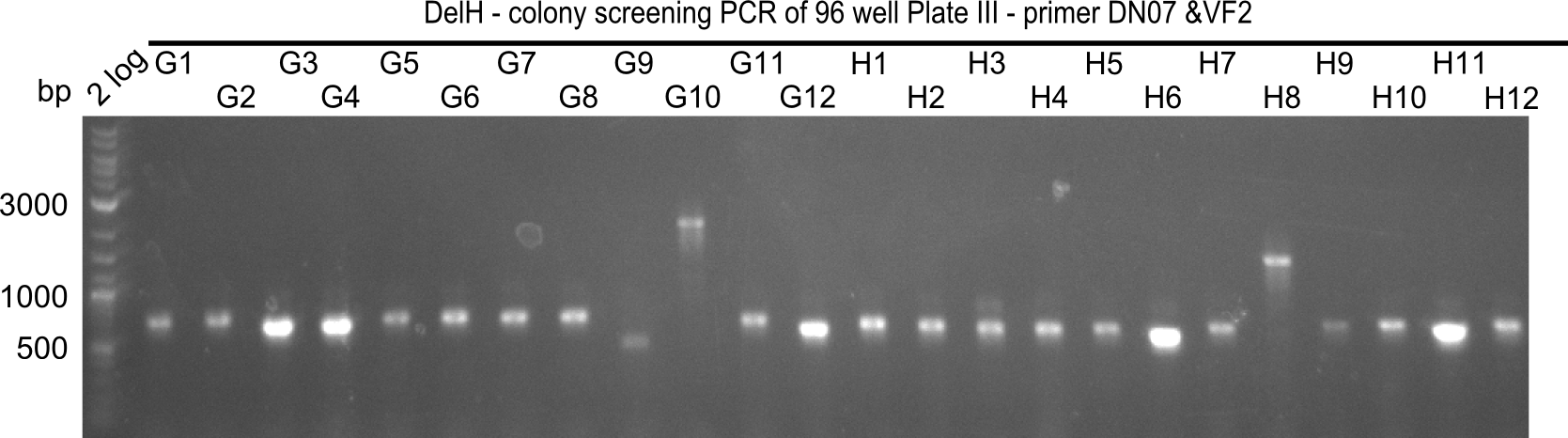
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since the reverse backbone primer DN08:AraCbb_PacI_rev did not work, a new one (DN06:AraCbb_PacI_rev2) was ordered, together with the colony PCR screening | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since the reverse backbone primer DN08:AraCbb_PacI_rev did not work, a new one (DN06:AraCbb_PacI_rev2) was ordered, together with the colony PCR screening primers DN07:Screen_DelH_rev and DN13:Screen_DelH_fw to check for correct ligation of the DelH fragment F1. |
</p> | </p> | ||
</div> | </div> | ||
| Line 86: | Line 87: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 4</h1> | <h1>Week 4</h1> | ||
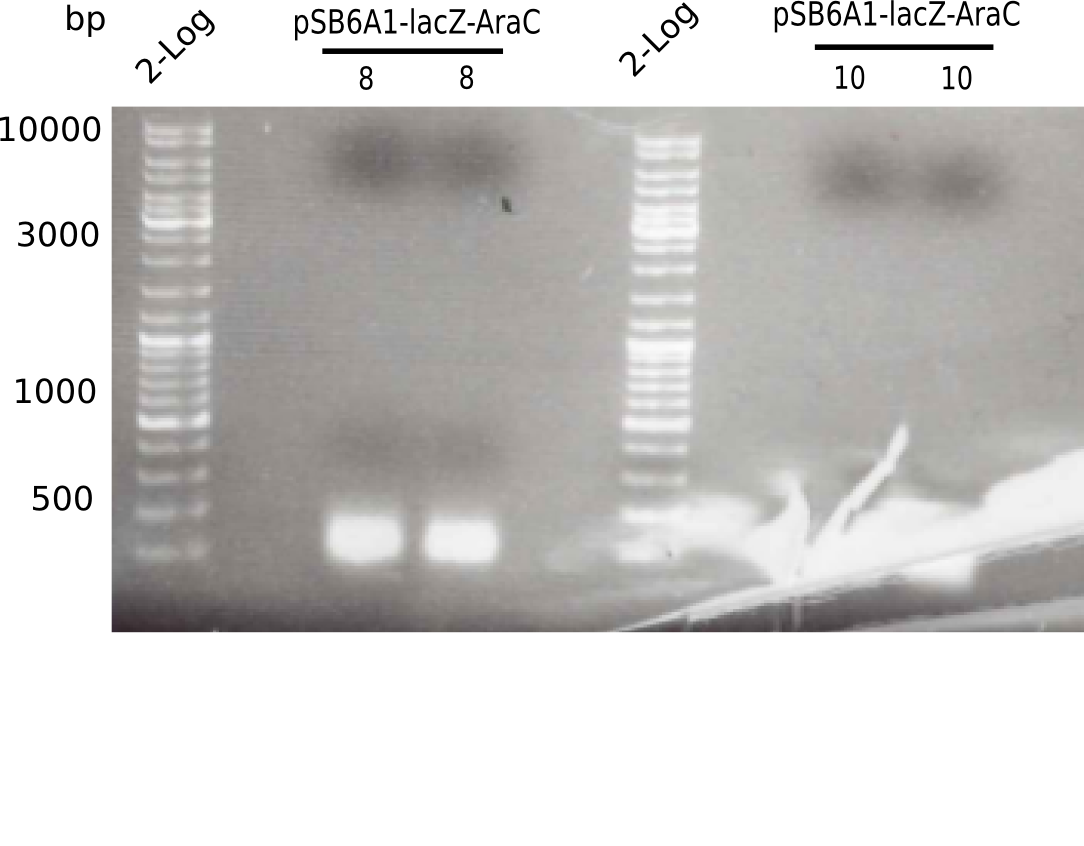
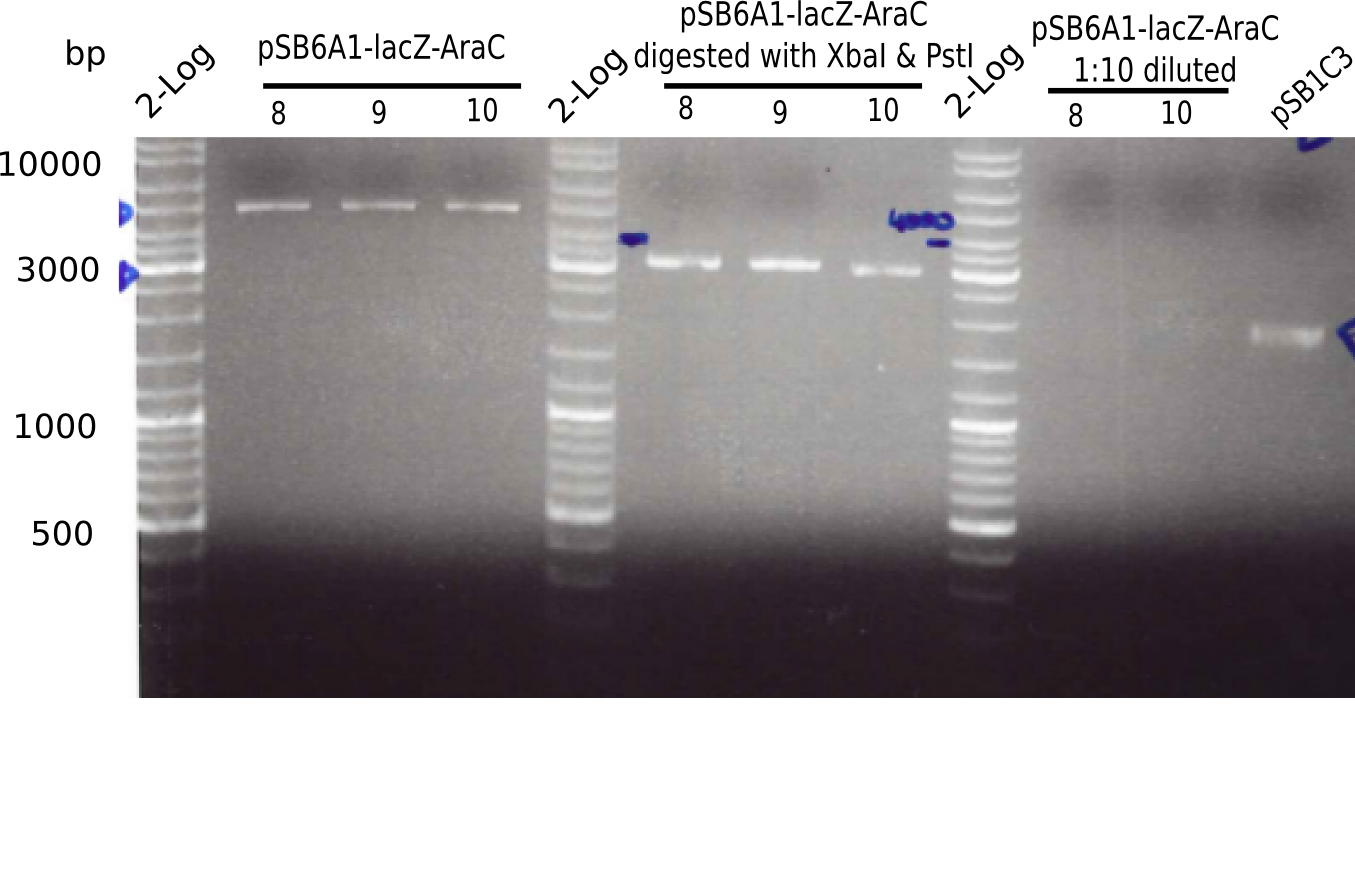
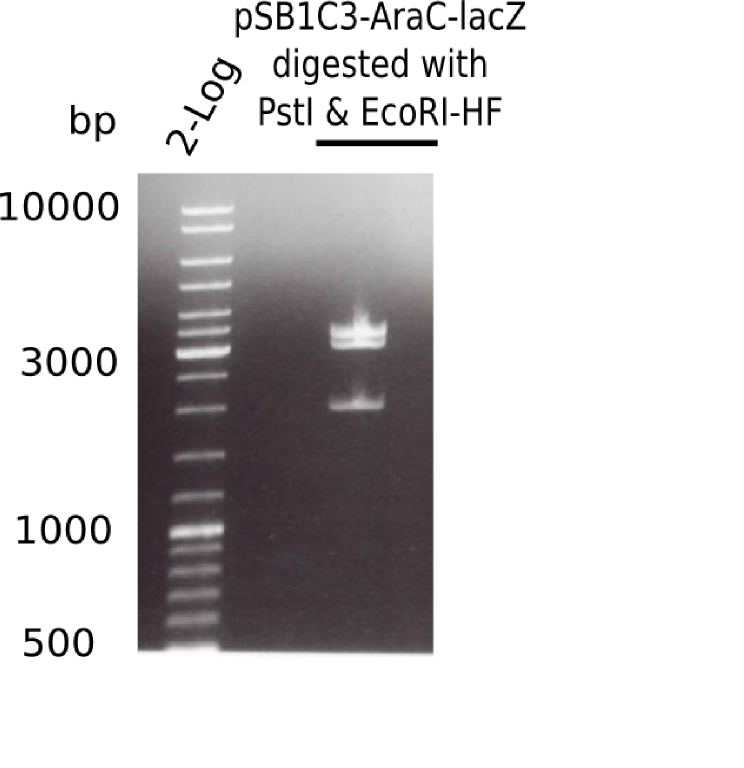
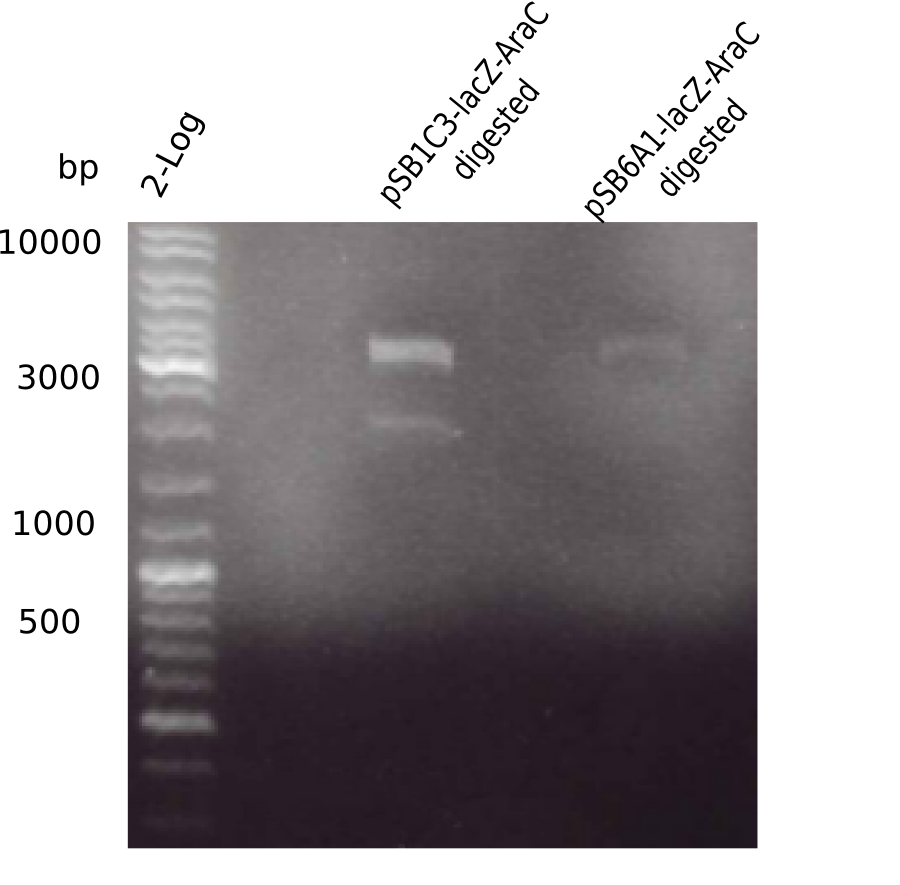
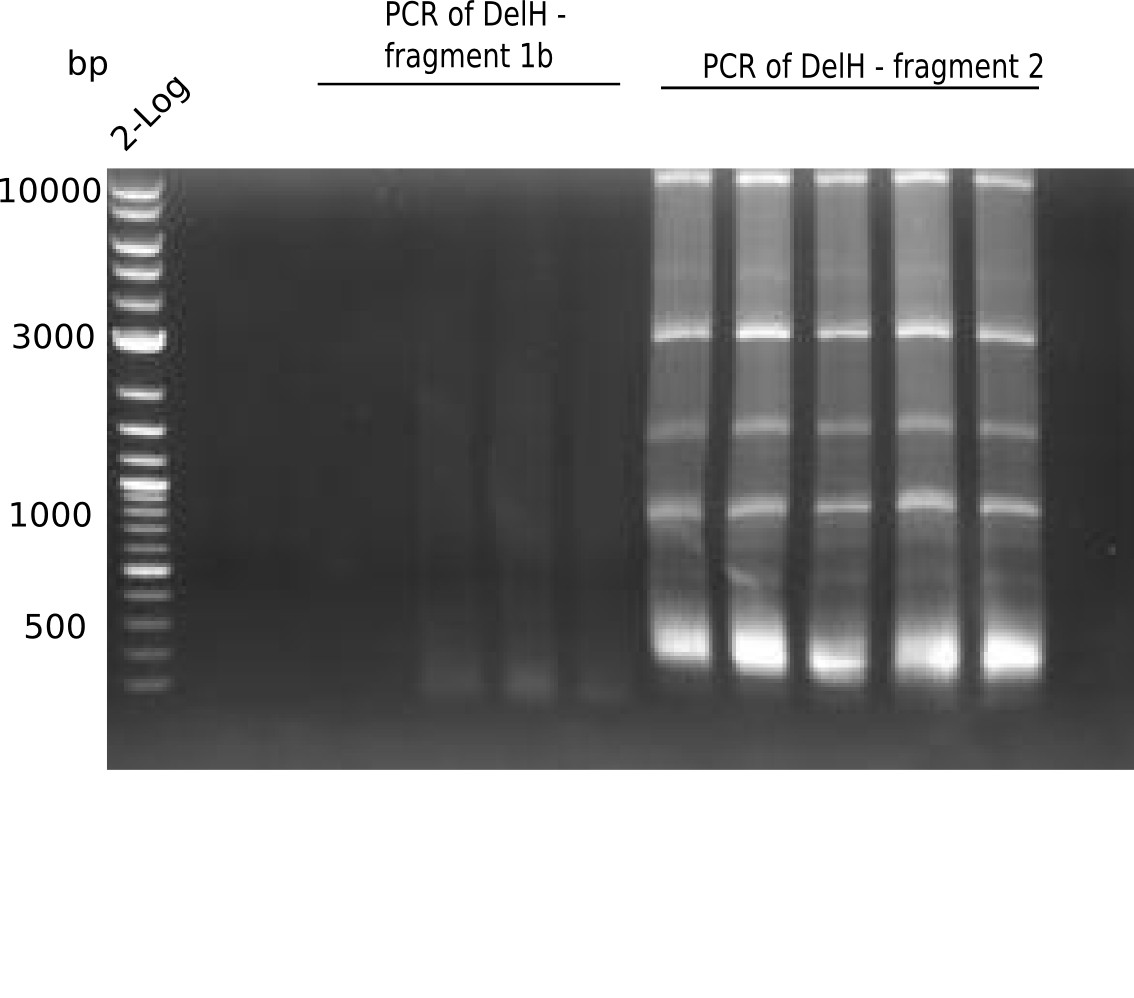
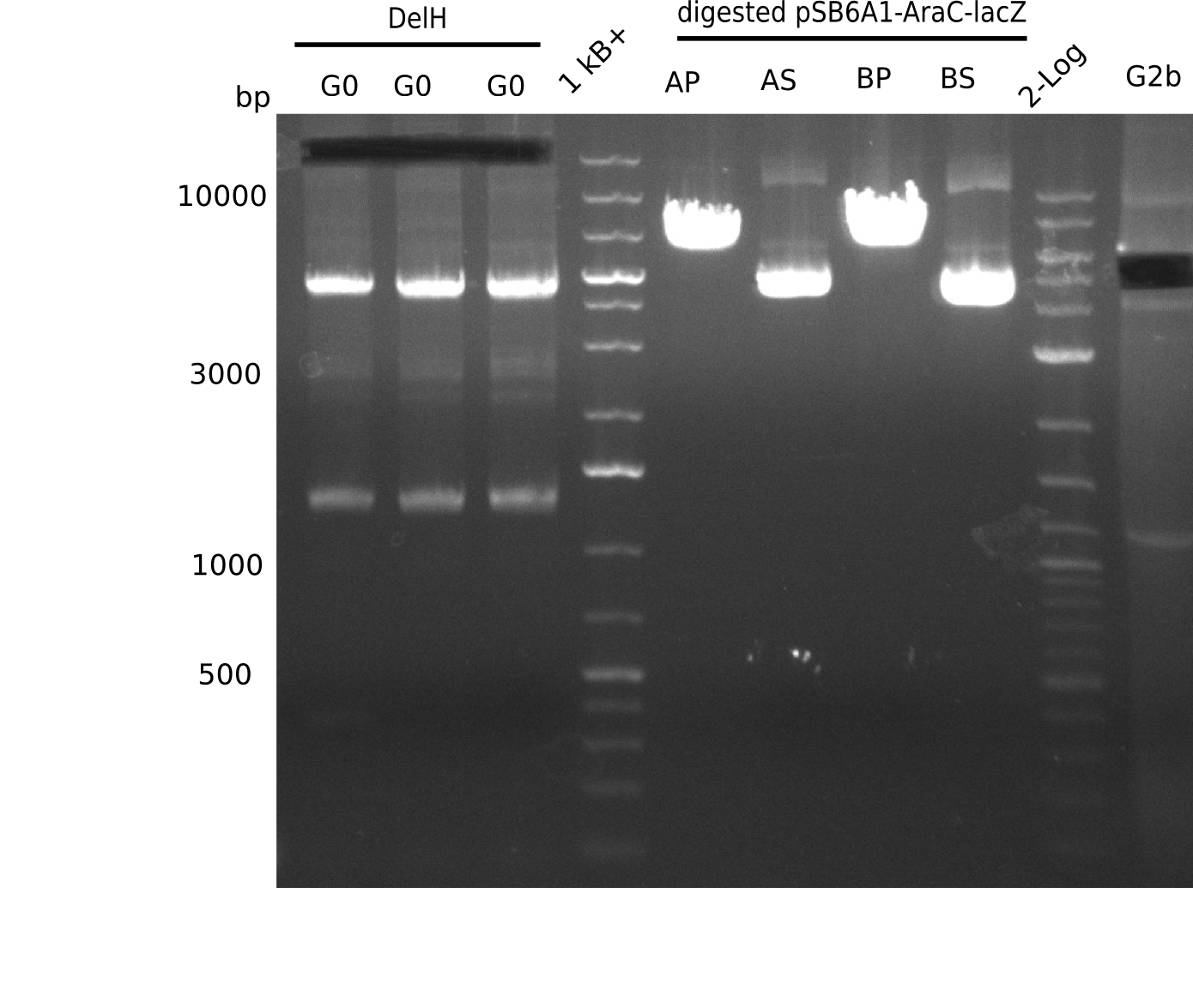
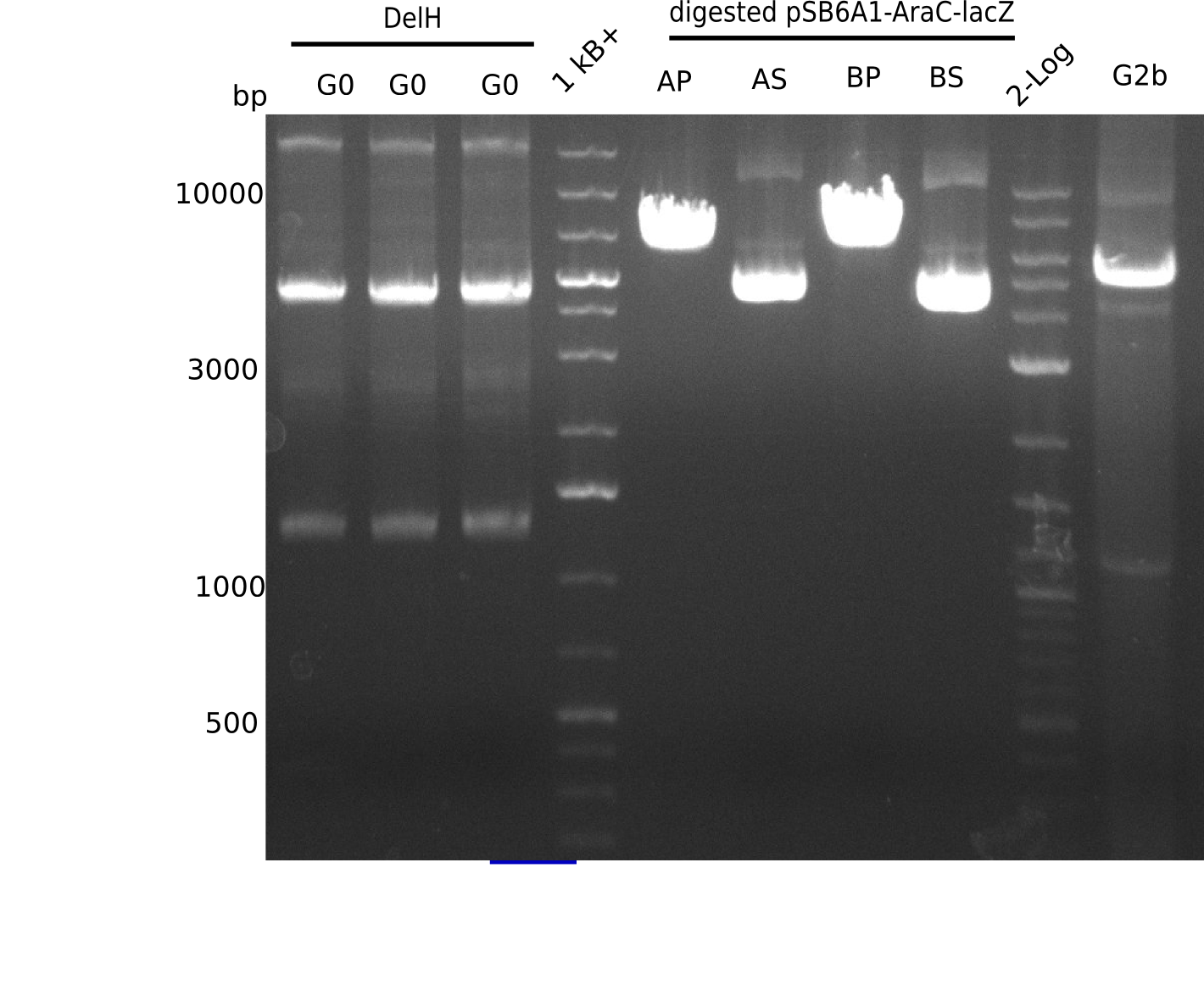
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The elongation of the pSB6A1-AraC-lacZ backbone | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The elongation of the pSB6A1-AraC-lacZ backbone turned out to be difficult. So in week 4, we performed different restriction digests both with our final backbone pSB6A1-AraC-lacZ and with the former construct pSB1C3-AraC-lacZ to verify the identity of the backbone. Besides, the amplification of the DelH fragment F1 was planned: it will be amplified in 2 subfragments - fragment F1a and fragment F1b (both 5 kb in size). </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 94: | Line 95: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 5</h1> | <h1>Week 5</h1> | ||
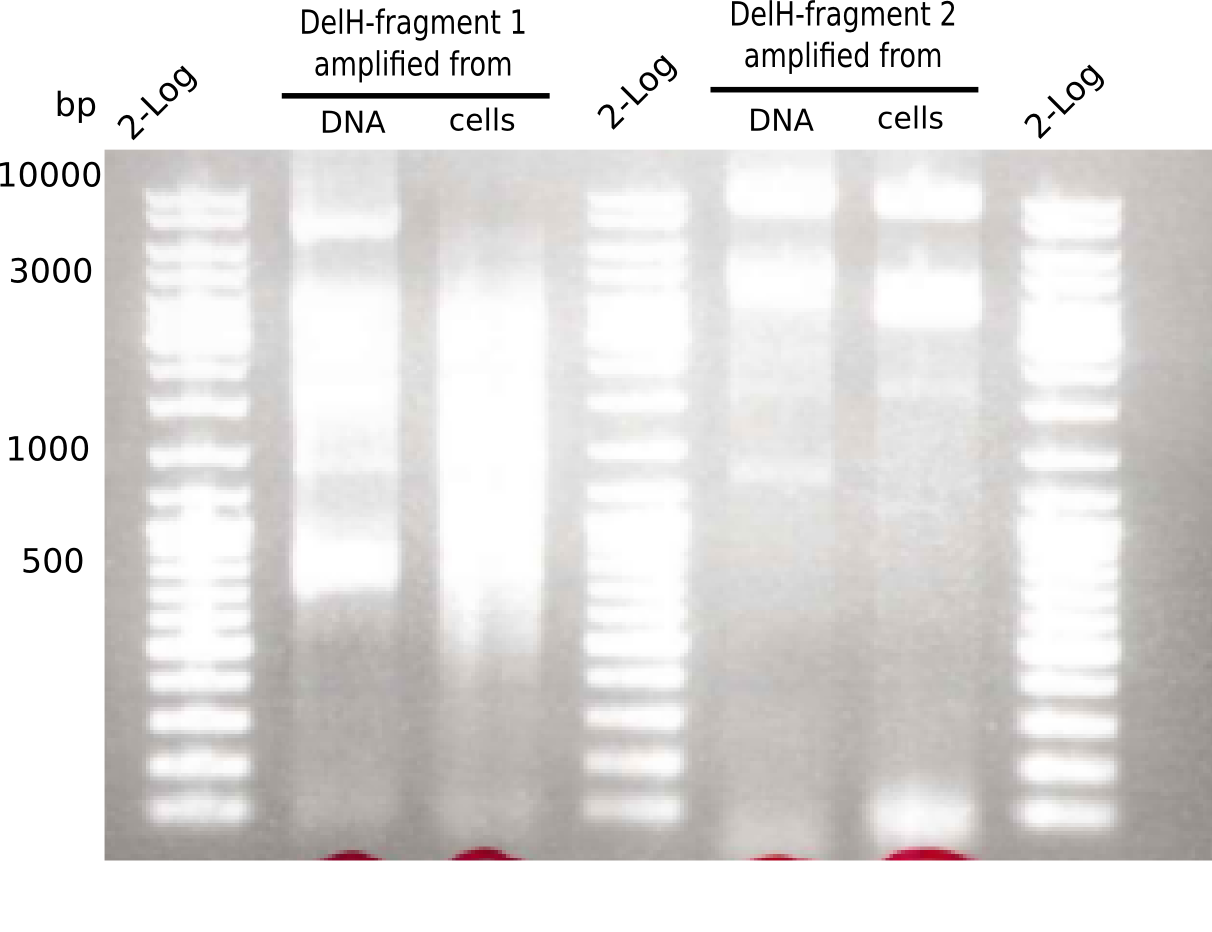
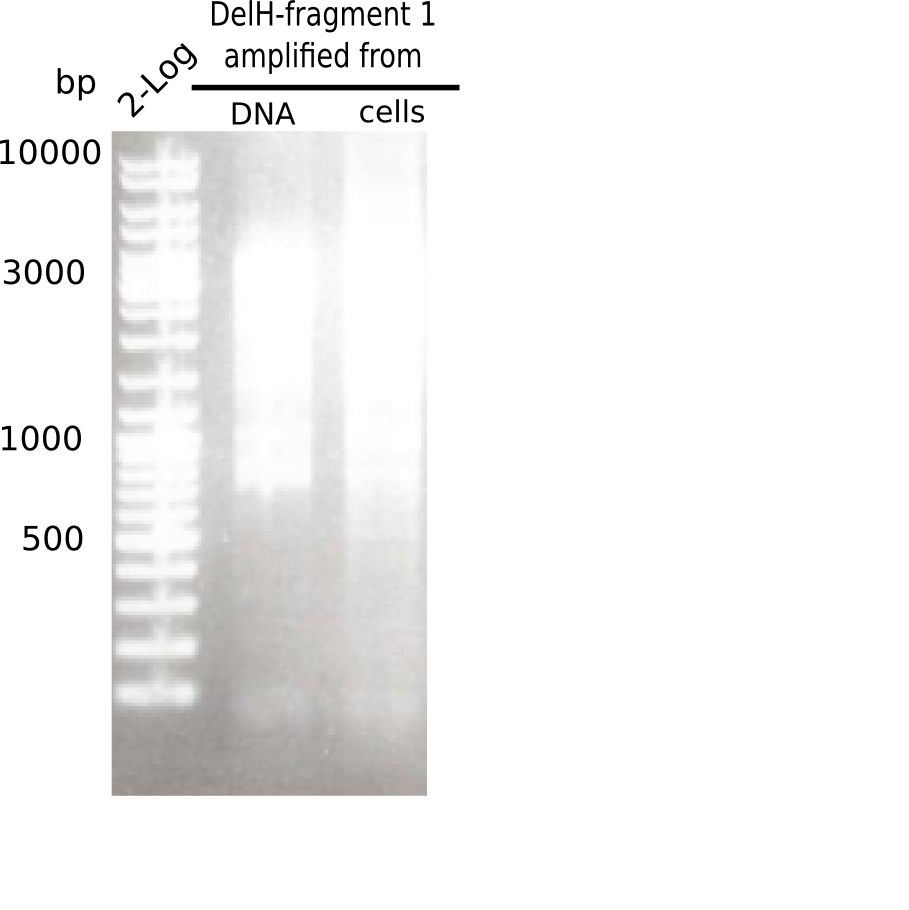
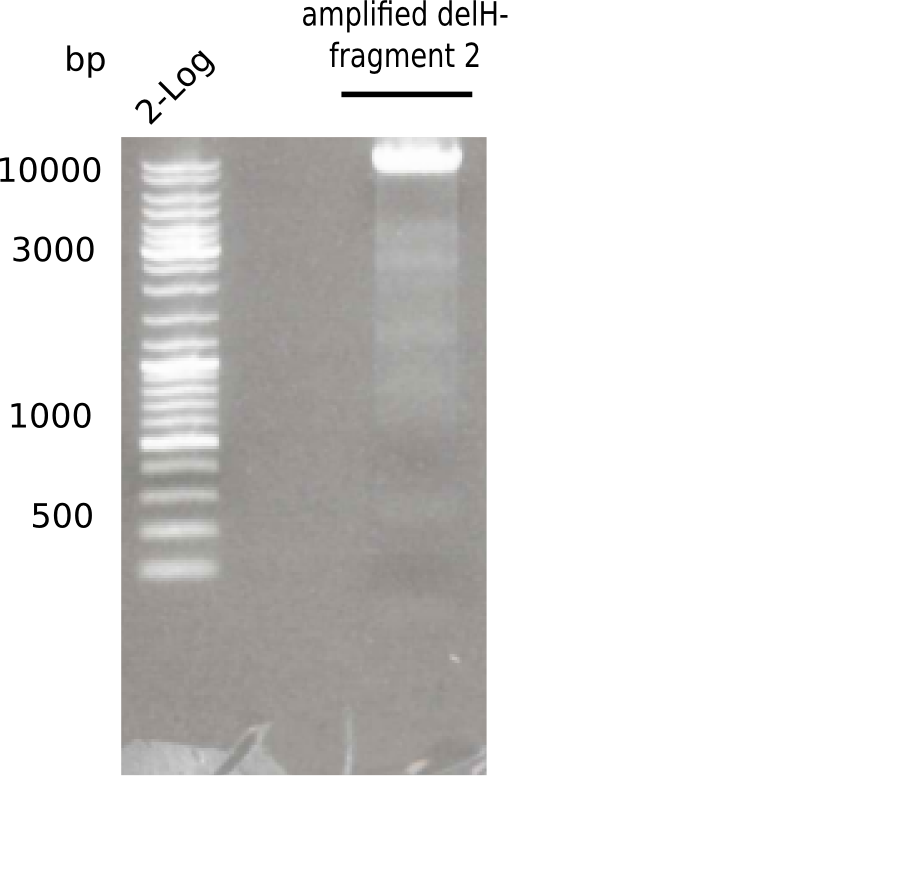
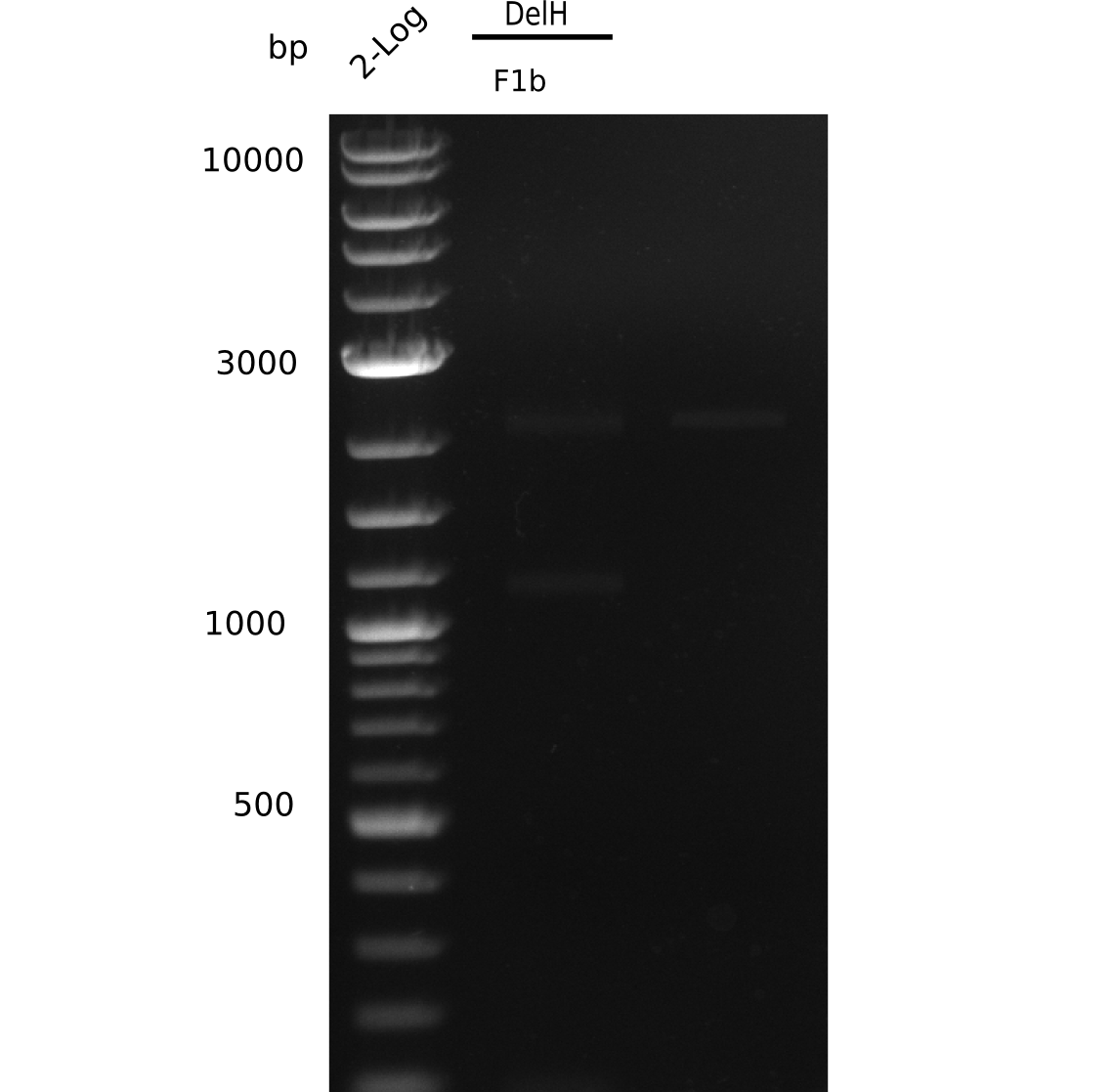
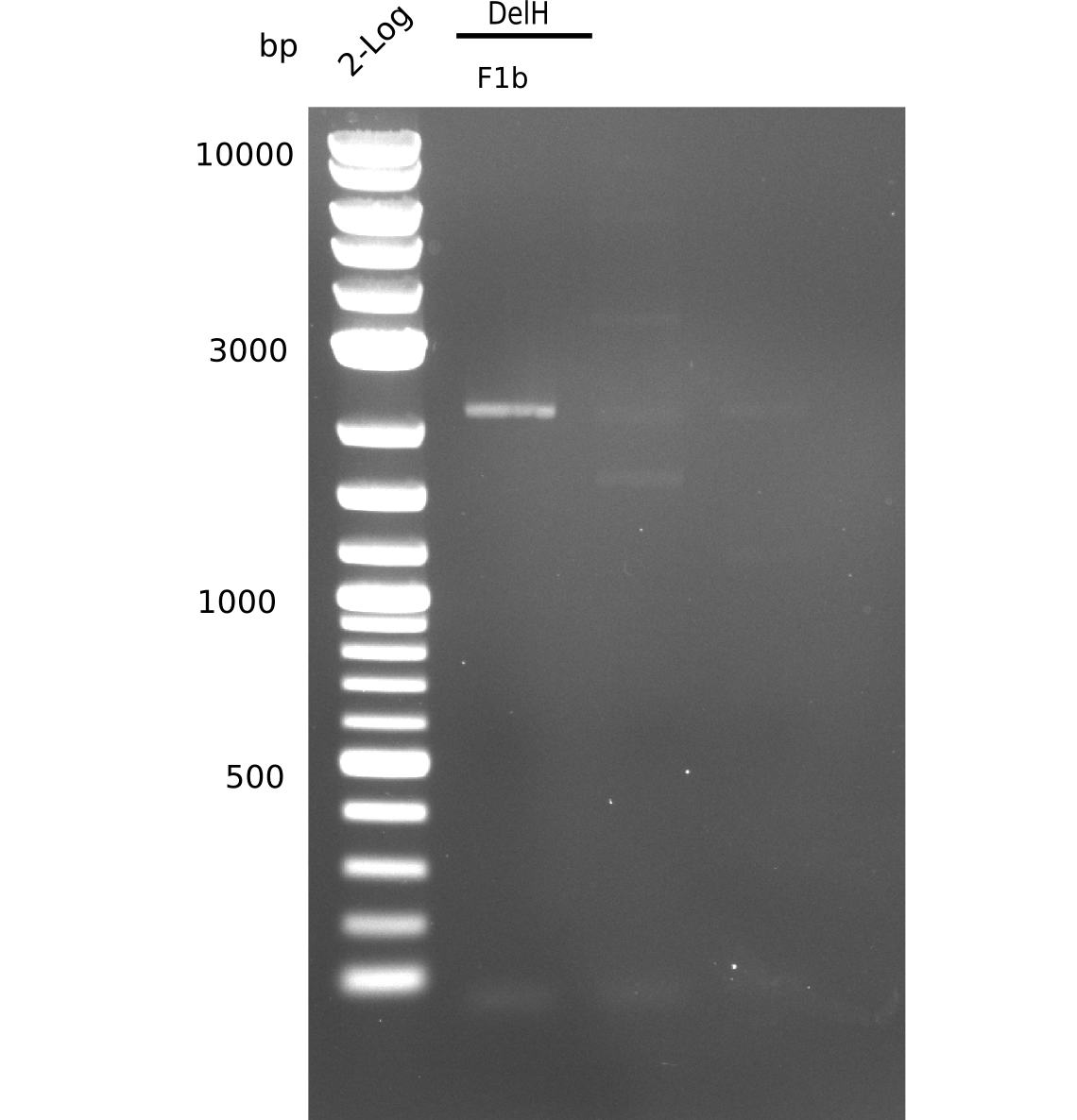
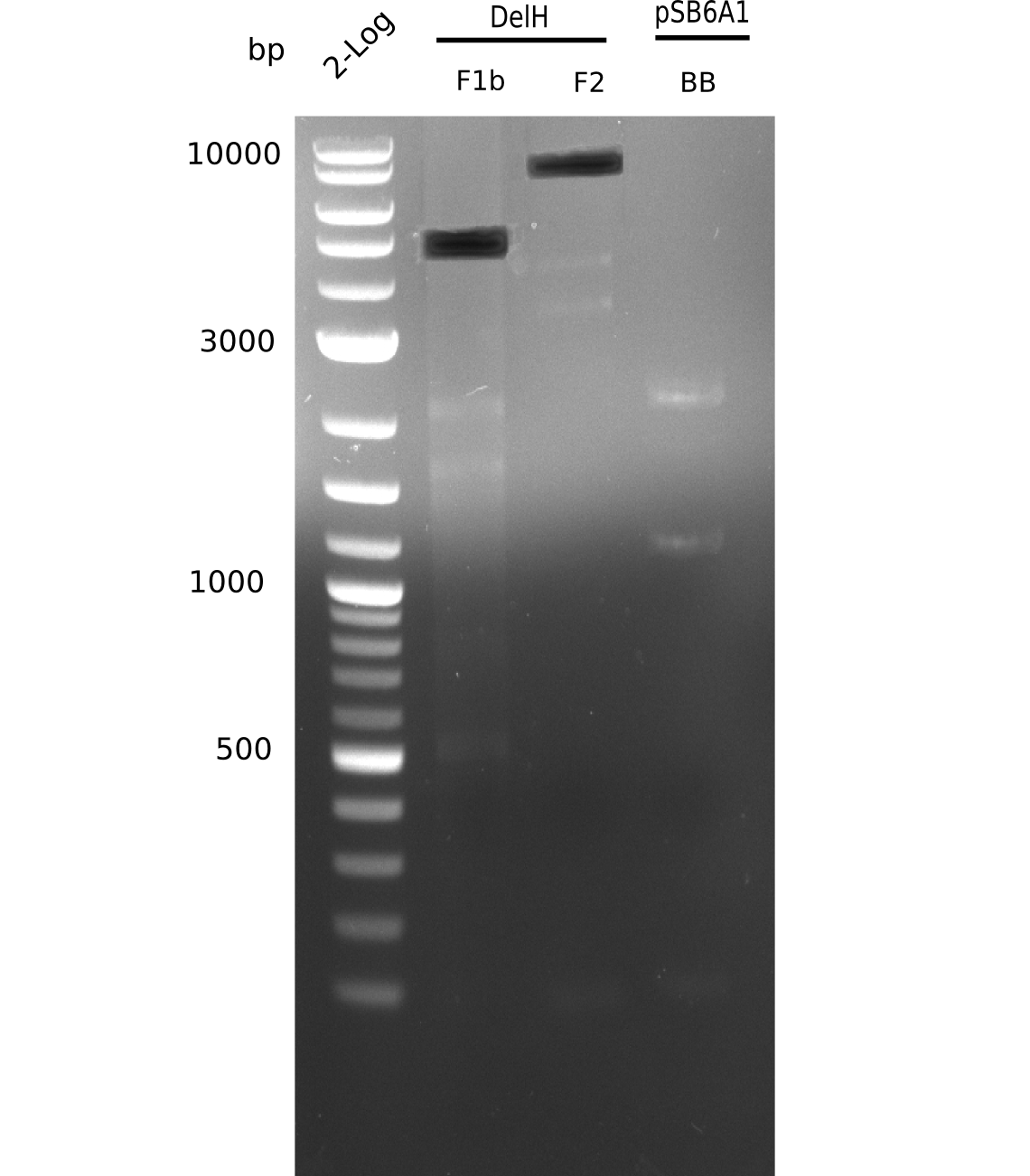
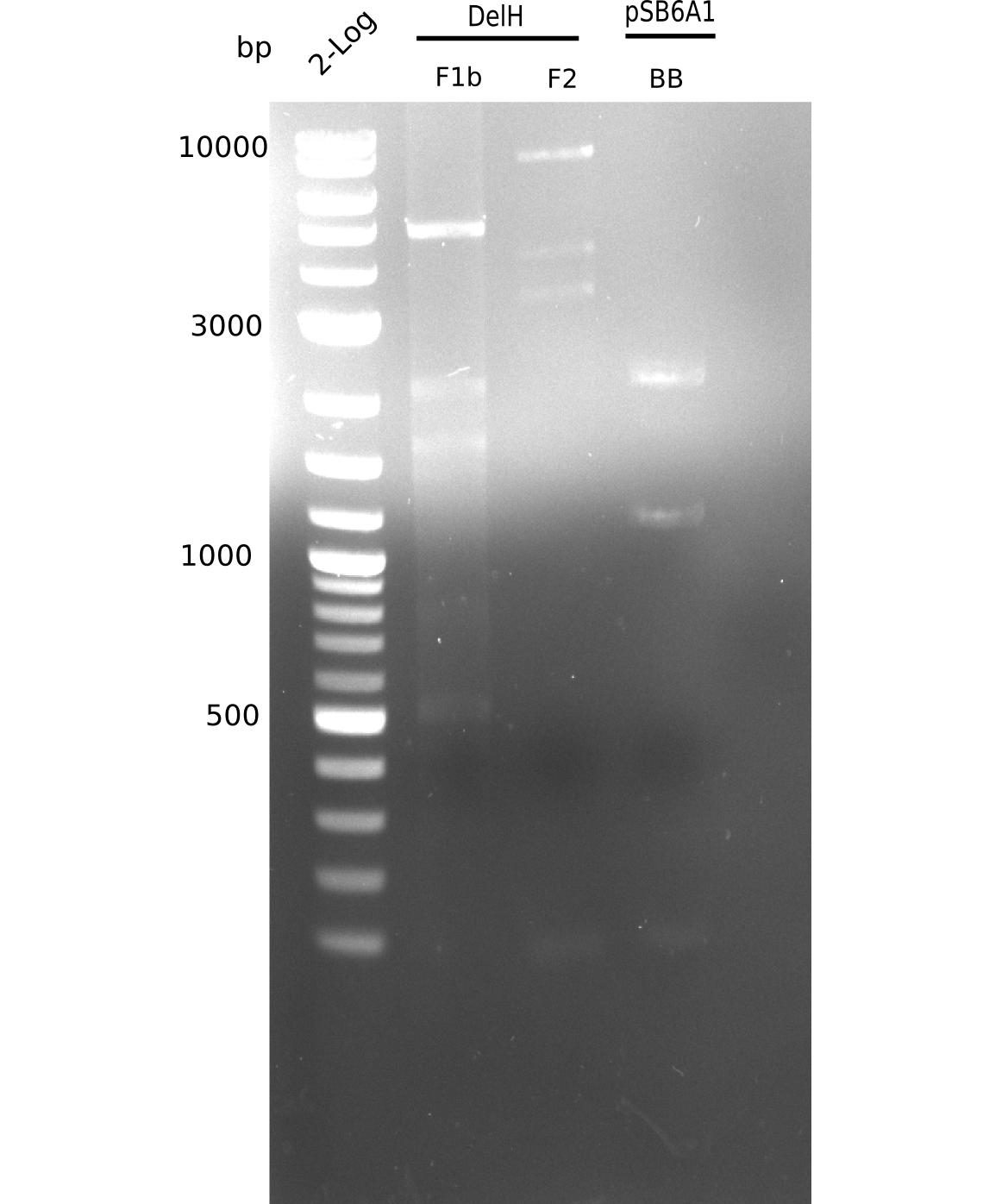
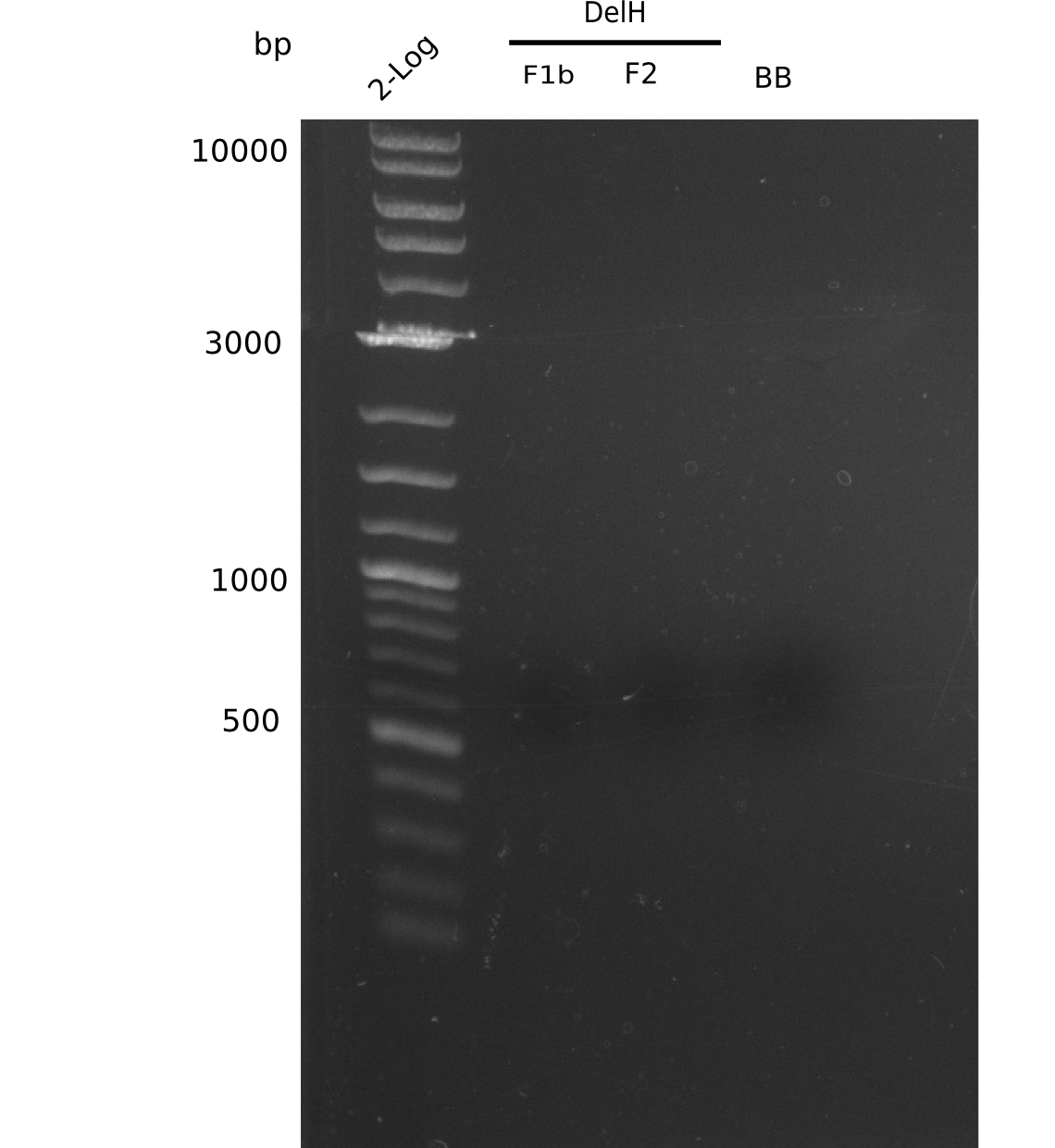
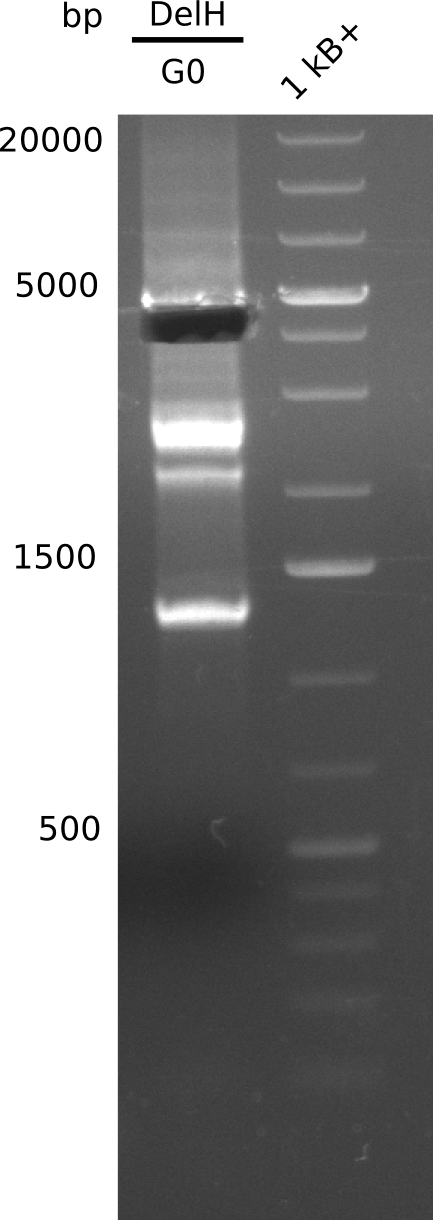
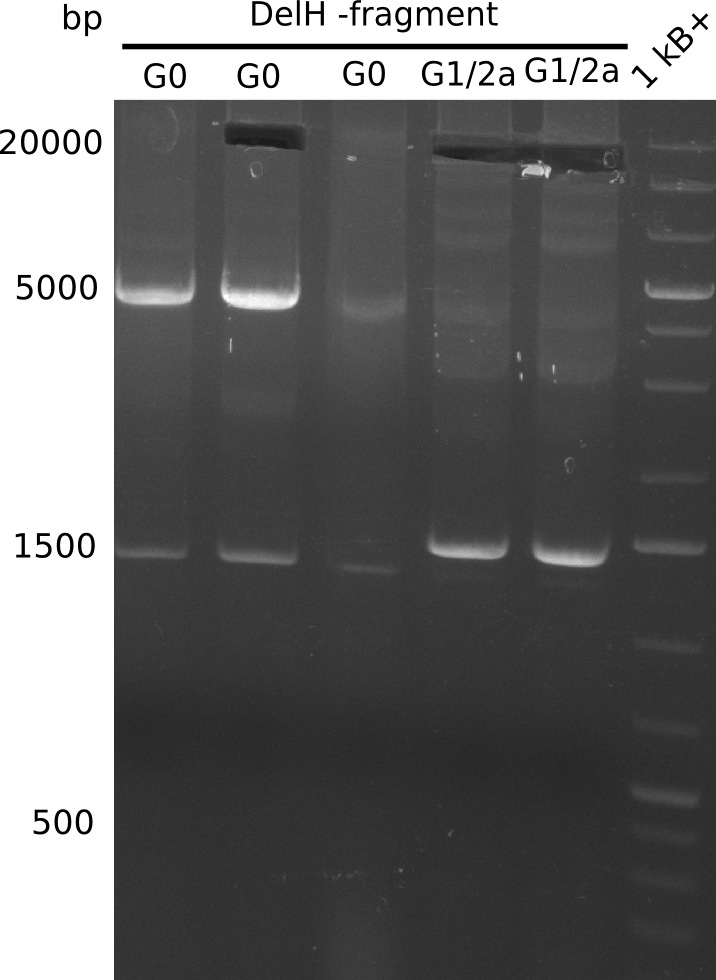
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week, we started over with the assembly of the backbone pSB6A1-AraC-lacZ: digesting AraC, lacZ and PSB6A1 based on the previously amplified fragments. The amplification of DelH was continued and we succesfully amplified all three fragments and gel-extracted them. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 103: | Line 104: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 6</h1> | <h1>Week 6</h1> | ||
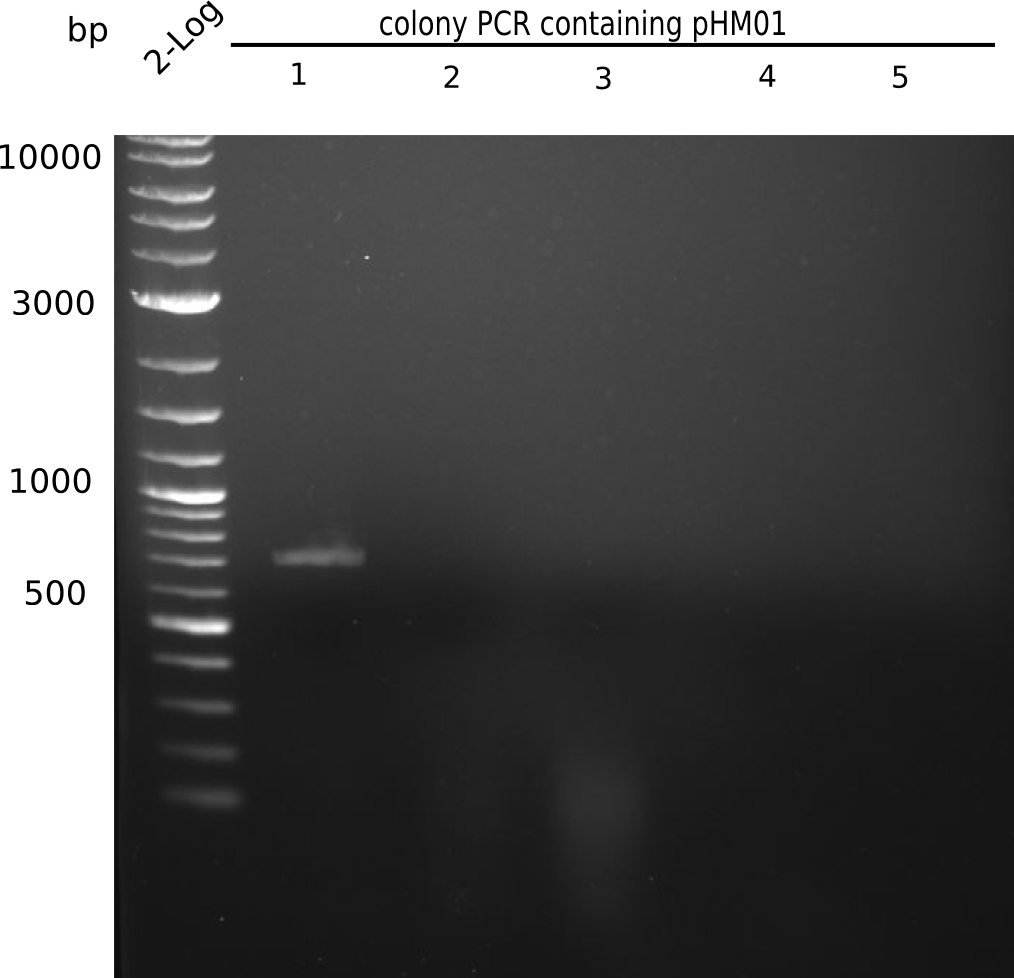
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since all DelH-fragments required for the final construct as well as the backbone were assembled in week 5, we tried to assemble the final plasmid pHM01. Therefore, every fragment was digested with two distinct enzymes, and then ligated. The ligated plasmid pHM01 was purified and electroporated in two separate DH10ß aliquots. The screening via colony-PCR was negative, so none of the transformed <i>E.coli</i> received the correct plasmid. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 111: | Line 112: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 7</h1> | <h1>Week 7</h1> | ||
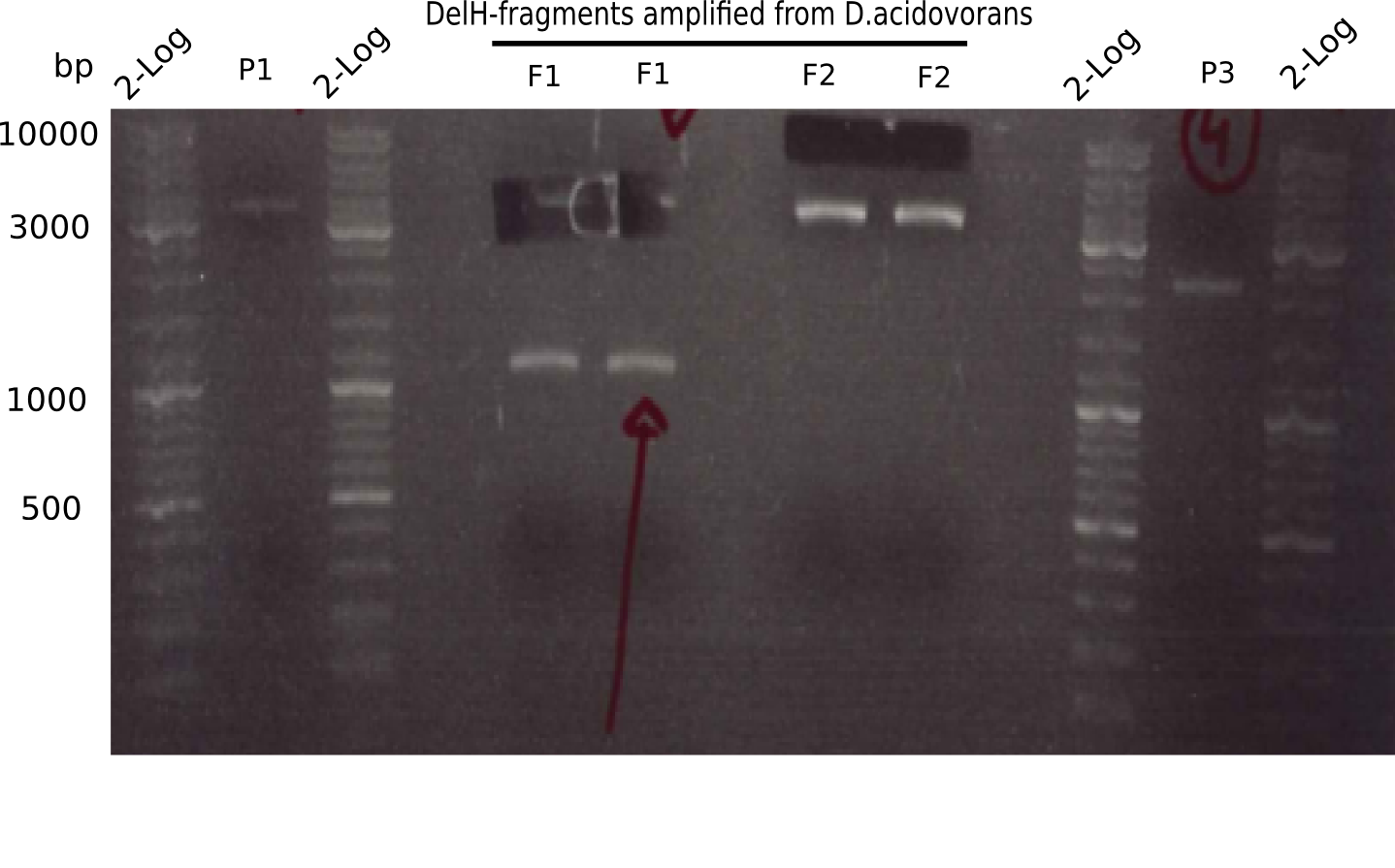
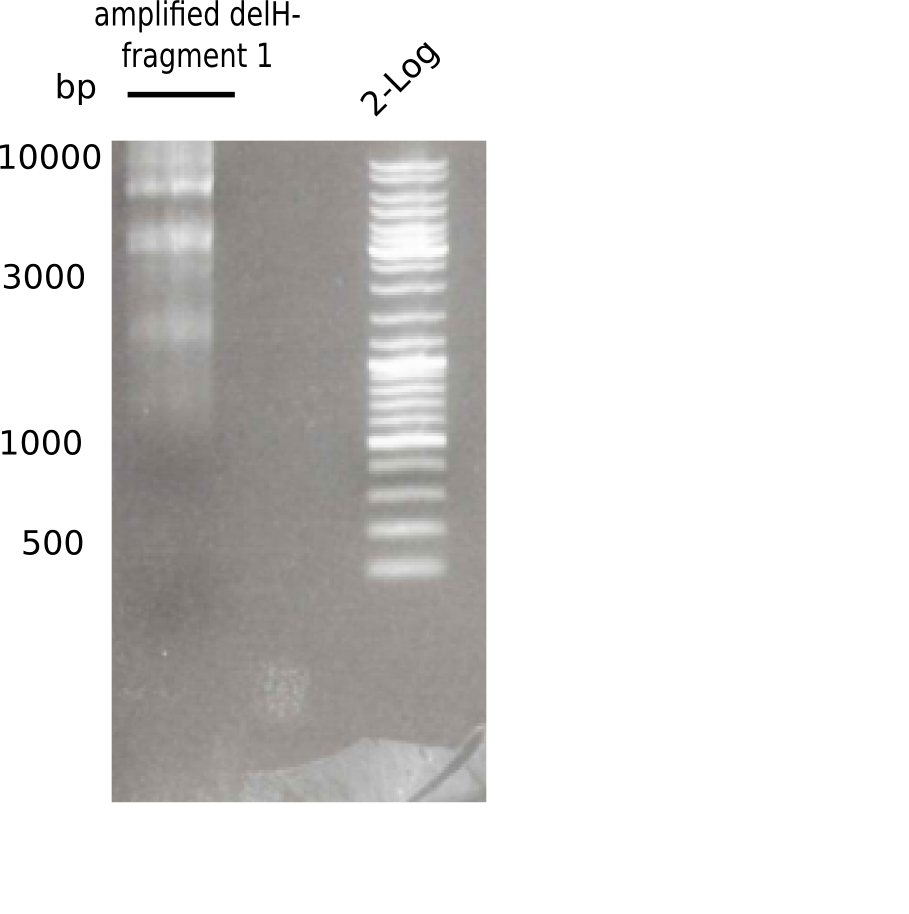
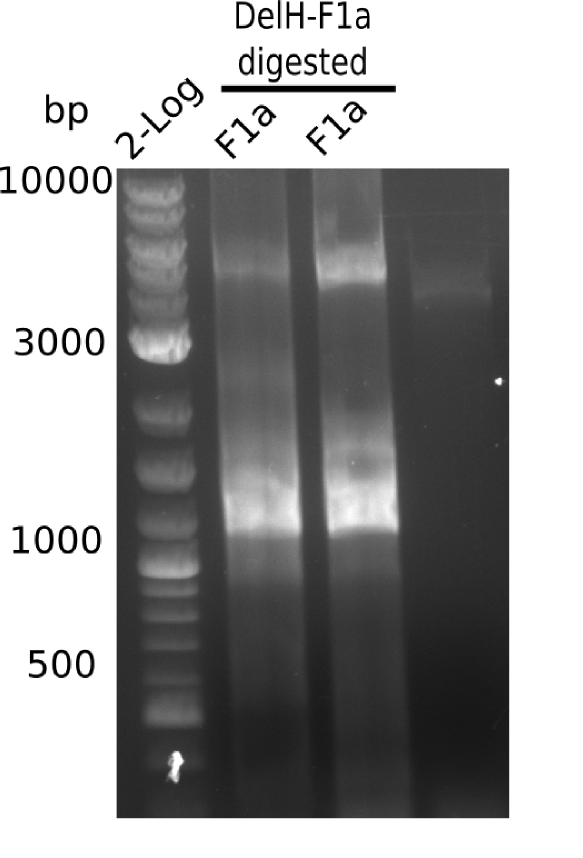
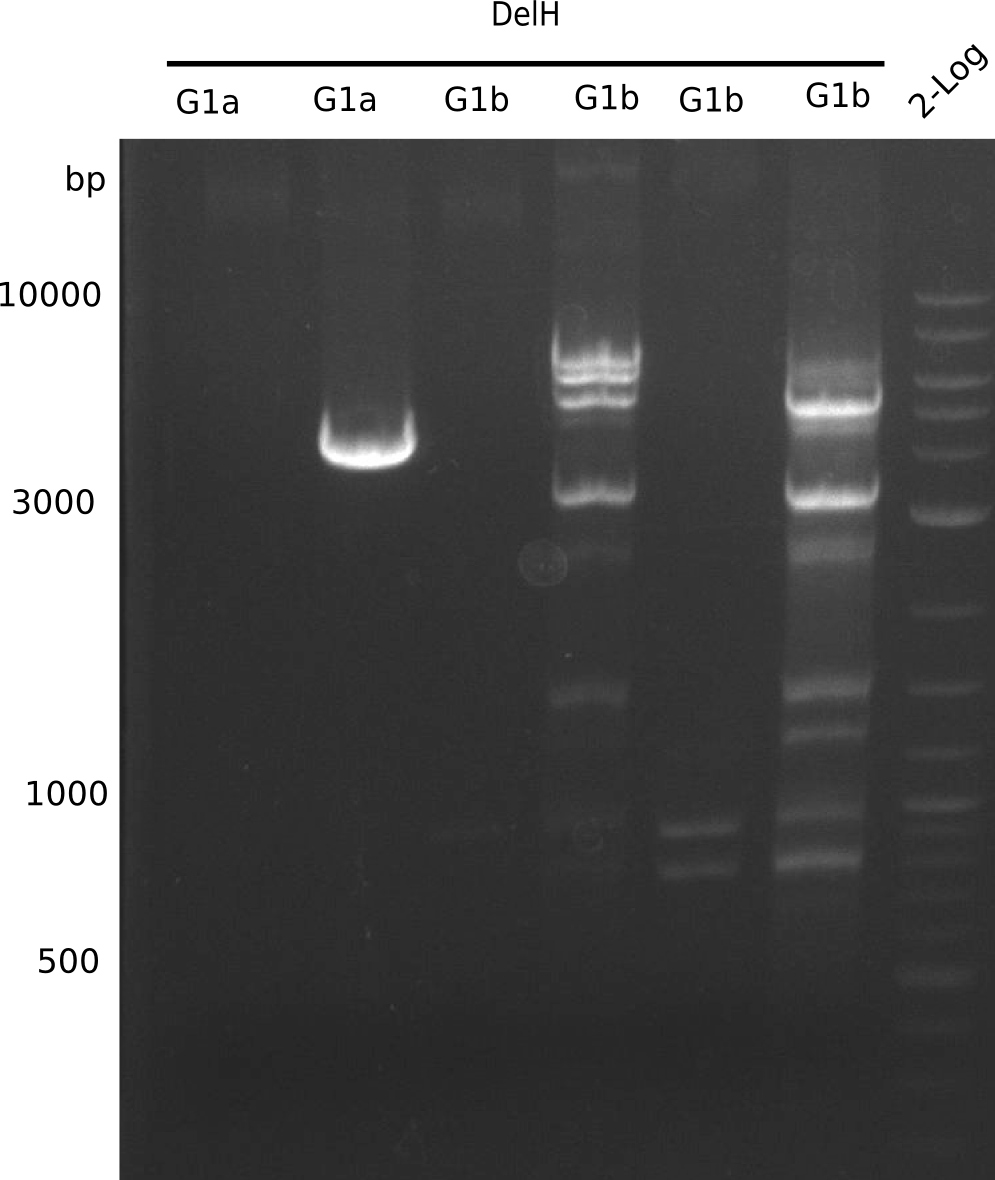
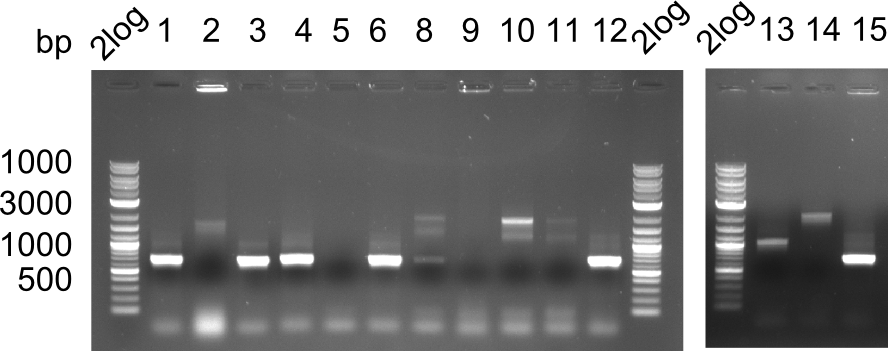
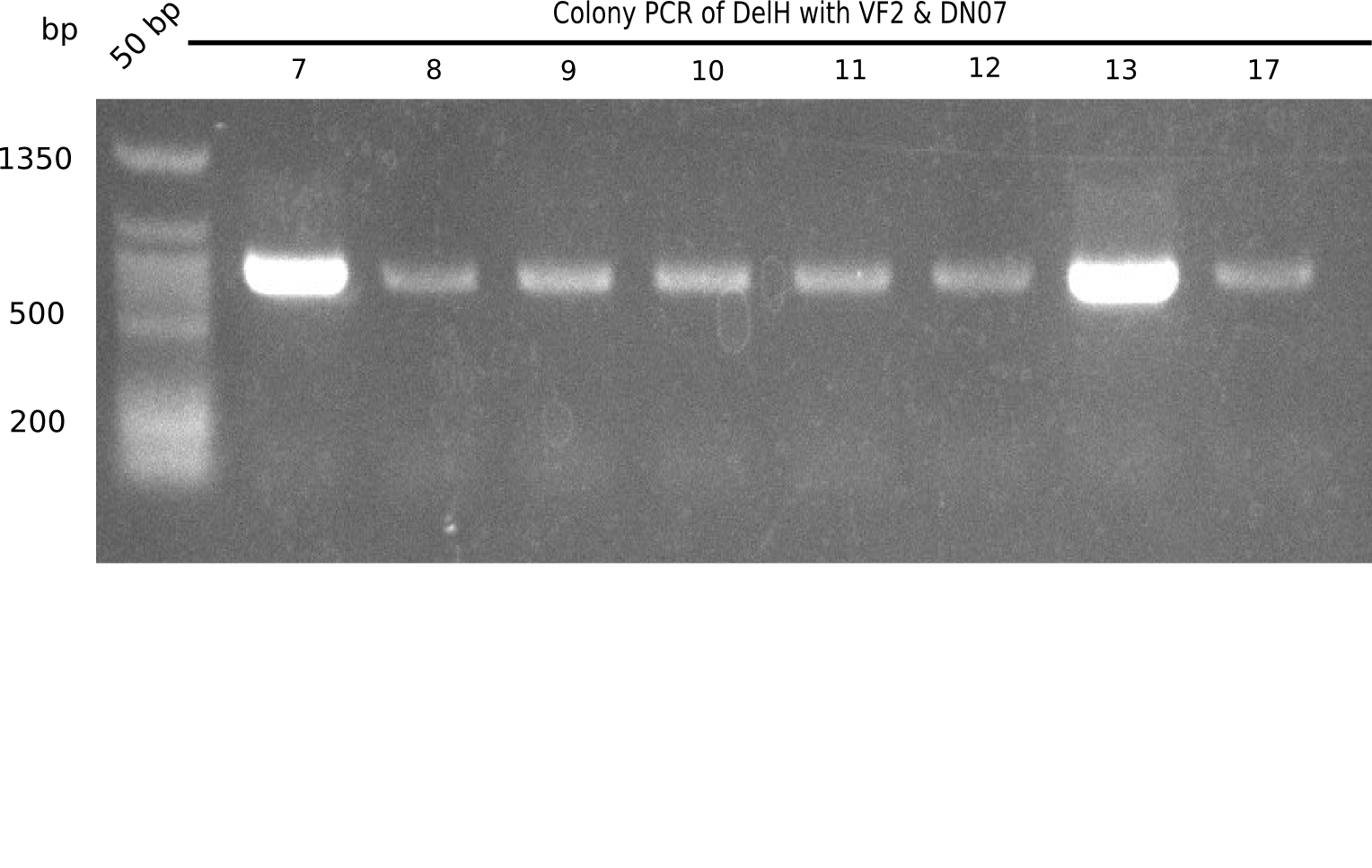
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 7, we reamplified the DelH fragments used for the transformation in week 6. We found | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 7, we reamplified the DelH fragments used for the transformation in week 6. We found that amplification of fragments F1a and F1b was not reproducible. Therefor, we designed new primers for DelH, which will allow to amplify the beginning of DelH in a more efficient and specific manner. The overview summarizes the primers we ordered and their performance in amplificating DelH F1a. Primer DN11 yielded best results and will be used in the next experiments. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 120: | Line 121: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 8</h1> | <h1>Week 8</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After improving the amplification of the | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After improving the amplification of the first subfragment of DelH (fragment 1a with the primer DN11), we used a higher concentration in the ligation assembling the pHM01 plasmid. The transformation of <i>E.coli</i> DH10ß cells was performed with electroporation. Four colonies were positive in the screening-PCR, but did not express lacZ (no blue color). To qualitatively determine DelH expression, we plan to conduct SDS-PAGEs of cell lysate derived from DelH transformed cells. Also, we will induce the transformed bacteria with higher concentrations of arabinose and X-Gal. |
</p> | </p> | ||
</div> | </div> | ||
| Line 127: | Line 128: | ||
<div class="item june last"> | <div class="item june last"> | ||
<div class="container"> | <div class="container"> | ||
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0 | + | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<h1>Week 9</h1> | <h1>Week 9</h1> | ||
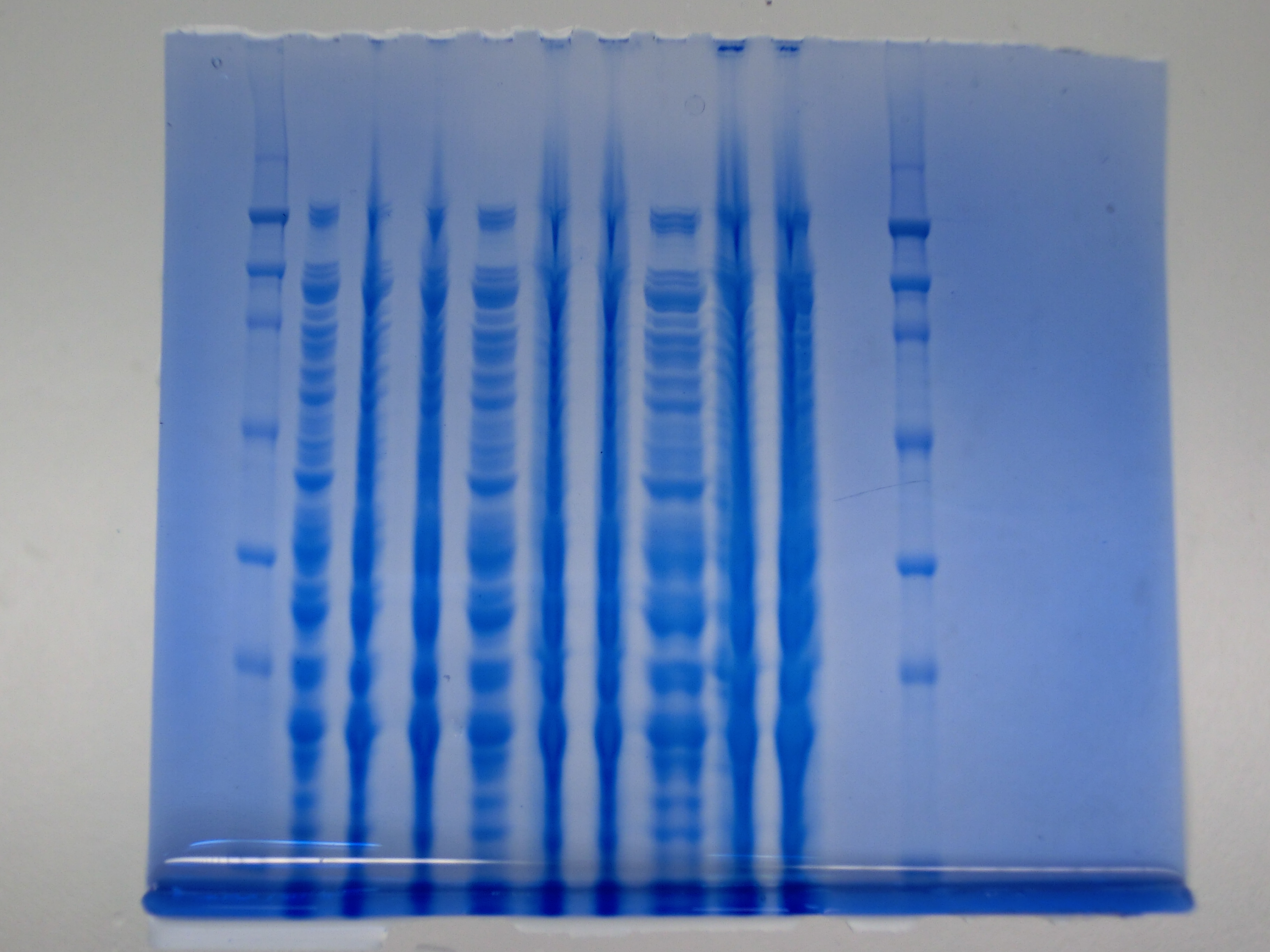
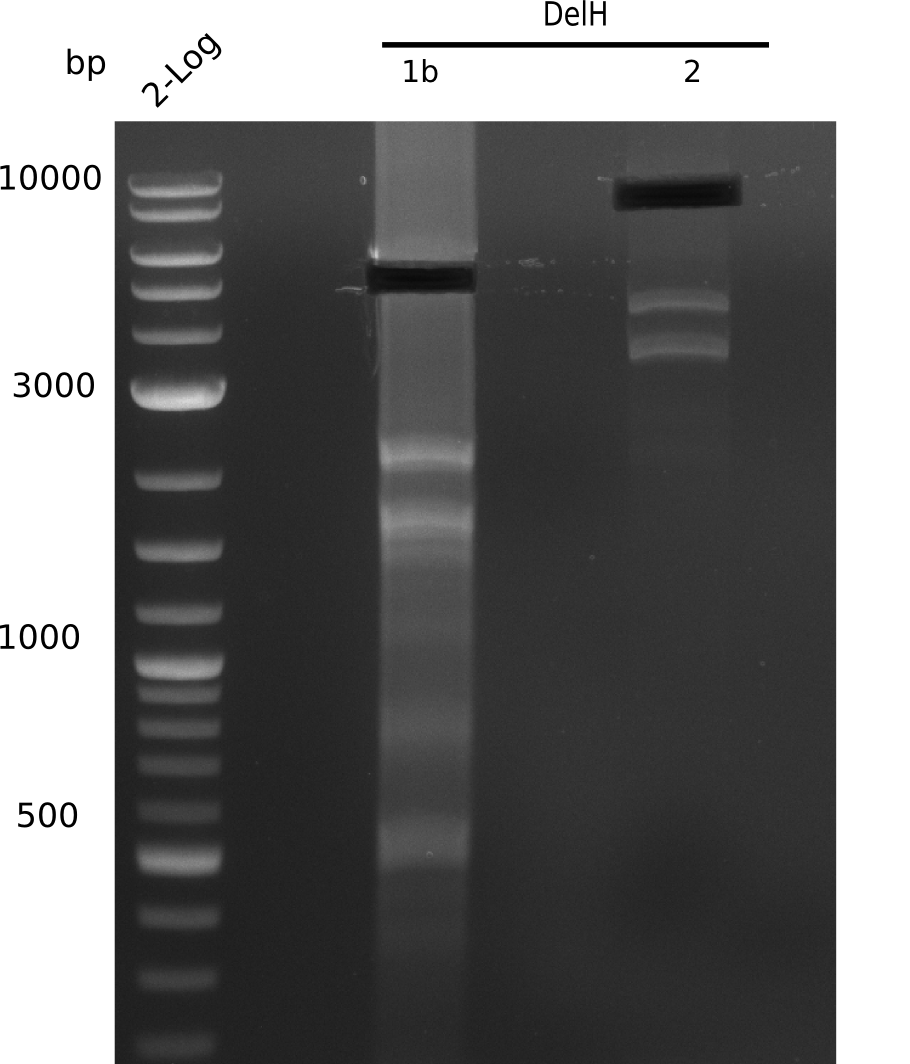
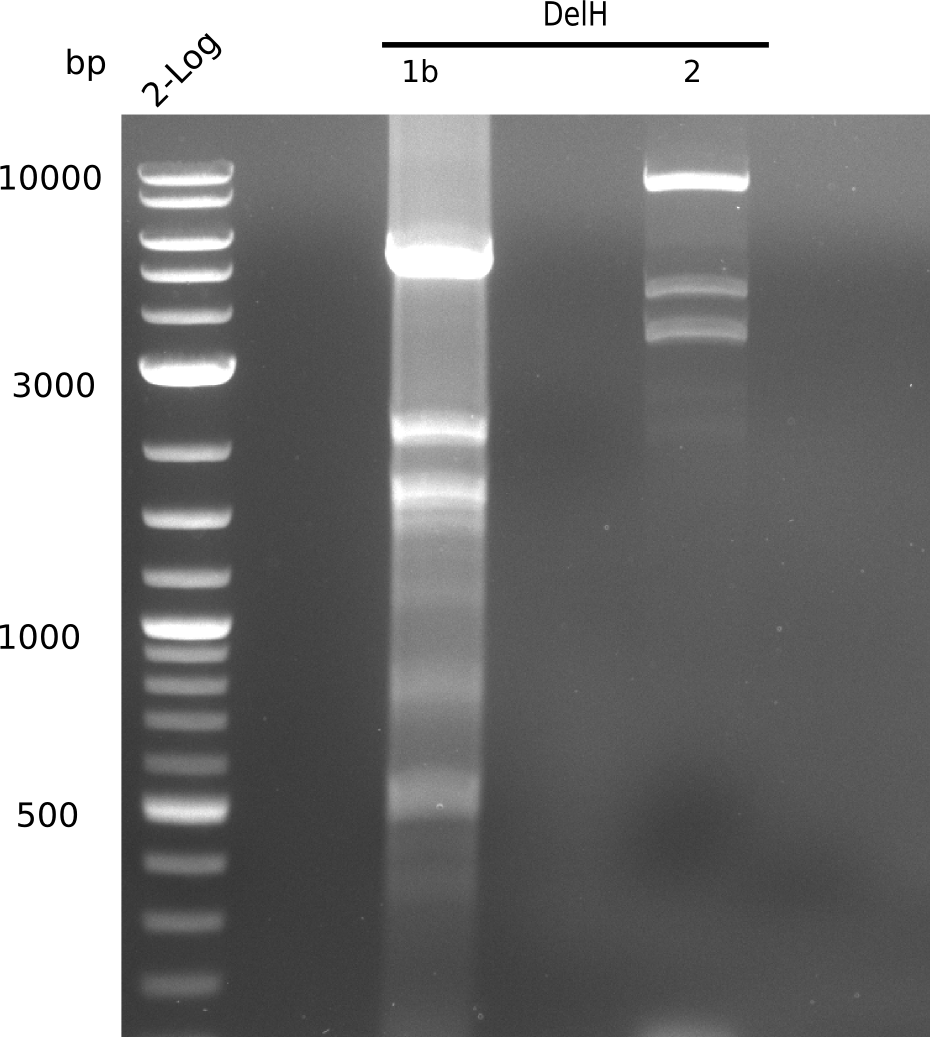
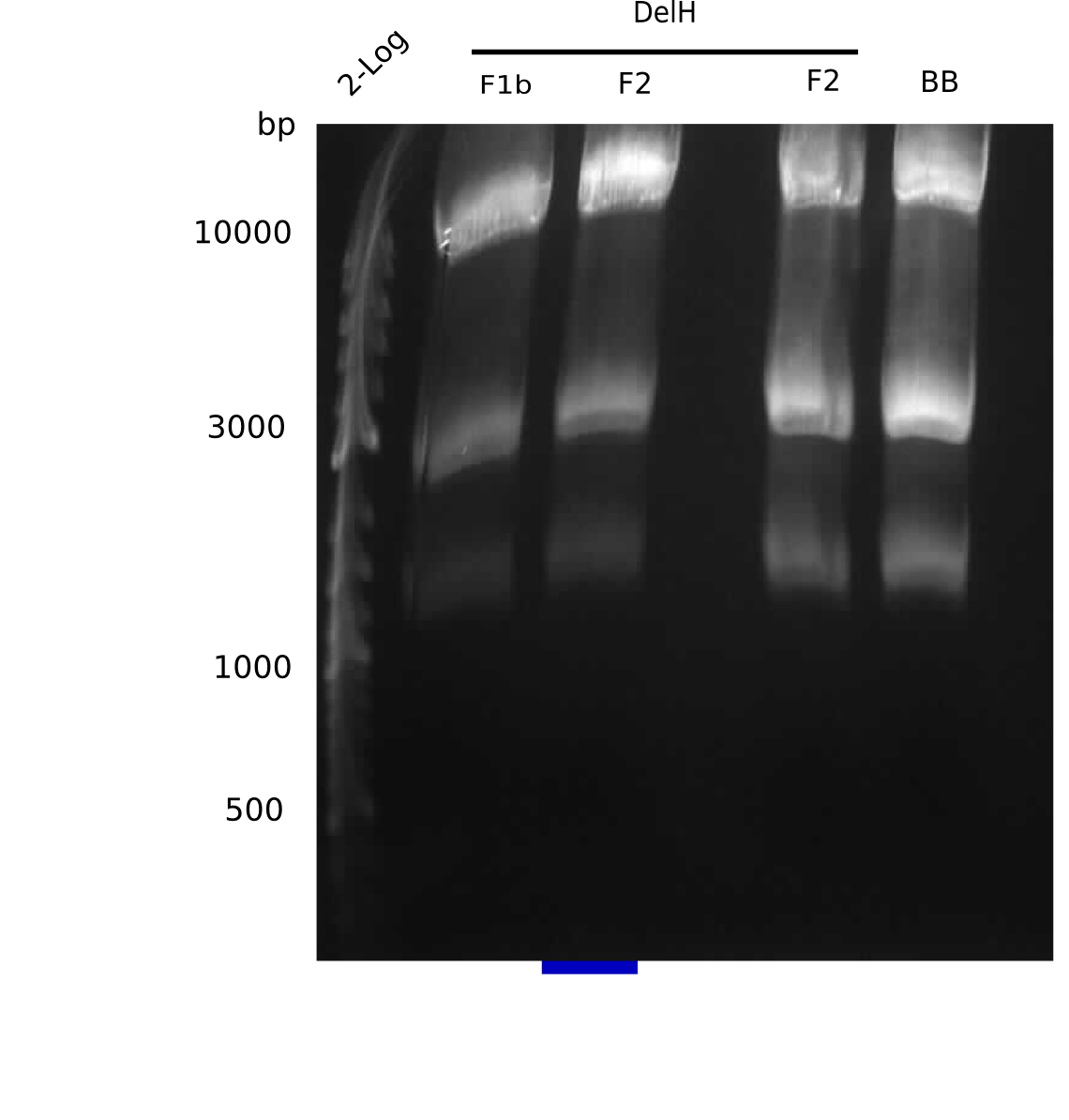
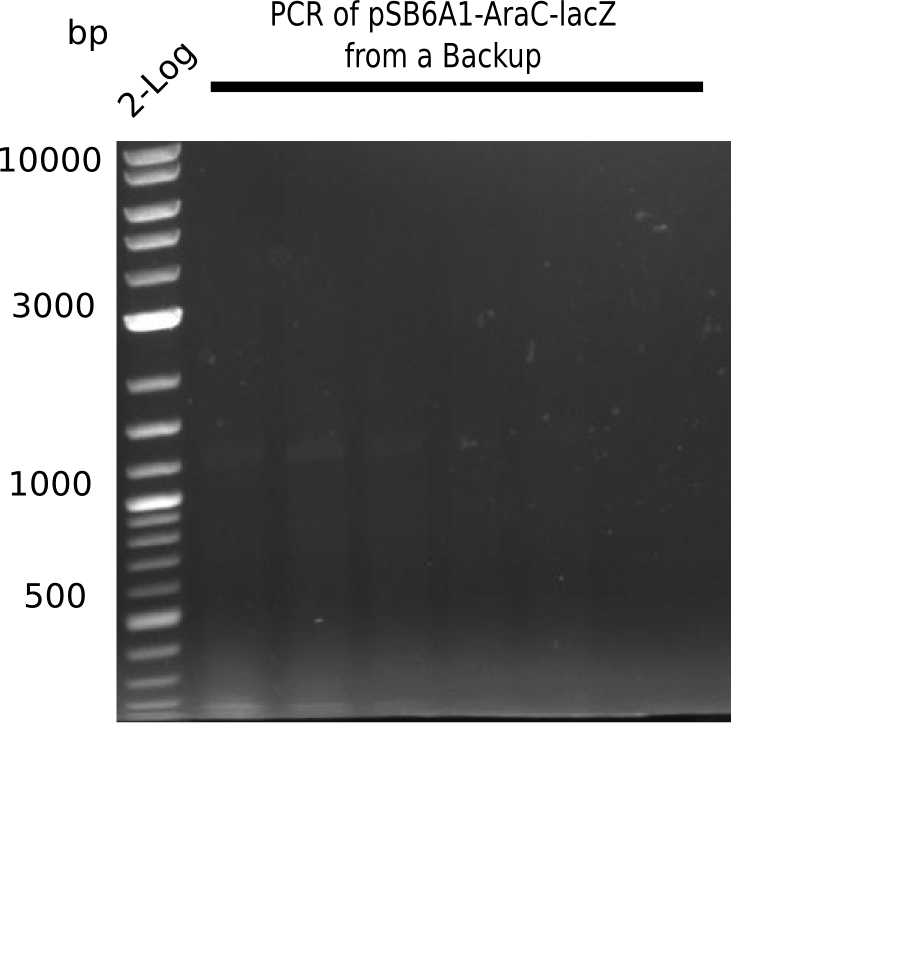
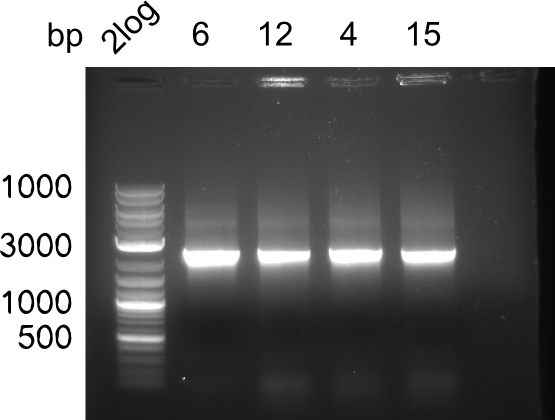
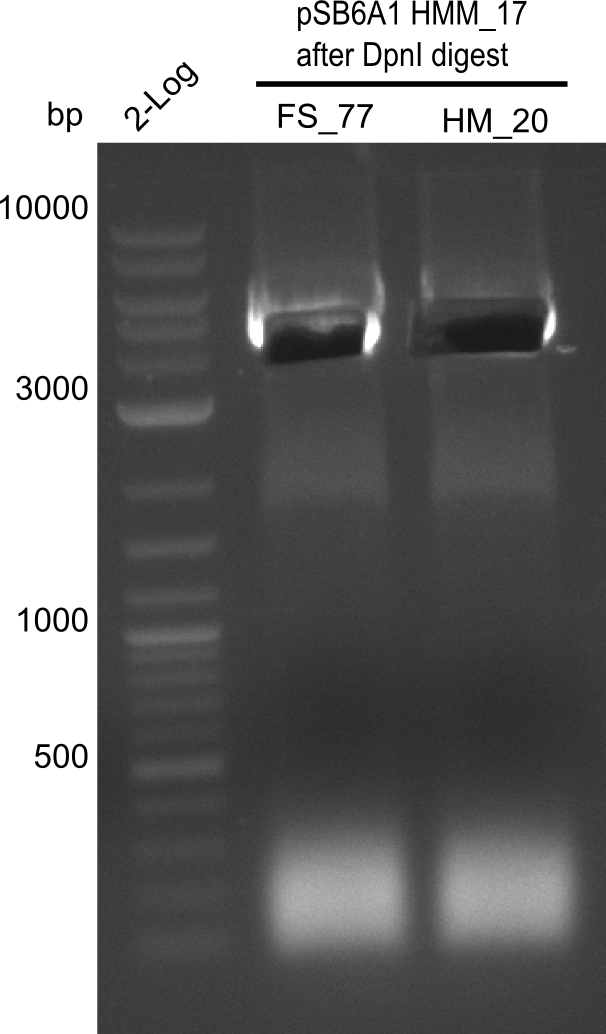
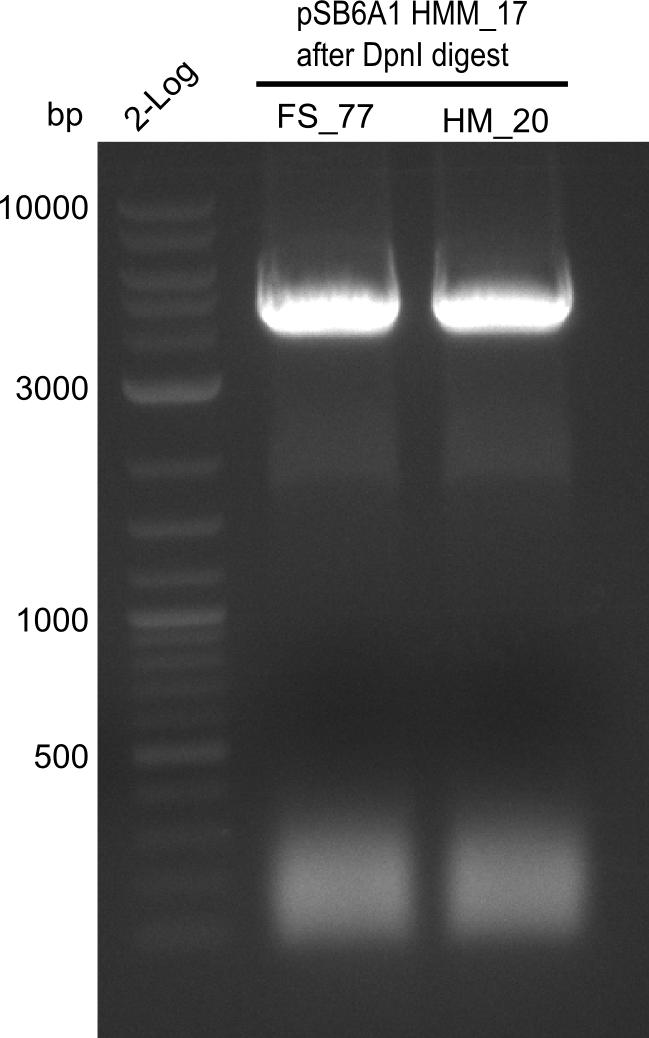
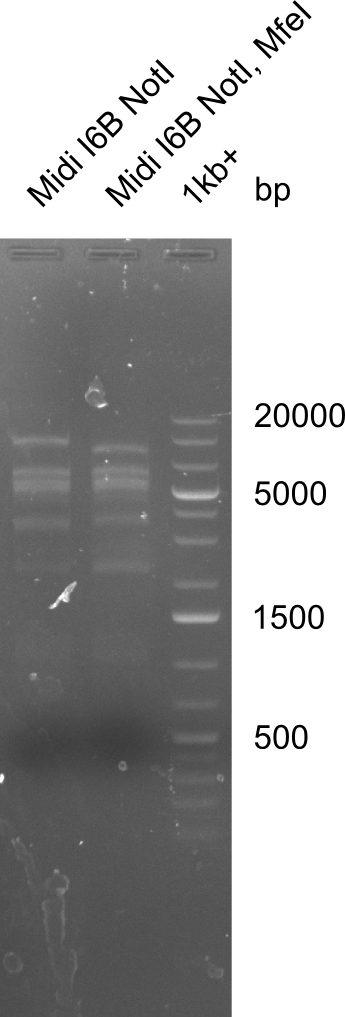
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> SDS-PAGE of cell lysate derived from DelH-transformed with subsequent coomassie staining did not yield a band at the expected protein size of ~600 kDa. The result was confirmed on DNA level, as the restriction digest of the DNA prepped from colonies of the transformed cells did not show the expected pattern. To perform a new assembly of plasmid pHM01, the fragments of DelH and the backbone had to be reamplified. The amplifications of the DelH fragments were successful (confirmed by gelelectrophoreses), but the backbone caused difficulties and we were not able to amplify it. |
</p> | </p> | ||
| Line 140: | Line 141: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 10</h1> | <h1>Week 10</h1> | ||
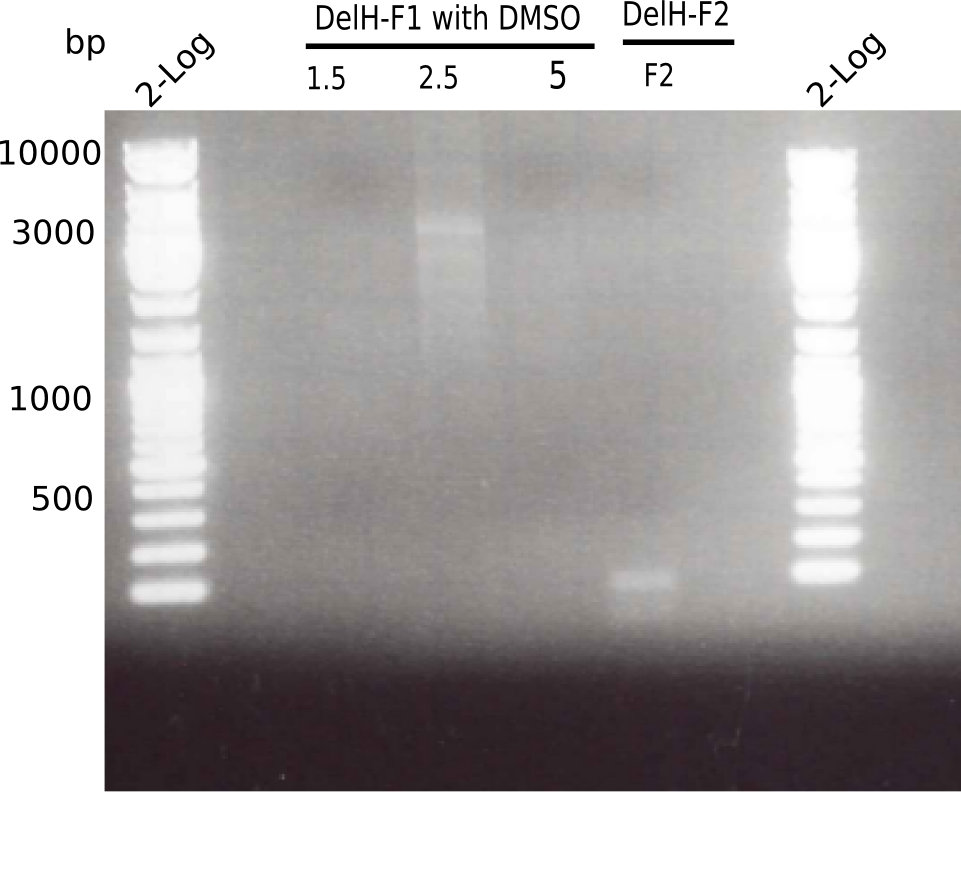
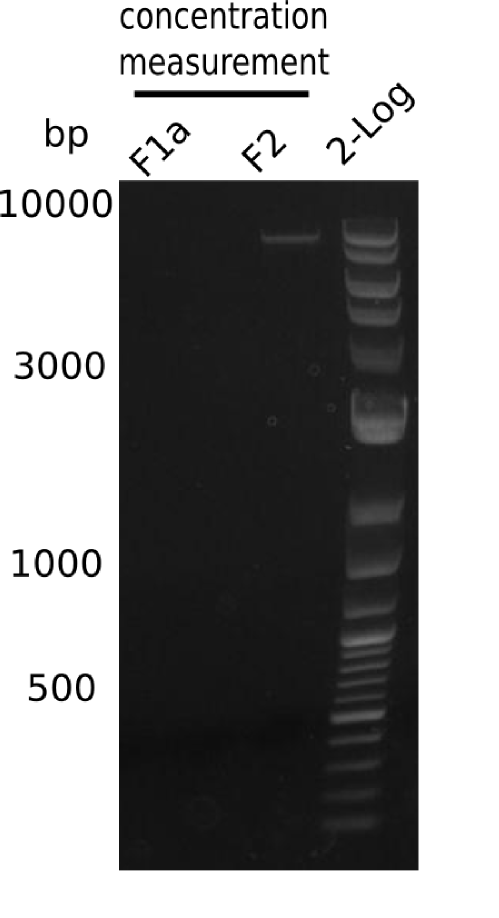
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After unsuccessful assembly attempts of pHM01, we tried to verify the previously amplified fragments. Two difficulties were encountered: On the one hand, the concentrations of the fragments were too low and could not be detected by gelelectrophoresis; on the other hand, a re-amplification of the fragments with the appropriate primers was not successful. For the DelH 1b fragment, the PCR products of the amplification from genomic DNA were slightly larger than the reamplifications. |
</p> | </p> | ||
</div> | </div> | ||
| Line 149: | Line 150: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 11</h1> | <h1>Week 11</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Based on this week's experiments and in silico analysis | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Based on this week's experiments and <i>in silico</i> analysis of the fragments DelH F1a, F1b and F2, we discarded the restriction digest and ligation strategy for the assembly of the DelH plasmid. Instead, we chose Gibson assembly as an alternative method, developed a strategy and designed primers accordingly. |
</p> | </p> | ||
</div> | </div> | ||
| Line 158: | Line 159: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 12</h1> | <h1>Week 12</h1> | ||
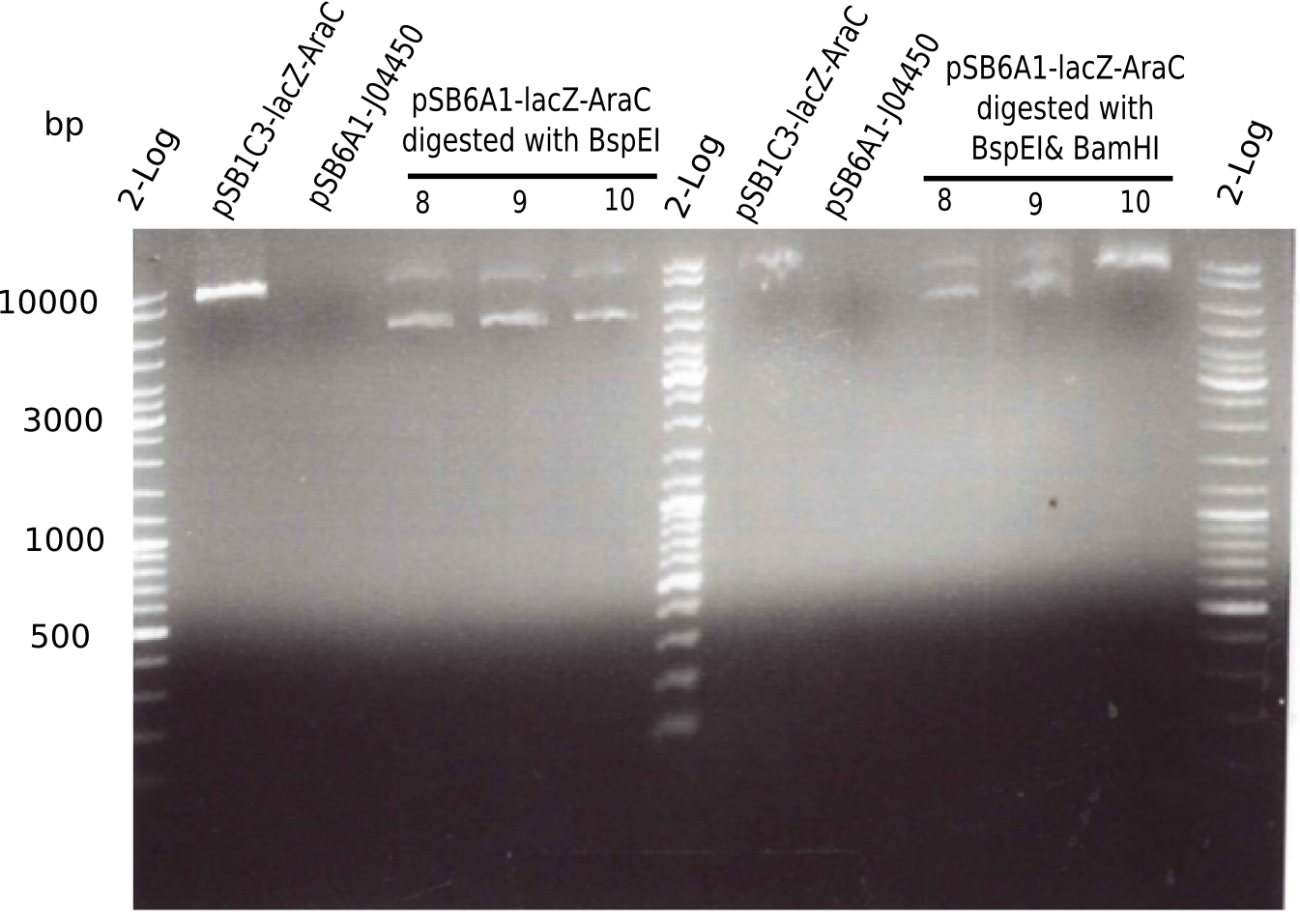
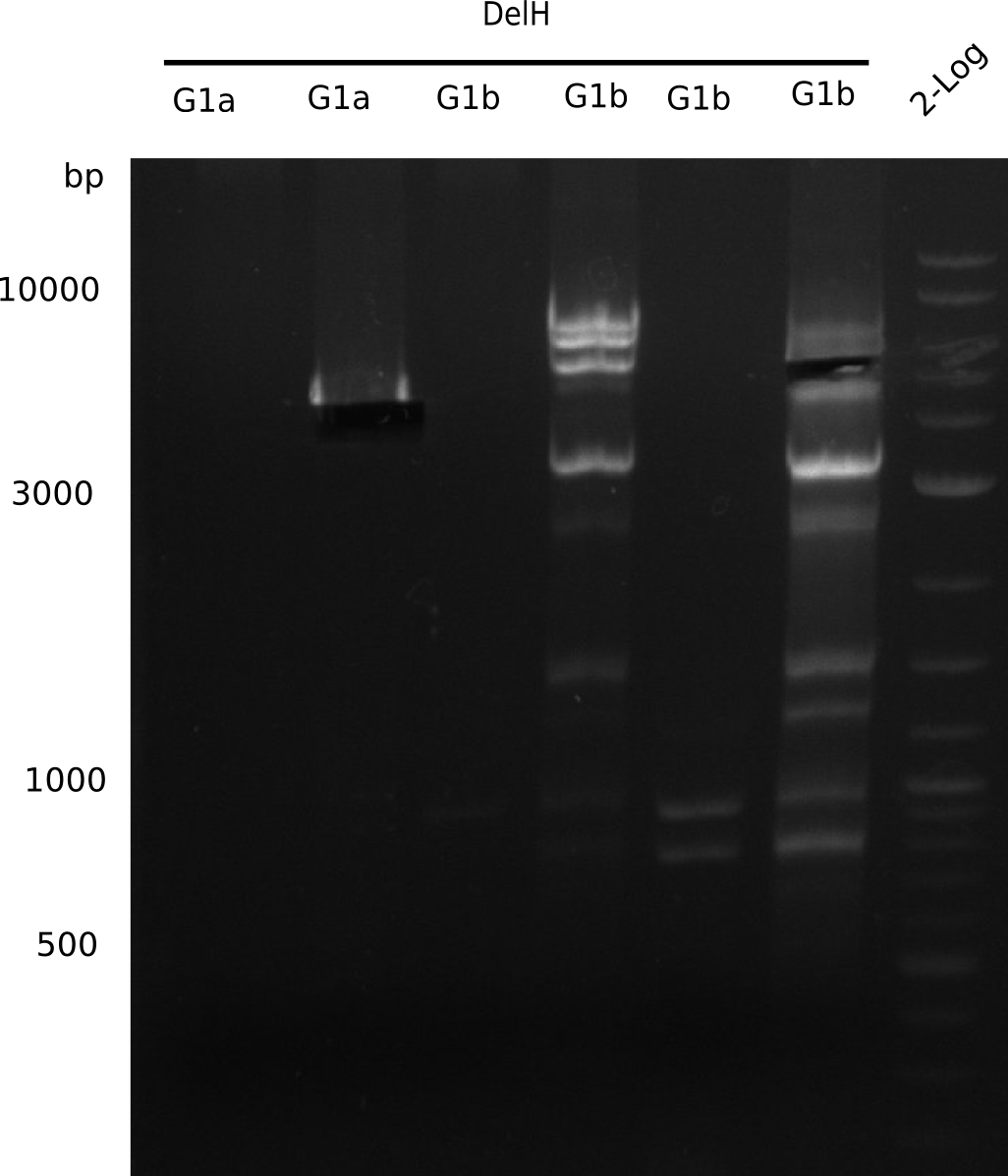
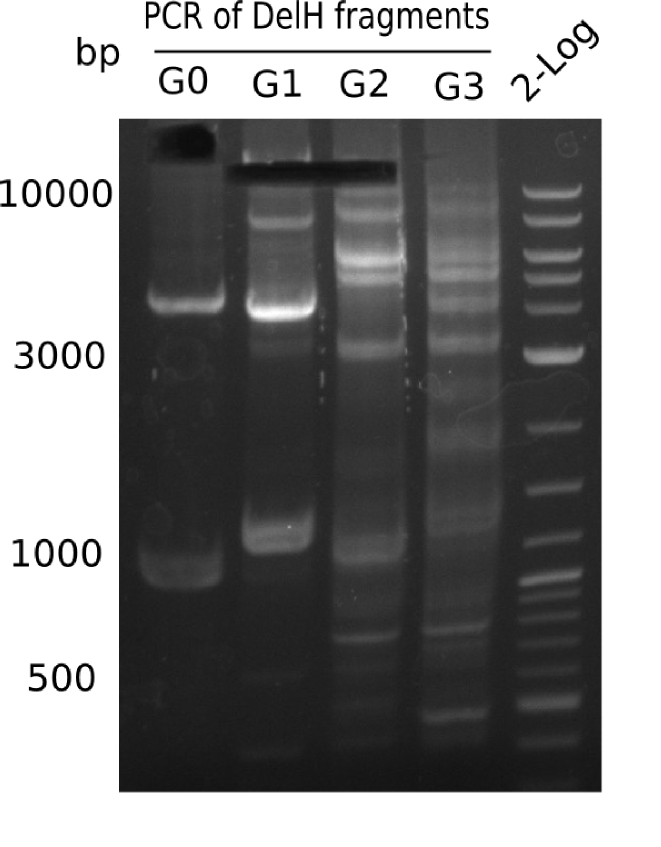
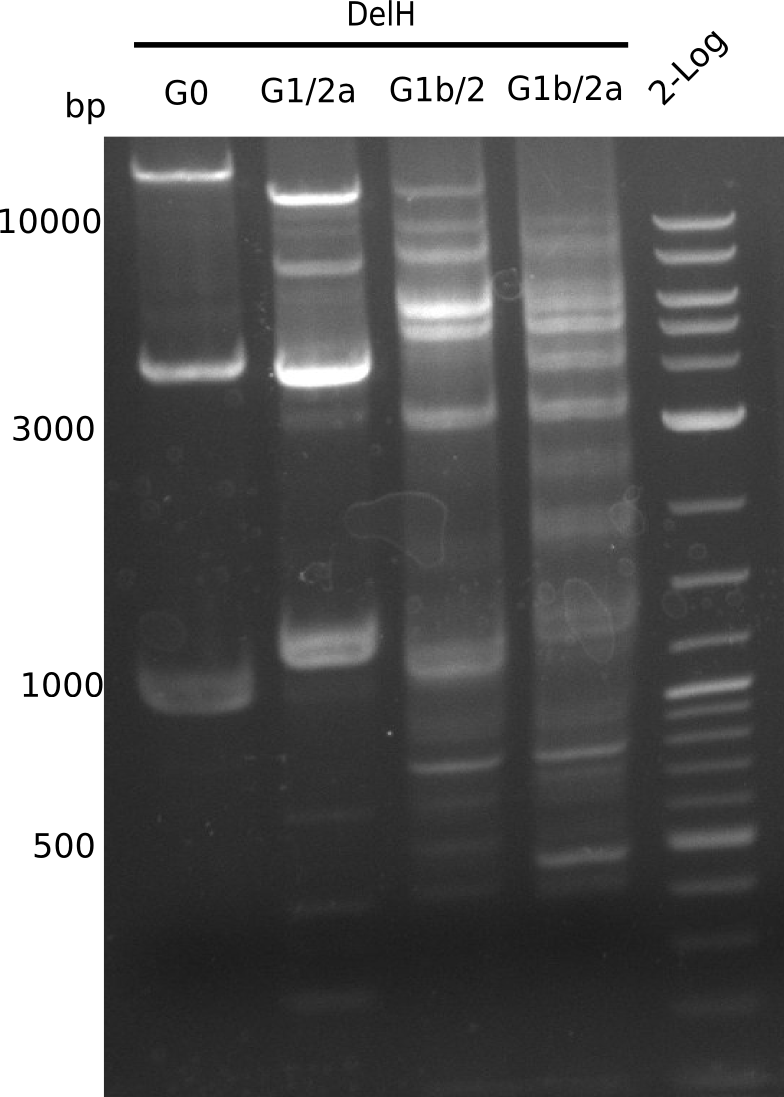
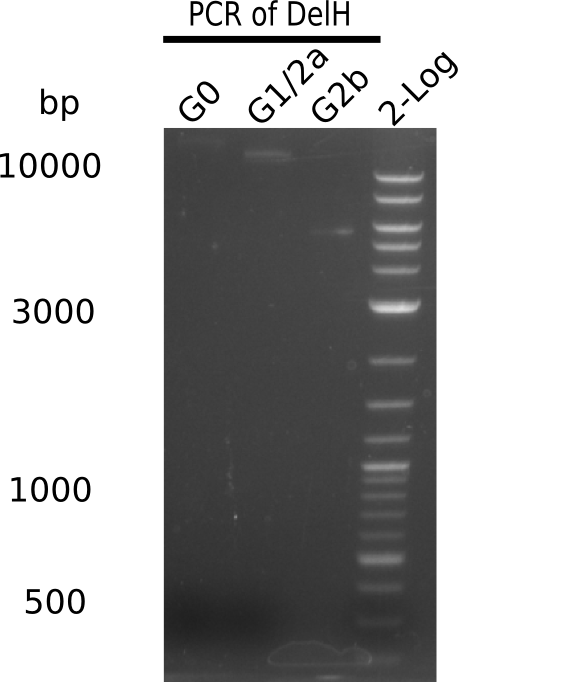
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We started amplifying the Gibson fragments using different PCR approaches. Since we were not able to amplify G1 by PCR, we decided to divide G1 into the subfragments G1a and G1b, for which we had already designed and ordered primers. The amplification of fragment G2 worked well and yielded sufficient DNA amounts for subsequent steps. Alternatively, we also produced the entire DelH G0 as one fragment, as well as further subfragments DelH G1/2a, G1b/2 and G2b. Amplification of G1b/2a did not work out. Due to the failure of the restriction digest strategy, we further analyzed the backbone pSB6A1-AraC-lacZ, which we decided to discard. Instead, we choose to use pSB6A1 and BBa_J04450, which is already available in the parts registry. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 167: | Line 168: | ||
<h1>Week 13</h1> | <h1>Week 13</h1> | ||
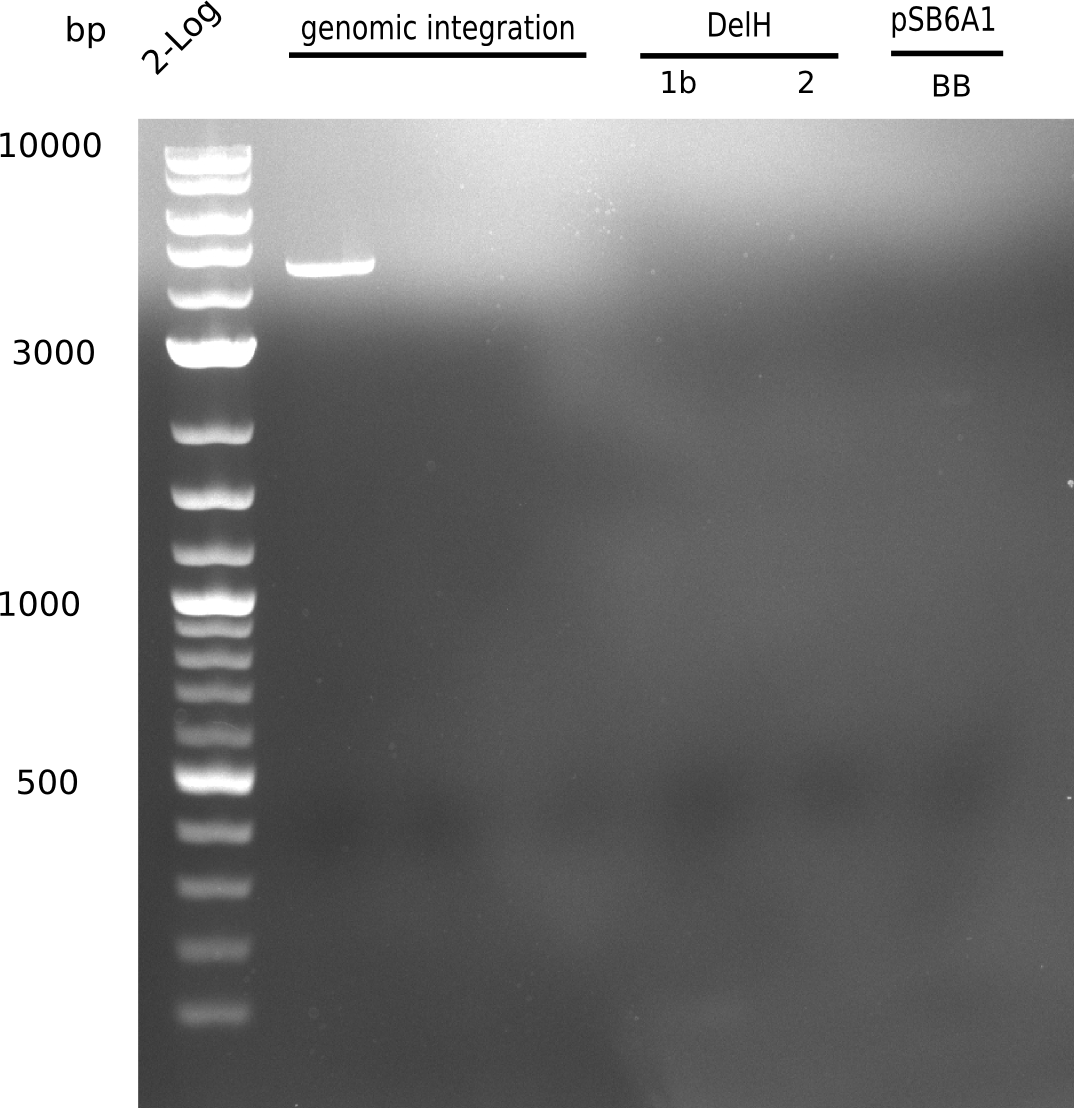
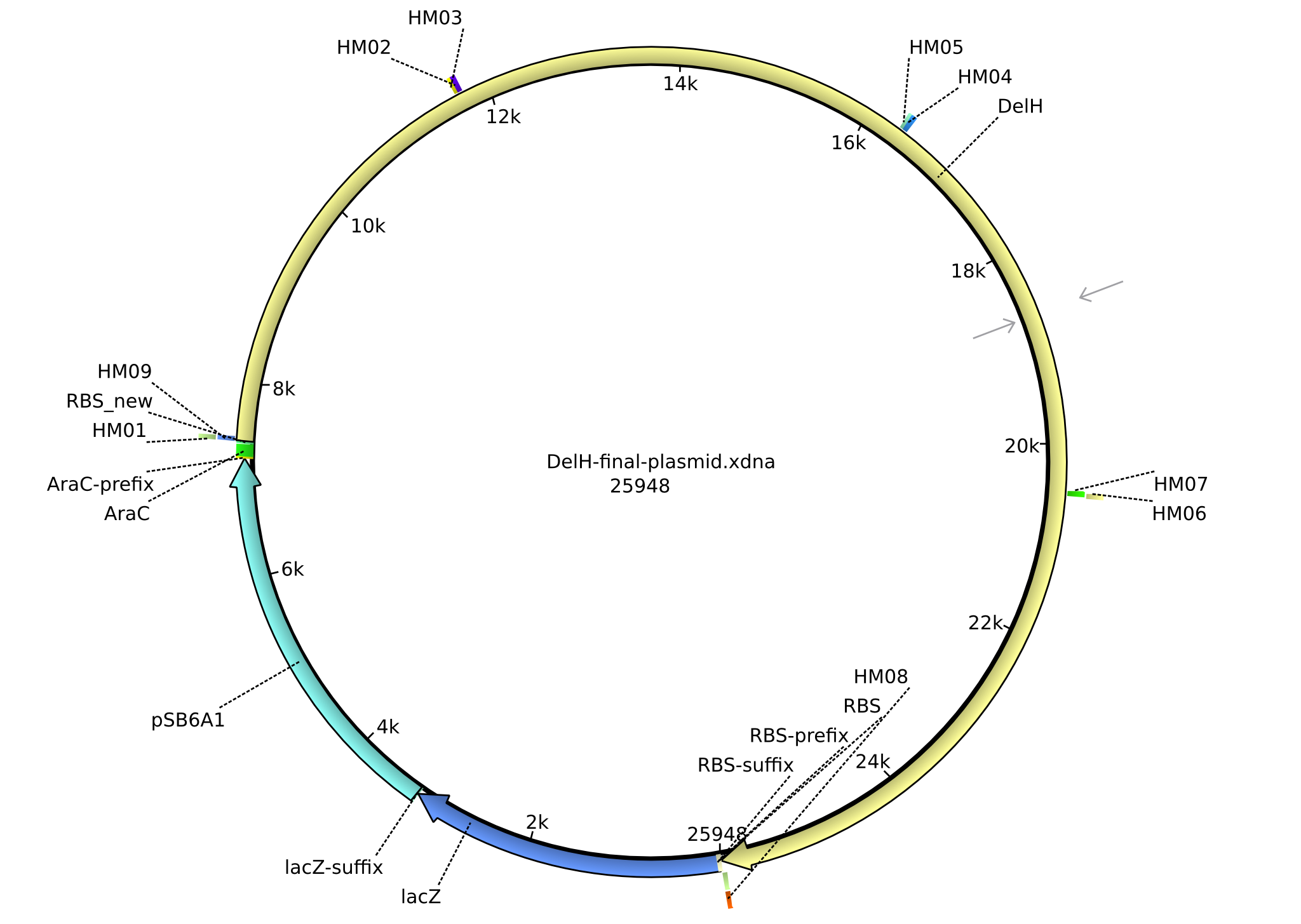
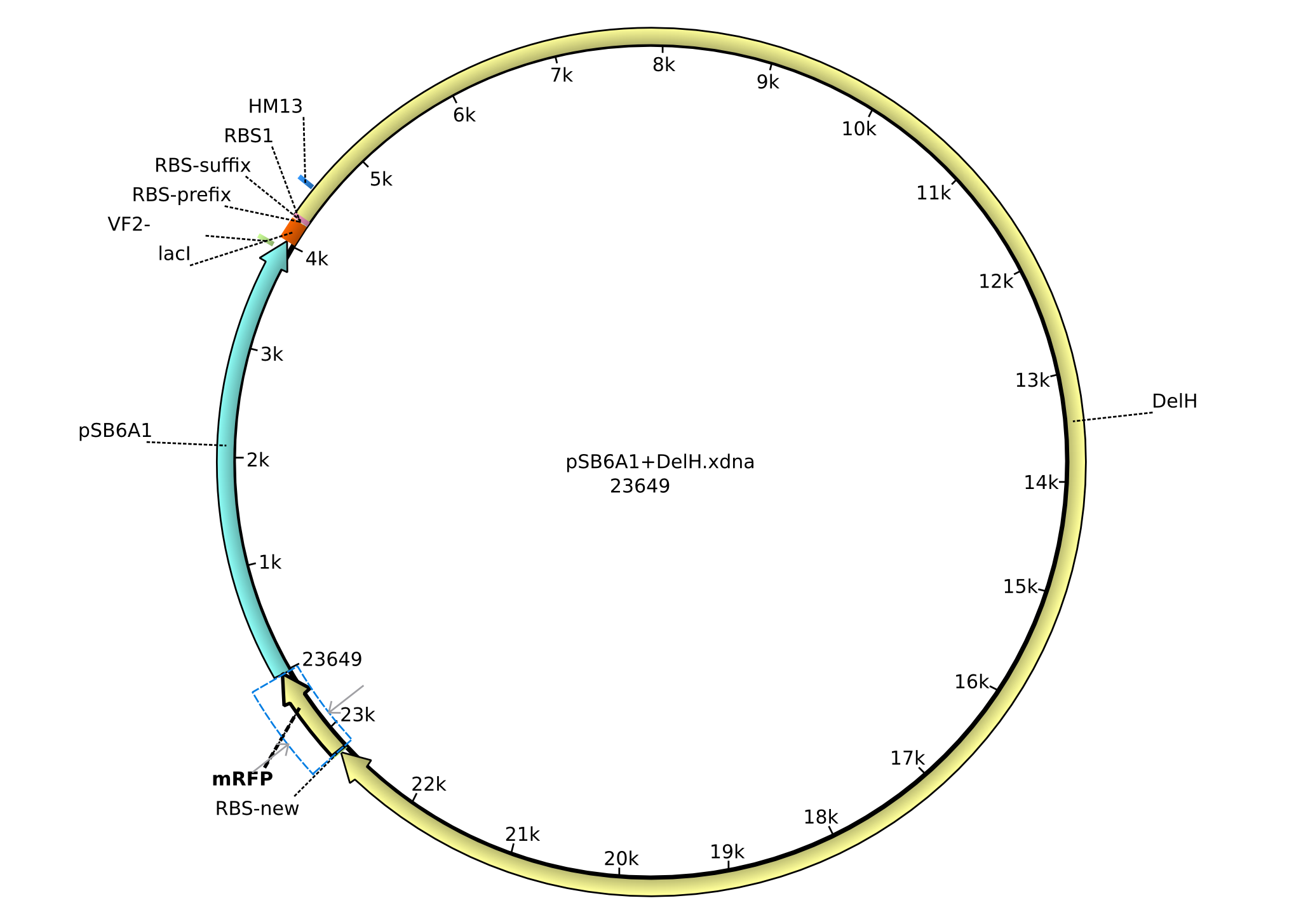
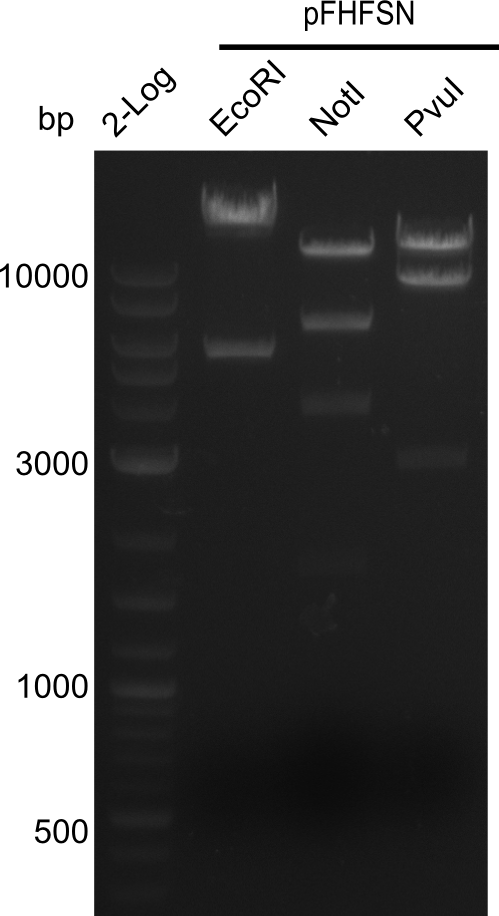
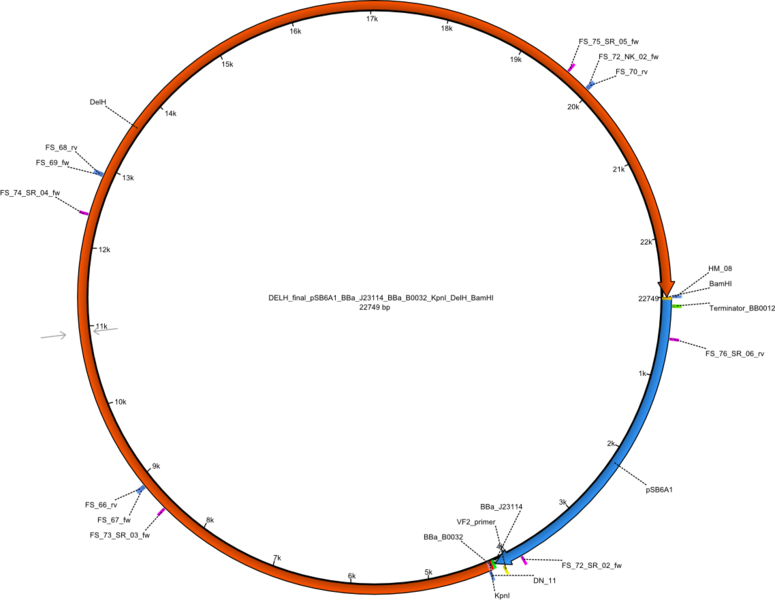
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to realize the new strategy using the already existing backbone from the parts registry pSB6A1-lacZ-mRFP, we designed two new primers for the backbone amplification and created a map of pHM03. | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to realize the new strategy using the already existing backbone from the parts registry pSB6A1-lacZ-mRFP, we designed two new primers for the backbone amplification and created a map of pHM03. | ||
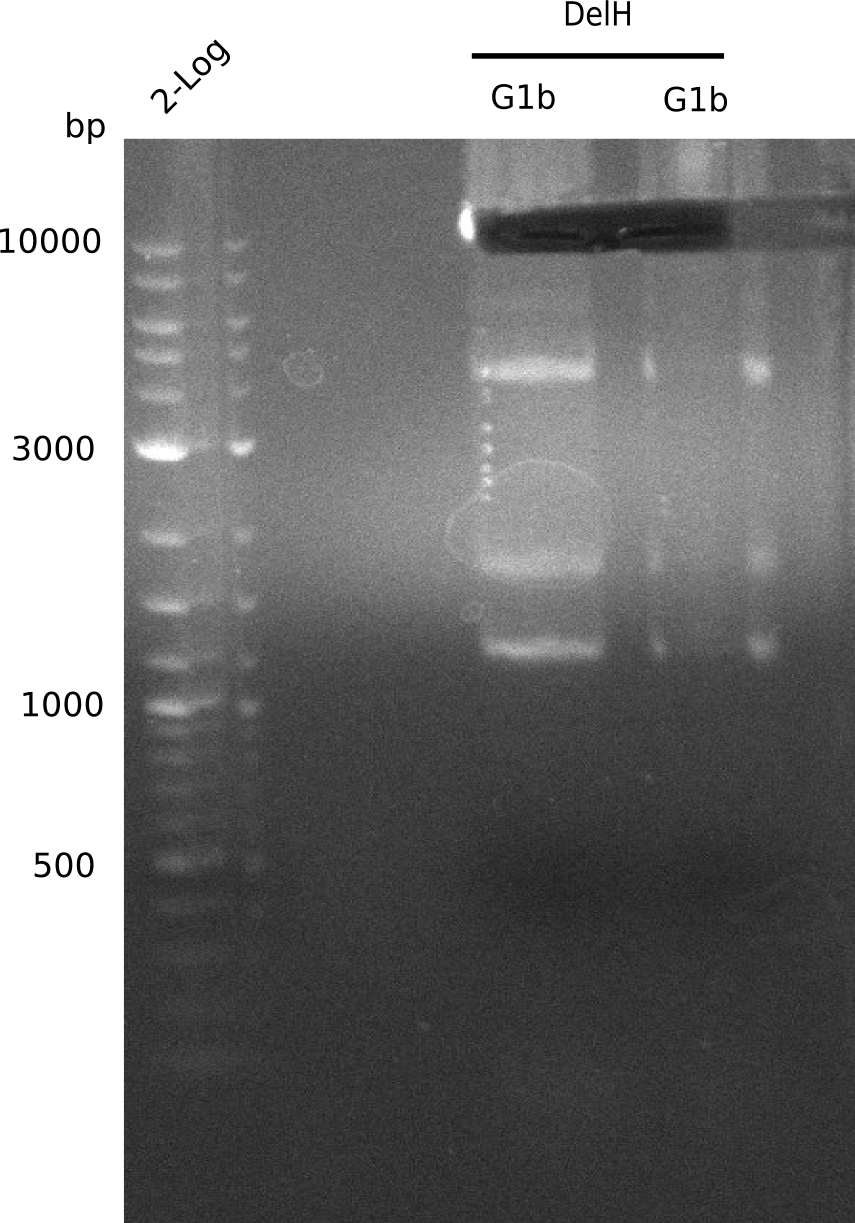
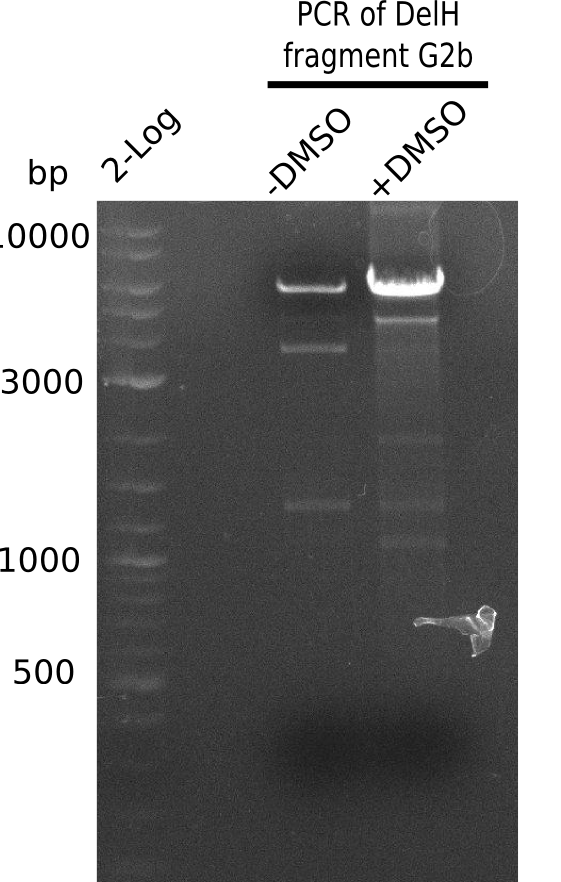
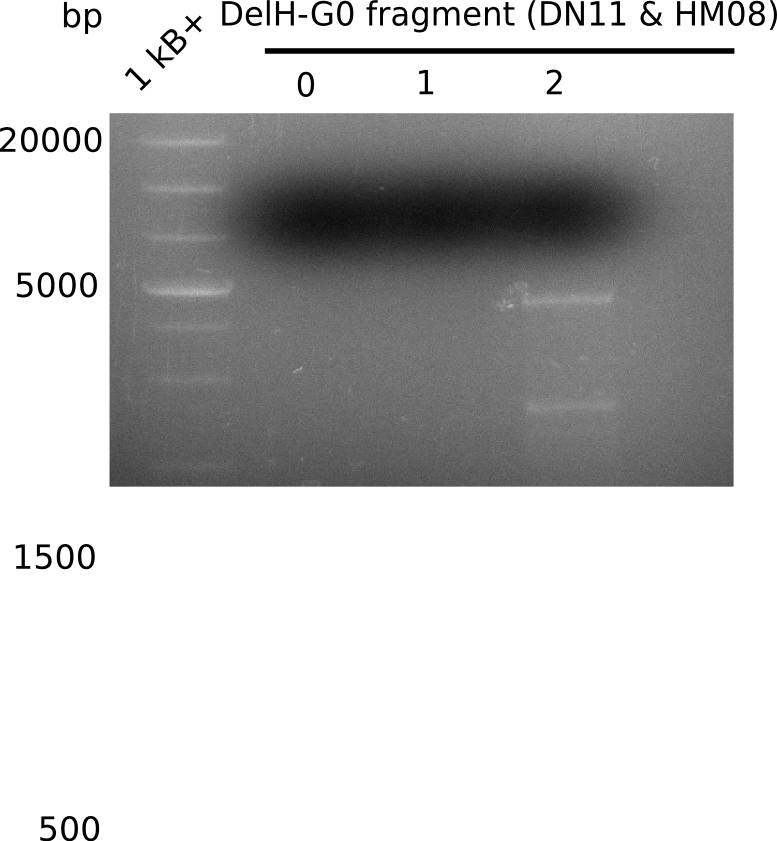
| - | The plasmid for the backbone was obtained from the registry, transformed | + | The plasmid for the backbone was obtained from the registry, transformed into <i>E.coli</i> and miniprepped. The Gibson fragment of the backbone was successfully amplified, together with Gibson fragments DelH G0, G1 and G2b. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 176: | Line 177: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 14</h1> | <h1>Week 14</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 14, we | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 14, we screened numerous colonies from last week's Gibson assembly (28-07) for plasmids containing DelH G0, G1/2a and 2b by screening-PCR. None of the analyzed colonies carried the correct plasmid. We performed another Gibson assembly (01-08) and screened numerous clones. We found few possibly correct ones that will be further analyzed next week. |
| - | Additionally, the new D. acidovorans strain SPH1, whose sequence is available in GenBank, was ordered. We will | + | Additionally, the new <i>D. acidovorans</i> strain SPH1, whose sequence is available in GenBank, was ordered. We will amplify all fragments from the genome of <i>D. acidovorans</i> SPH1 as soon as it arrives. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 185: | Line 186: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 15</h1> | <h1>Week 15</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We screened colonies from last weeks Gibson assemblies (01-08) for plasmids containing DelH G0 as well as G1/2a and 2b. None of the screening-PCRs yielded the expected DNA bands. Therefore, we amplified the Gibson fragments again and performed further Gibson assemblies. Yet again, we could not detect positive colonies by colony-PCR. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 194: | Line 195: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 16</h1> | <h1>Week 16</h1> | ||
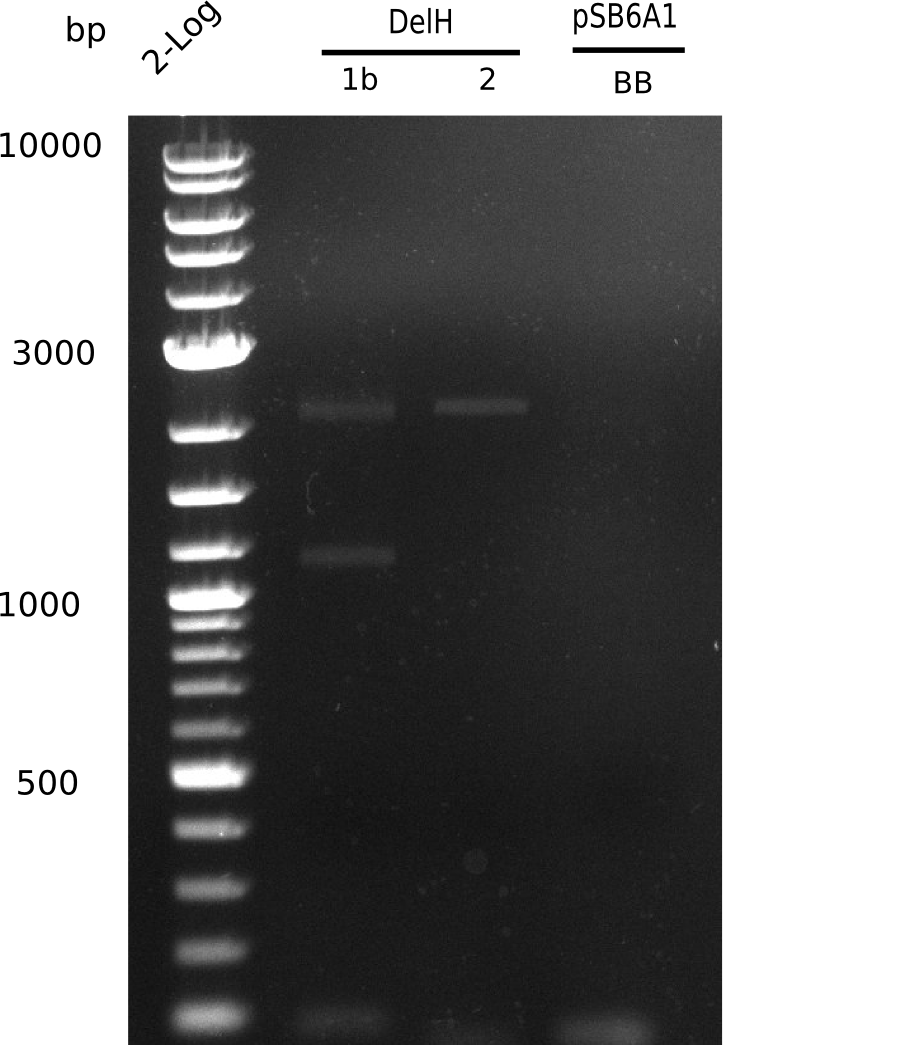
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We further characterized the DelH plasmid created | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We further characterized the DelH plasmid created by Gibson assembly. None of the screened colonies yielded definit positive results. Selection of red colonies was not clear, and PCR screened colonies were all negative. |
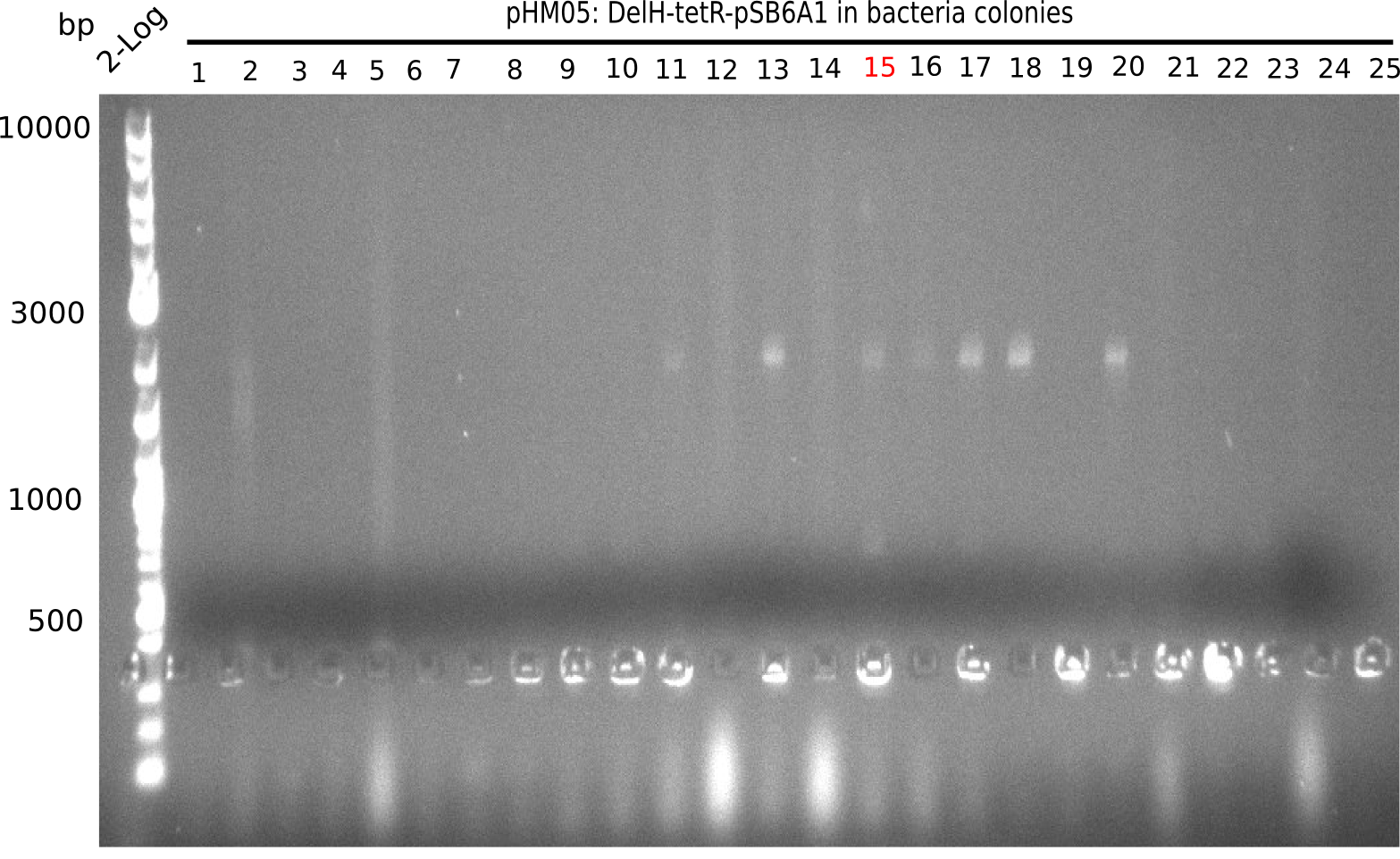
| - | In order to avoid high background during screening of the colonies, we decided to run two different strategies. | + | In order to avoid high background during screening of the colonies, we decided to run two different strategies. First, we will amplify the backbone without mRFP, avoiding backbone reassembly due to ribosome binding site homology (pHM04). In the second strategy (pHM05), we will additionally introduce a tetracycline resistance to select for the insert via antibiotic resistance. |
In addition to the primers for the new strategies, we designed a new screening primer at the end of DelH. </p> | In addition to the primers for the new strategies, we designed a new screening primer at the end of DelH. </p> | ||
</div> | </div> | ||
| Line 205: | Line 206: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 17</h1> | <h1>Week 17</h1> | ||
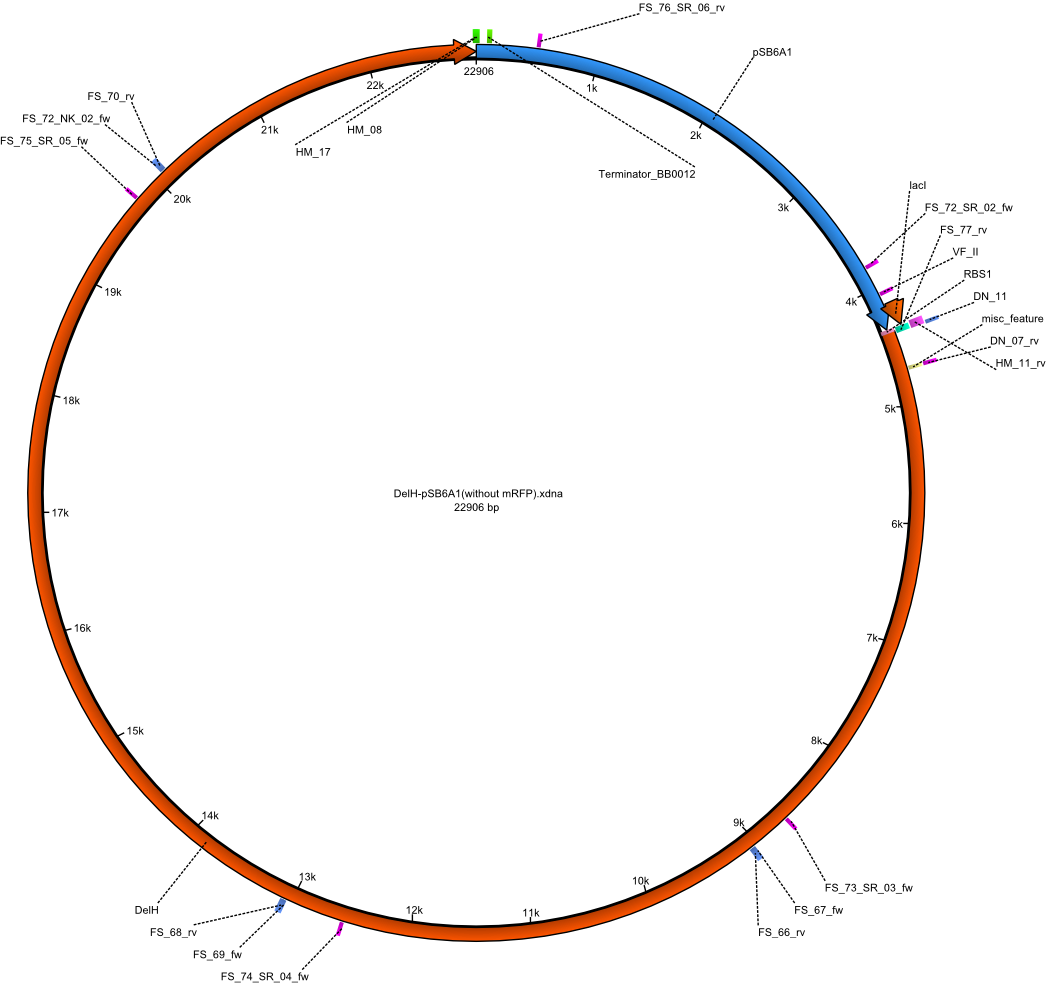
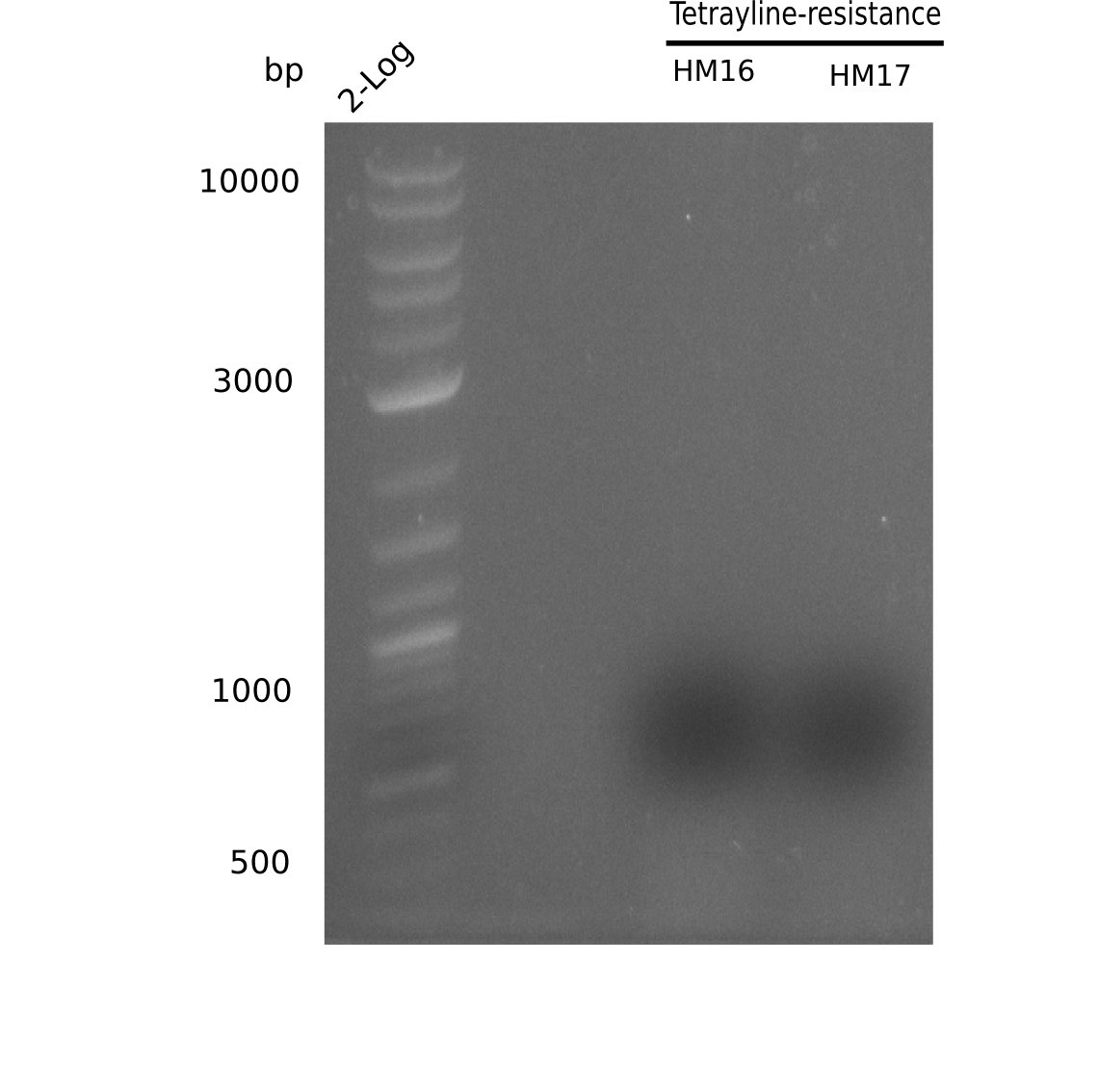
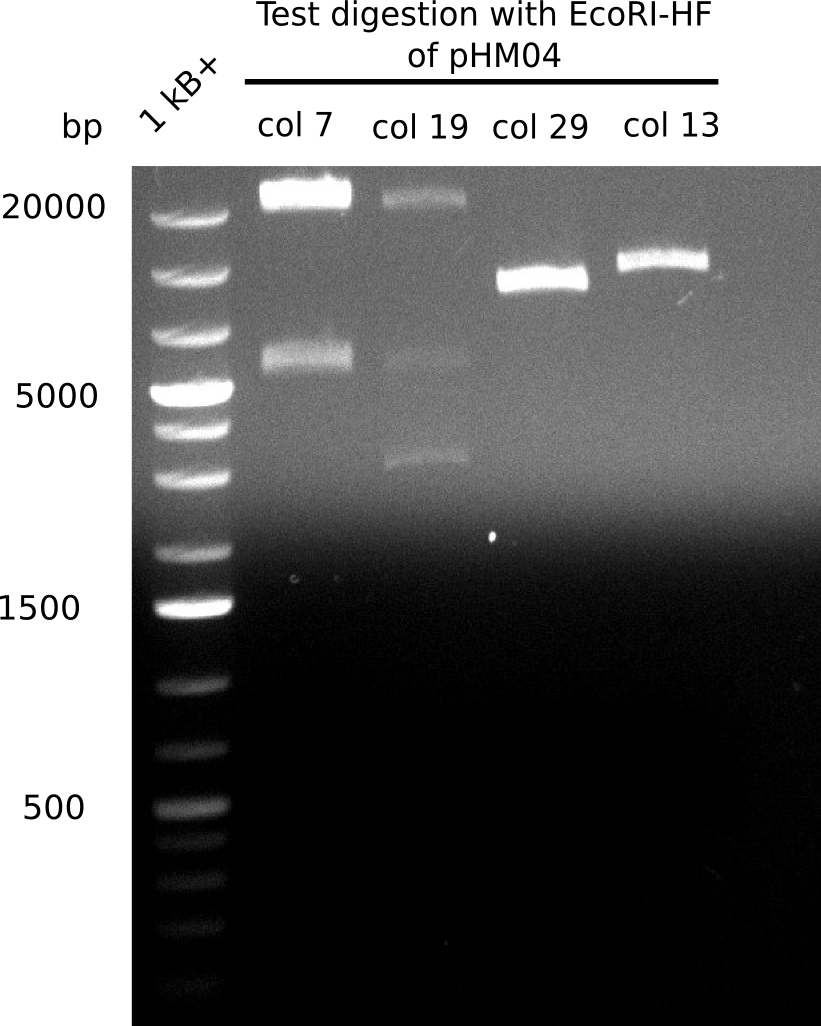
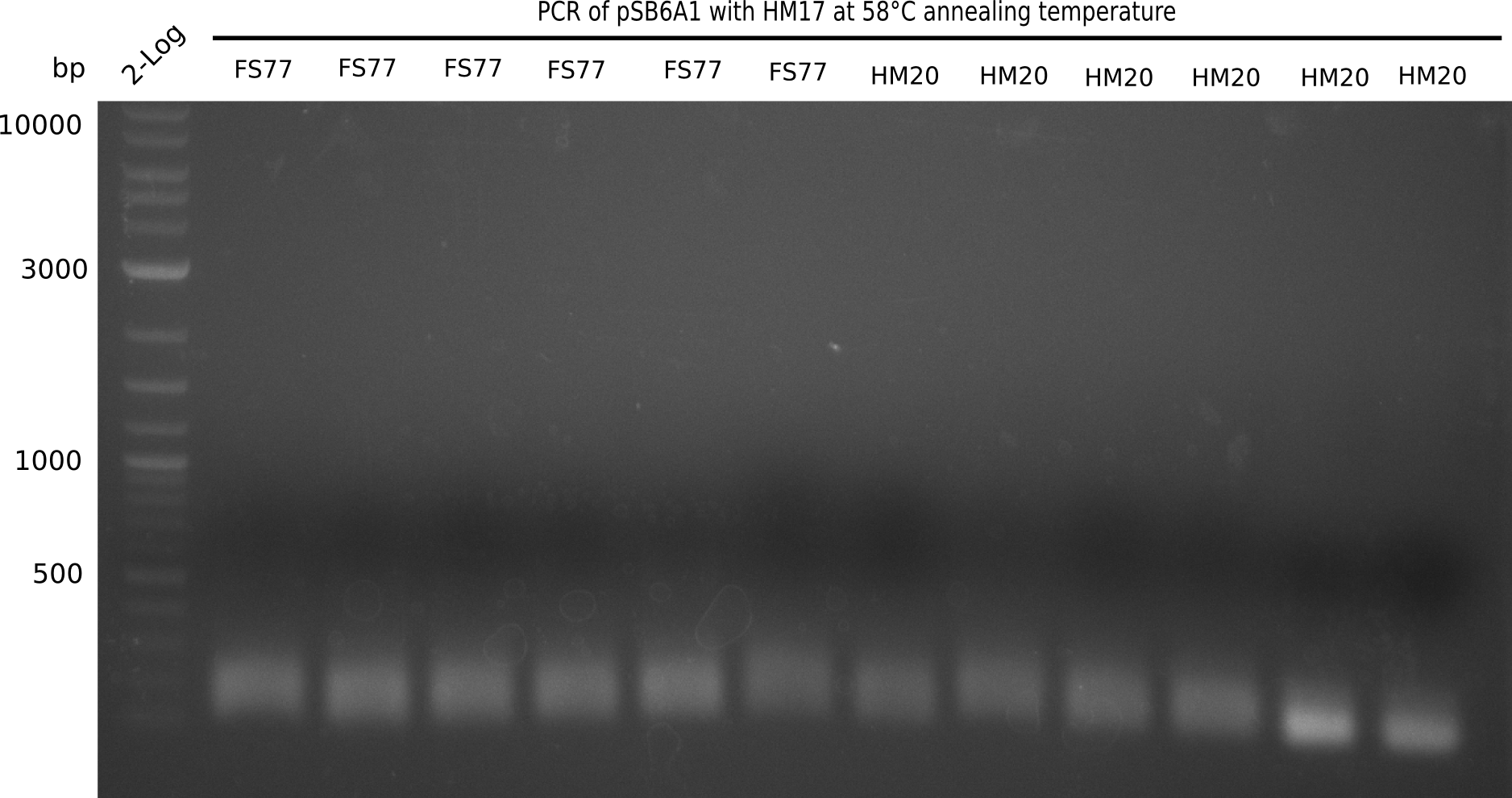
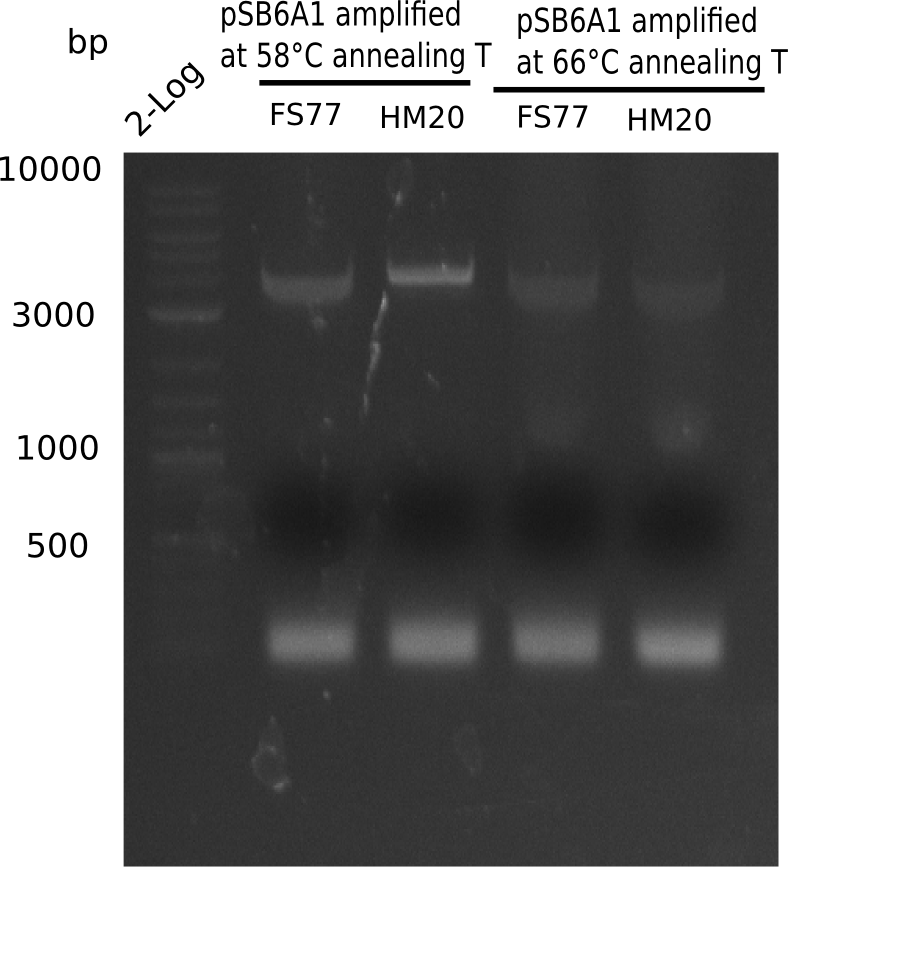
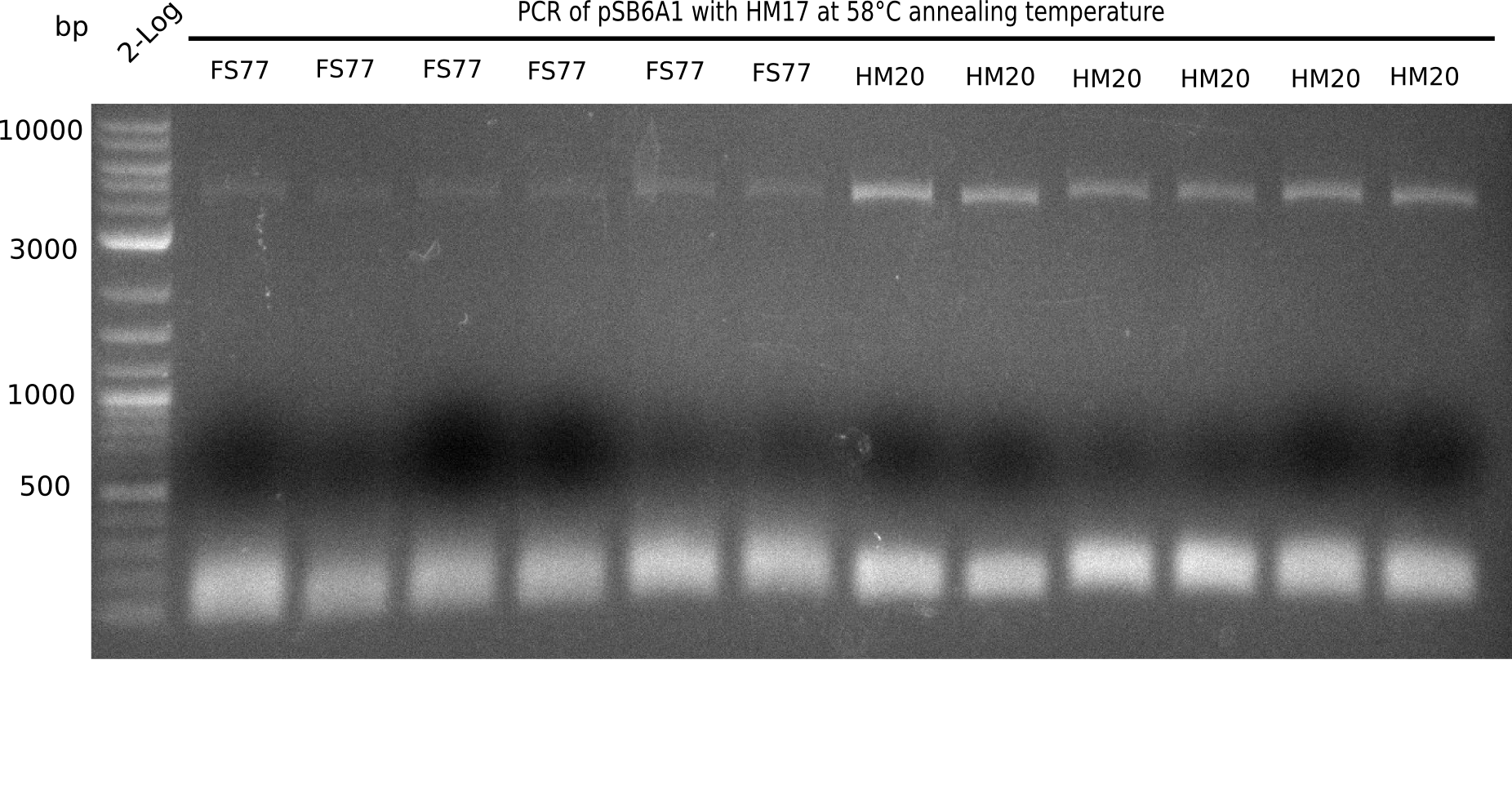
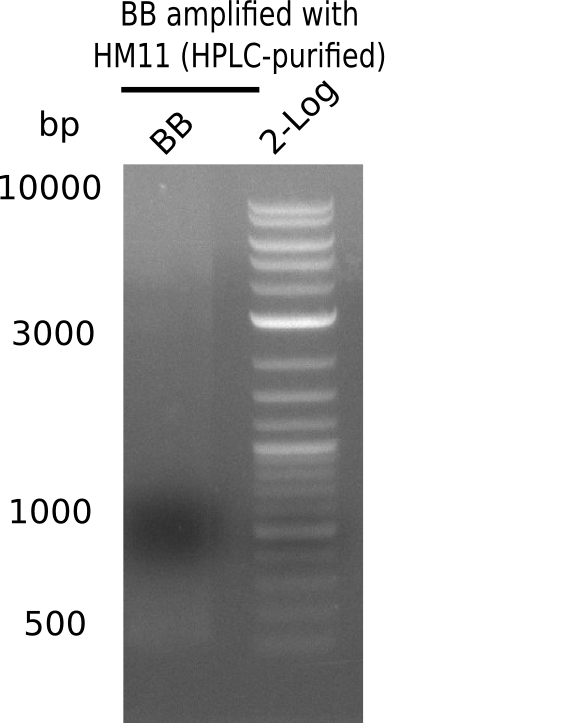
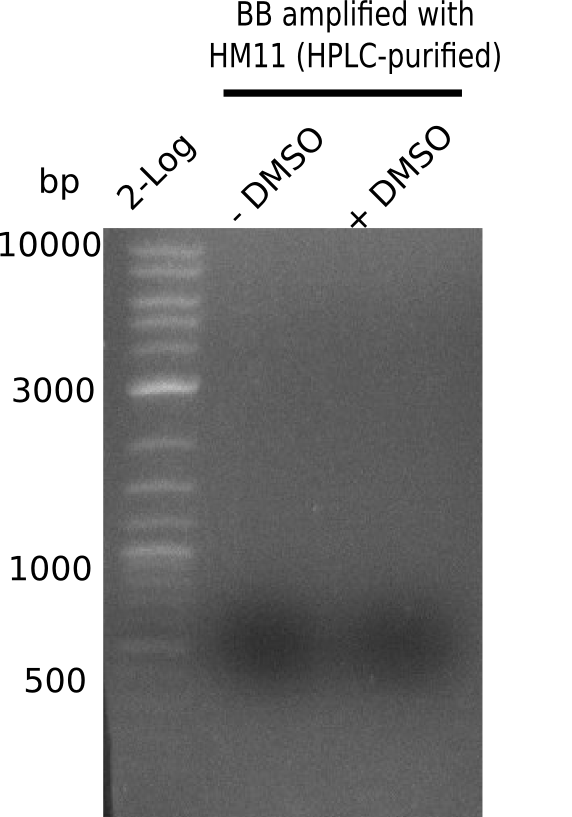
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">A possible explanation for the failed Gibson assemblies could be a religation of the backbone fragment pSB6A1 allowing <i>E.coli</i> to survive. In this case, transformed bacteria will still express mRFP. This week, our aim is to design a new construct without mRFP (pHM04), by which we will be able to exclude red colonies from the screening. For this strategy, we are going to use a new reverse primer for the backbone still including the terminator of mRFP, but omitting mRFP itself. The primers for the backbone amplification are HM11 & HM17.</p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 214: | Line 215: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 18</h1> | <h1>Week 18</h1> | ||
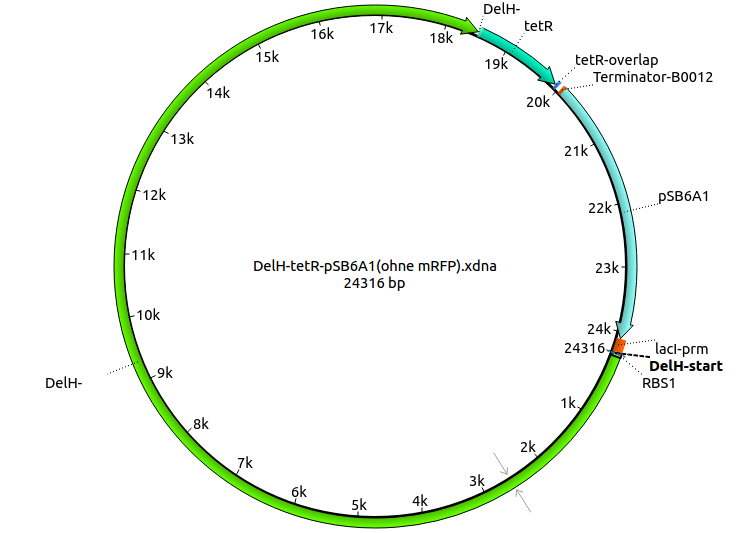
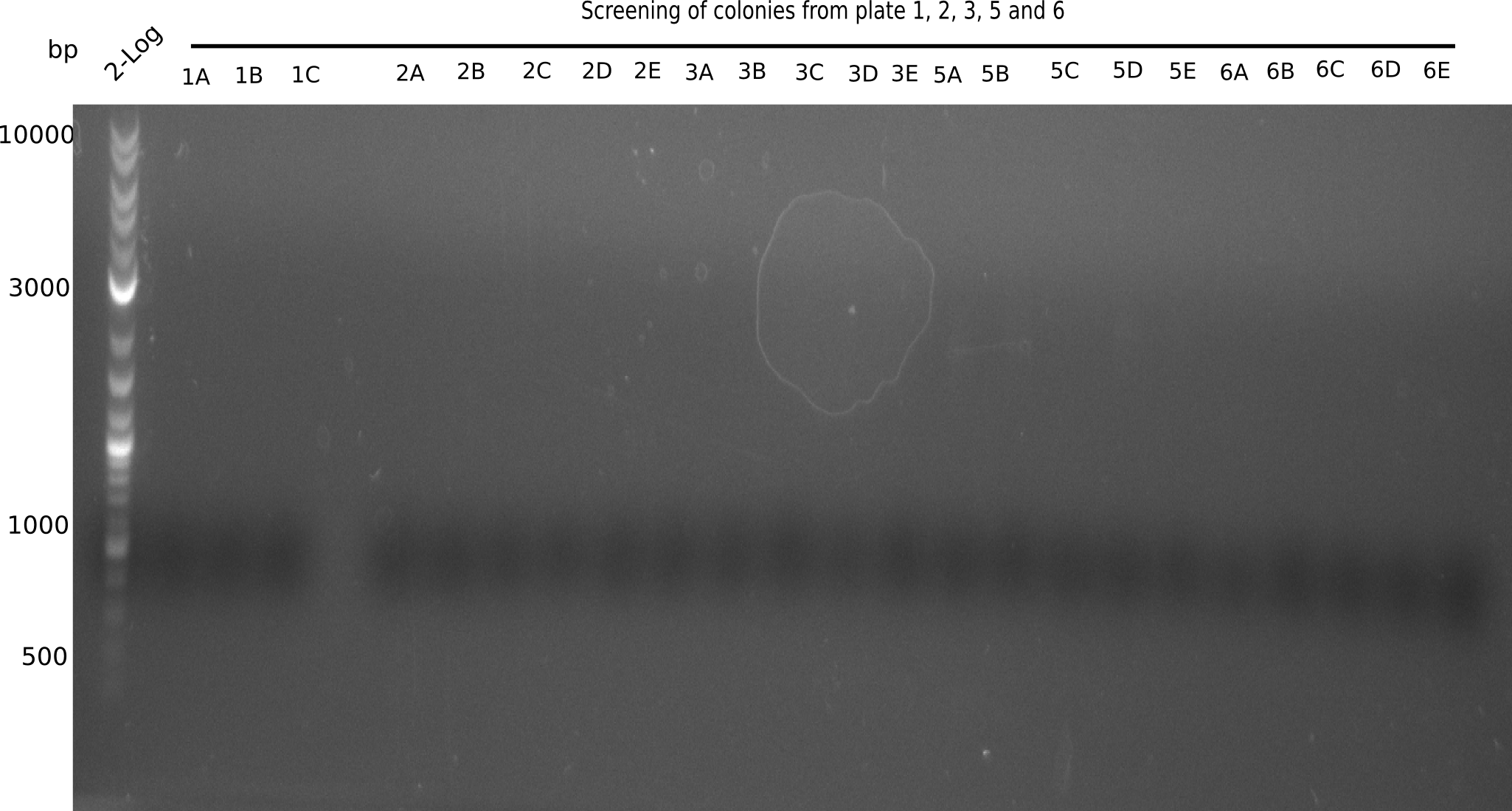
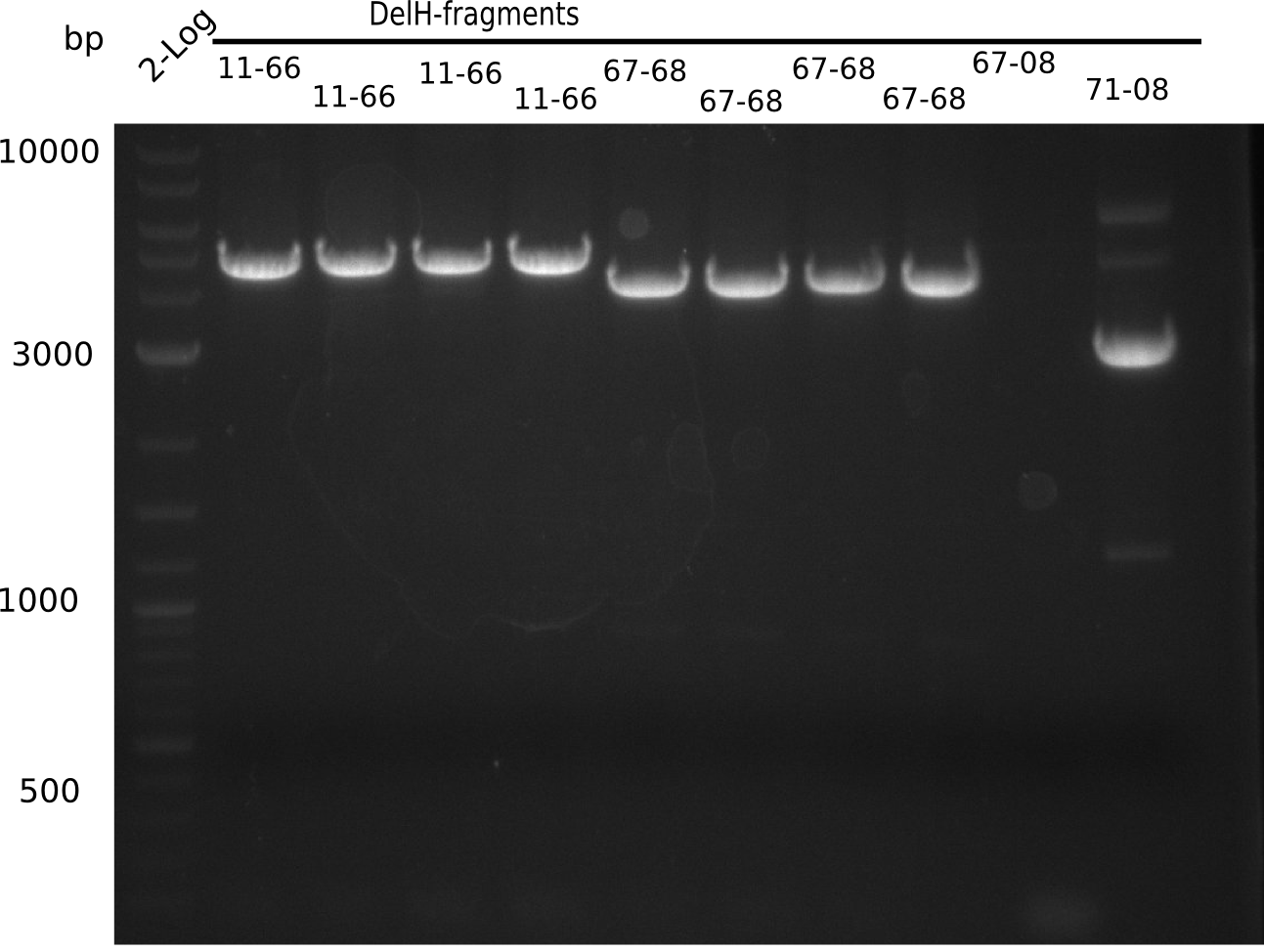
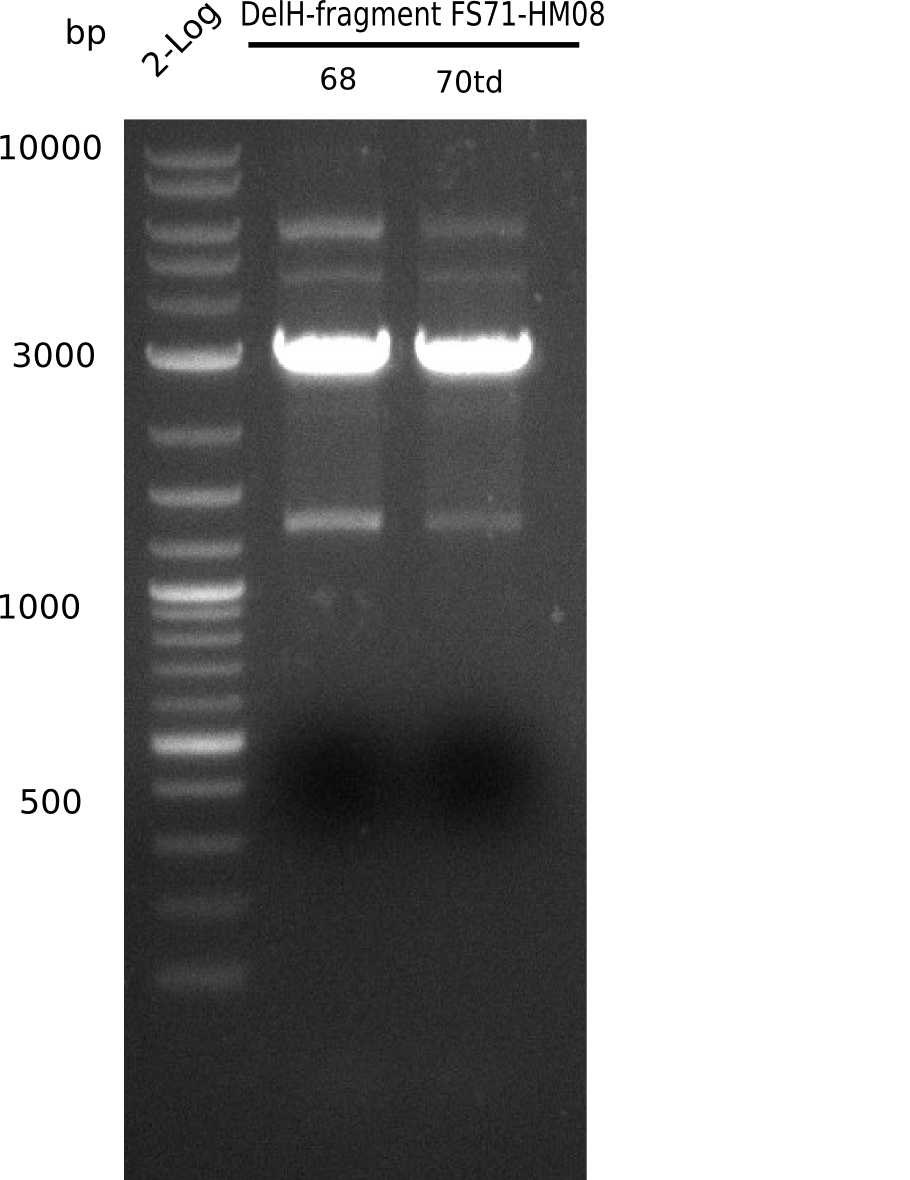
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since the assembly strategy for pHM04 did not yield positive clones (see experiments week 17), we will follow the idea to introduce a tetracycline resistance to the ampicillin backbone as an additional selection marker for successful assembly. With this approach, positive clones can be easily determined by their white phenotype (exclusion of mRFP) and their ability to grow on plates containing tetracycline. Furthermore, we decided to amplify DelH in various fragments to increase Gibson assembly efficiency. Therefore, we also ordered new screening primers. The primers were designed as shown in the following table. </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 223: | Line 224: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
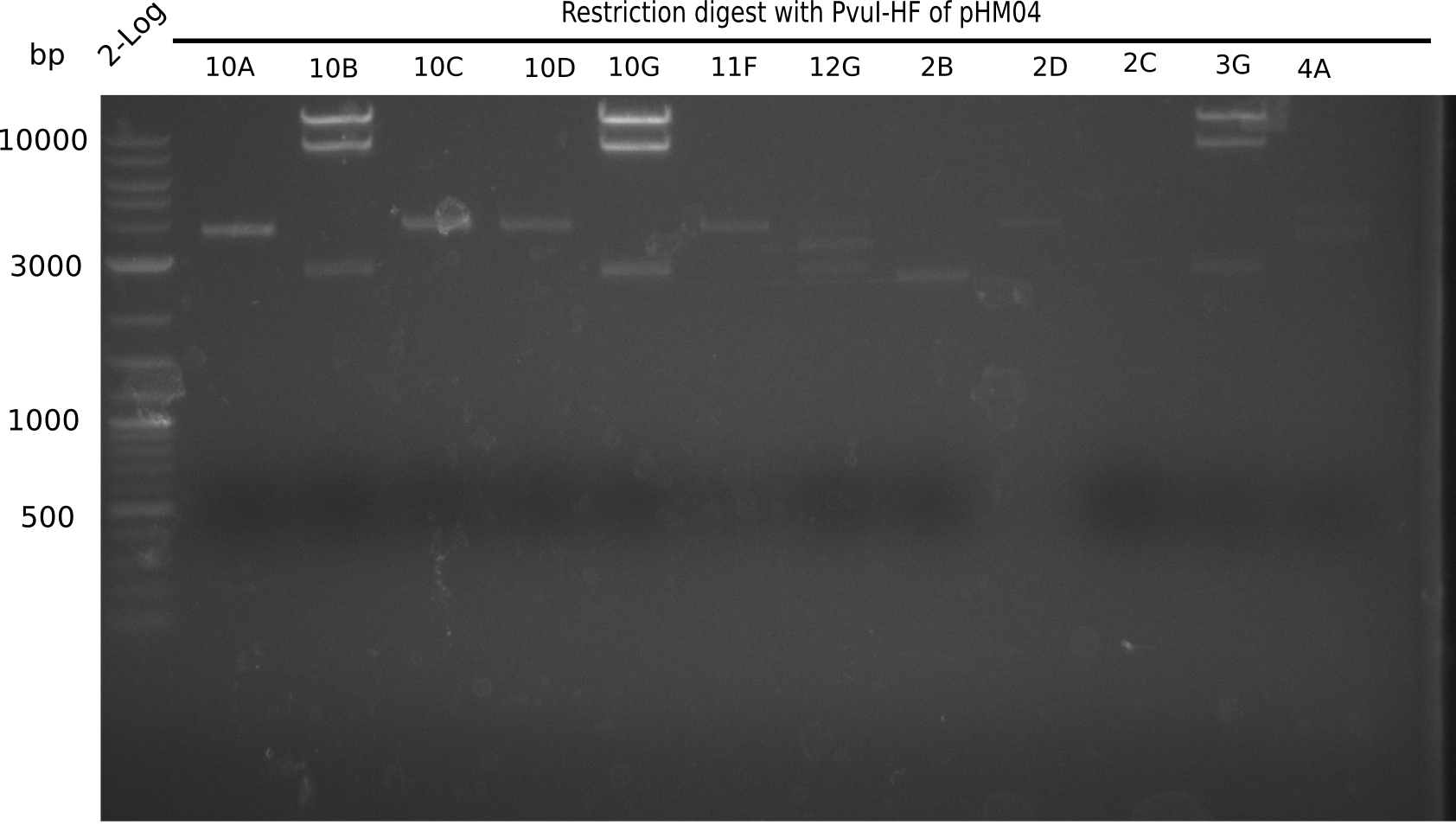
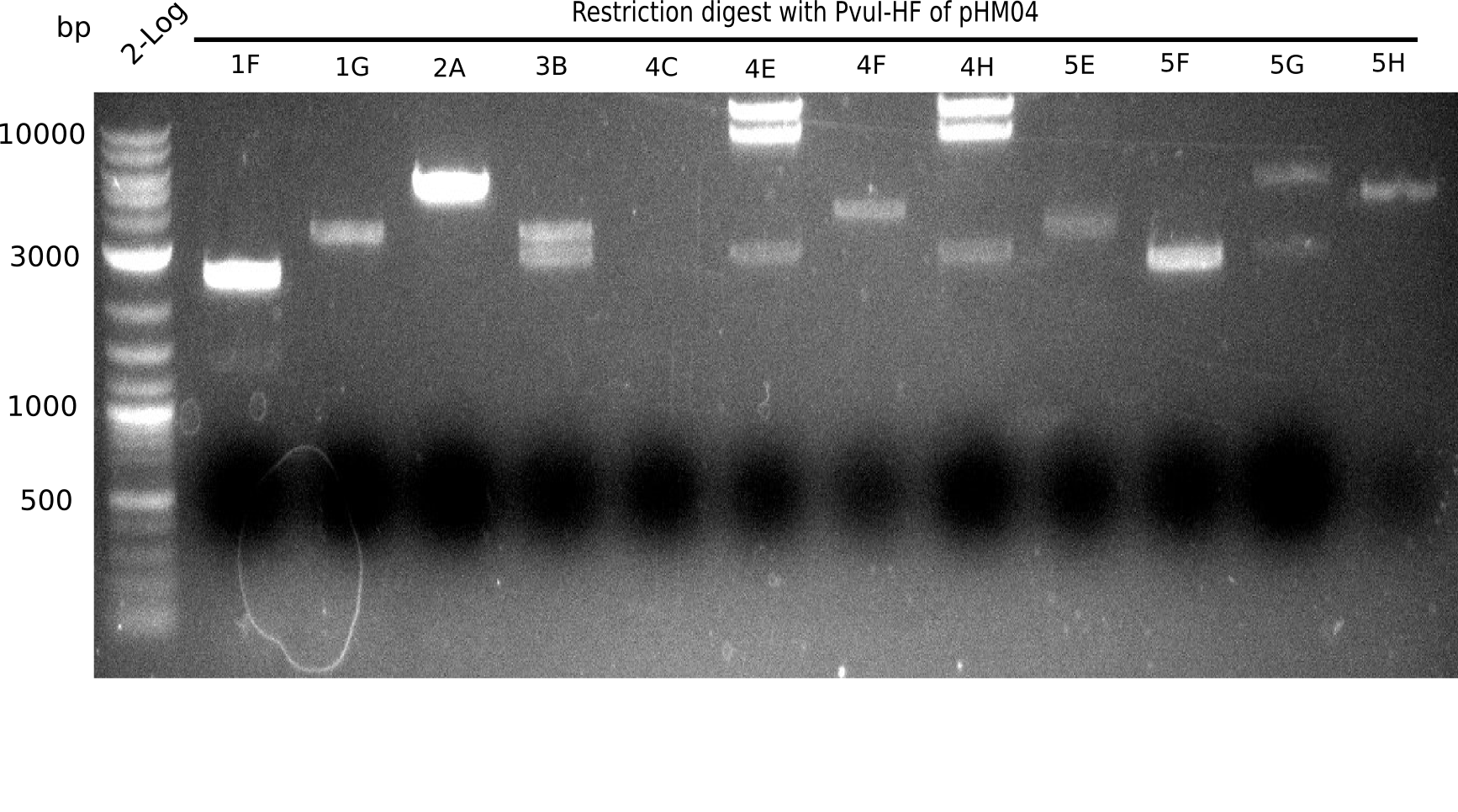
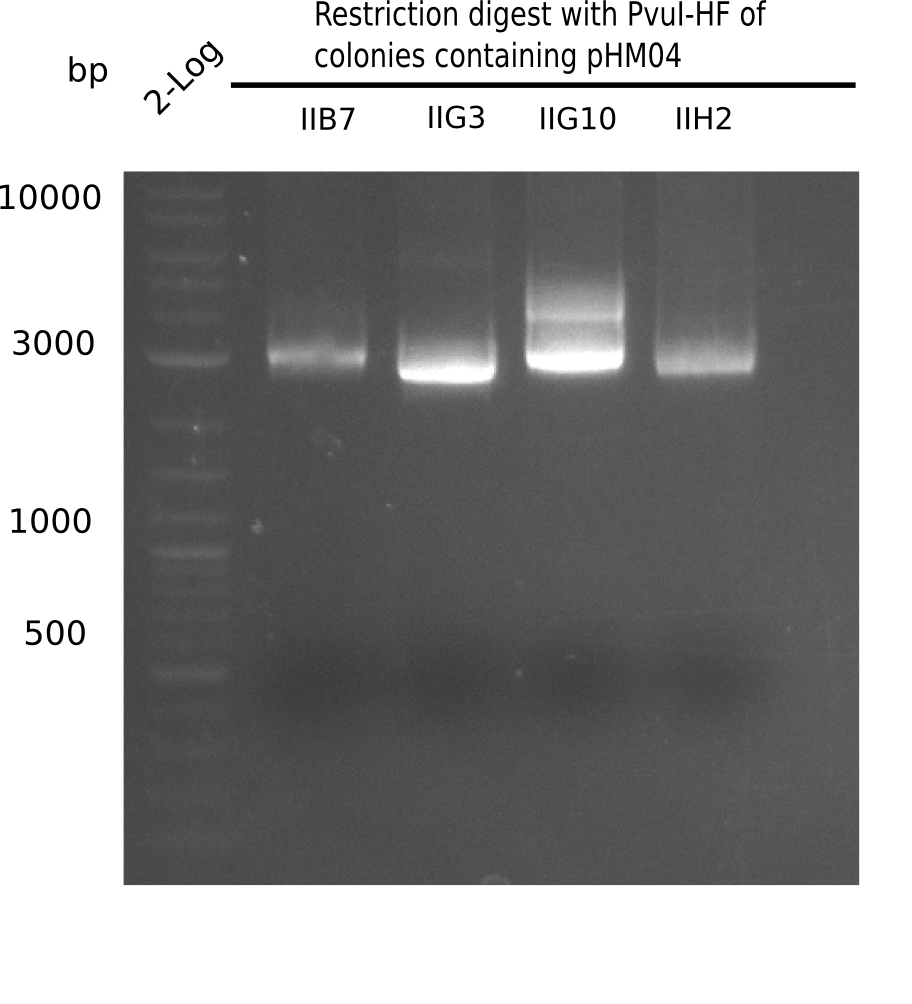
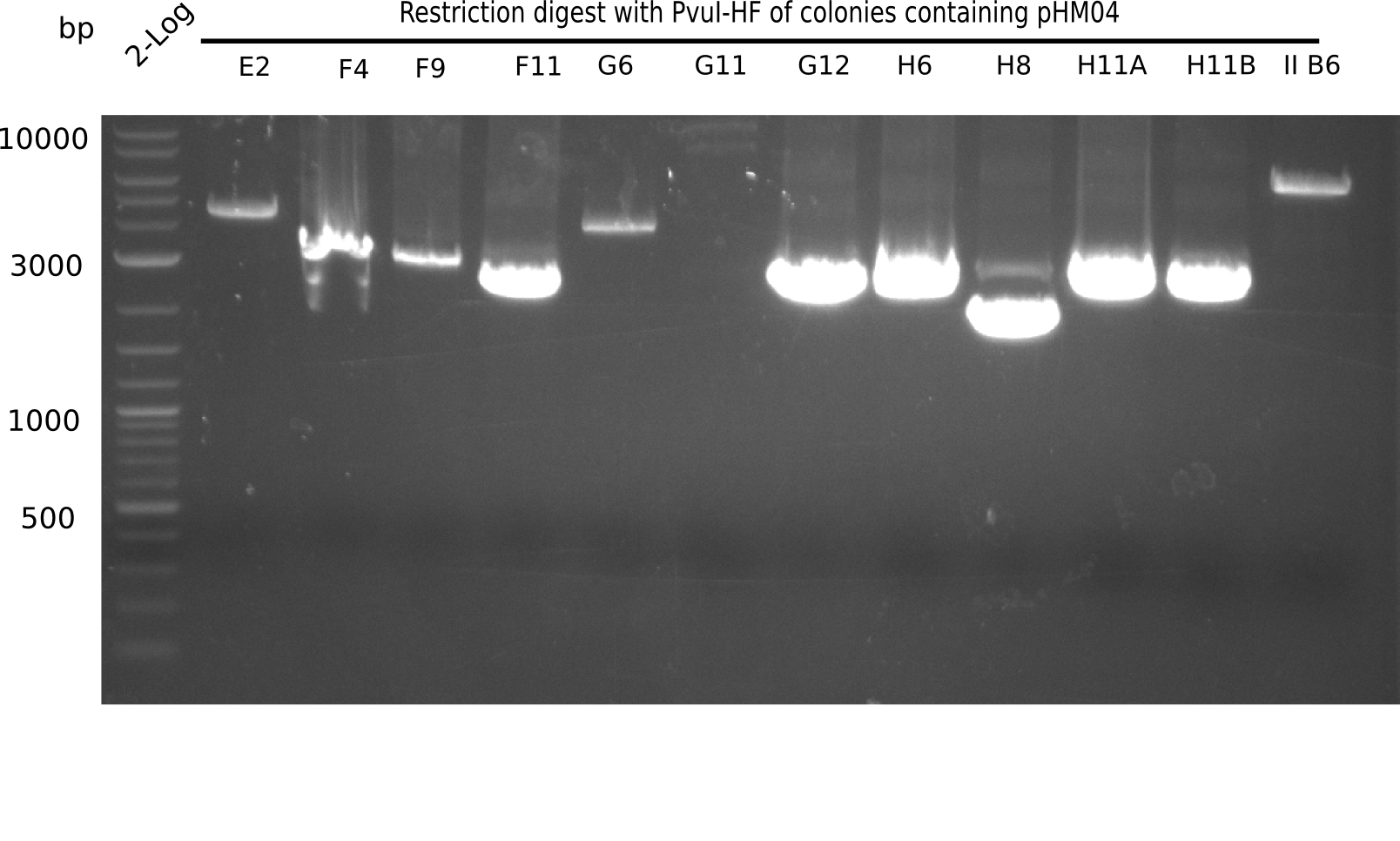
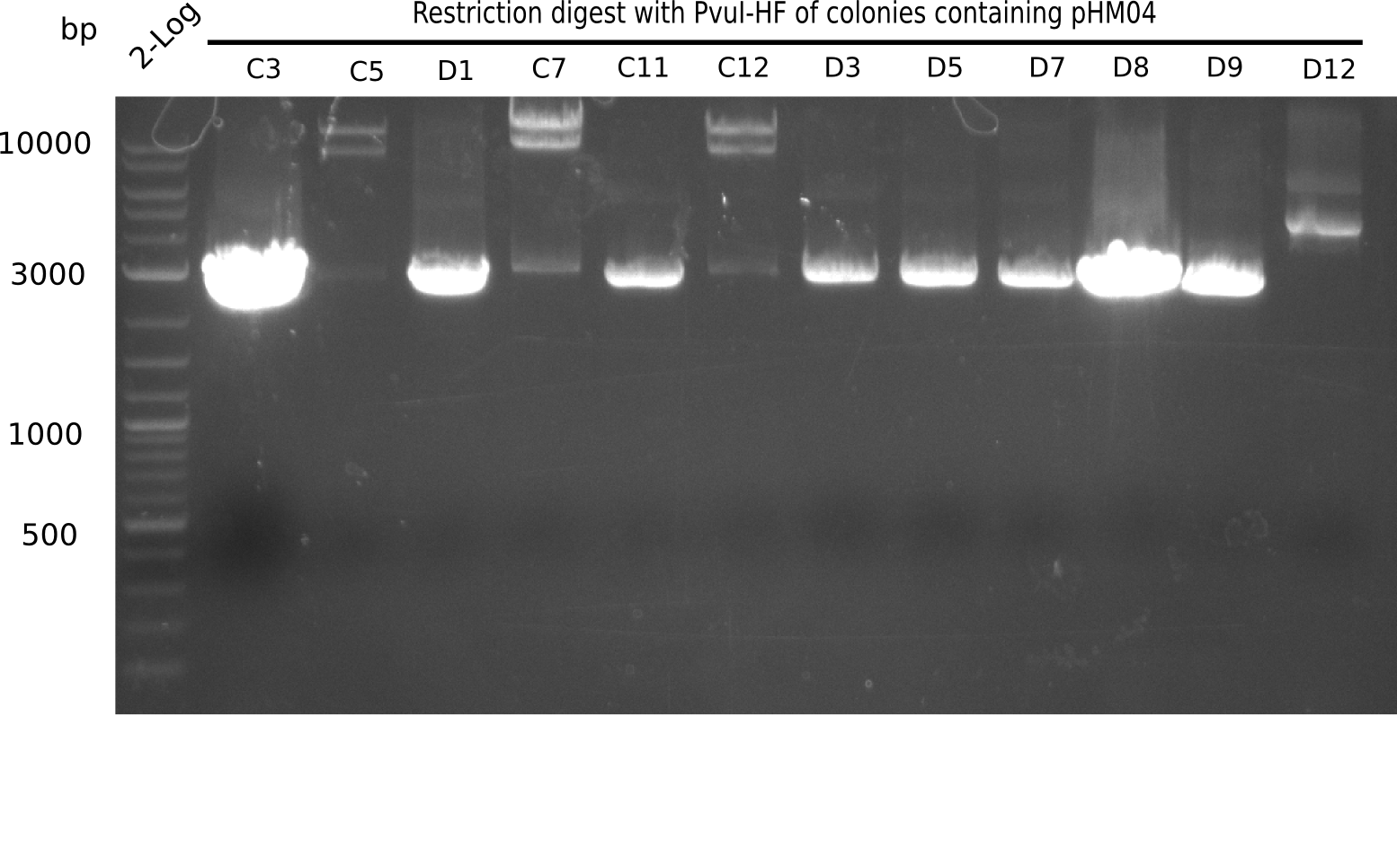
<h1>Week 19</h1> | <h1>Week 19</h1> | ||
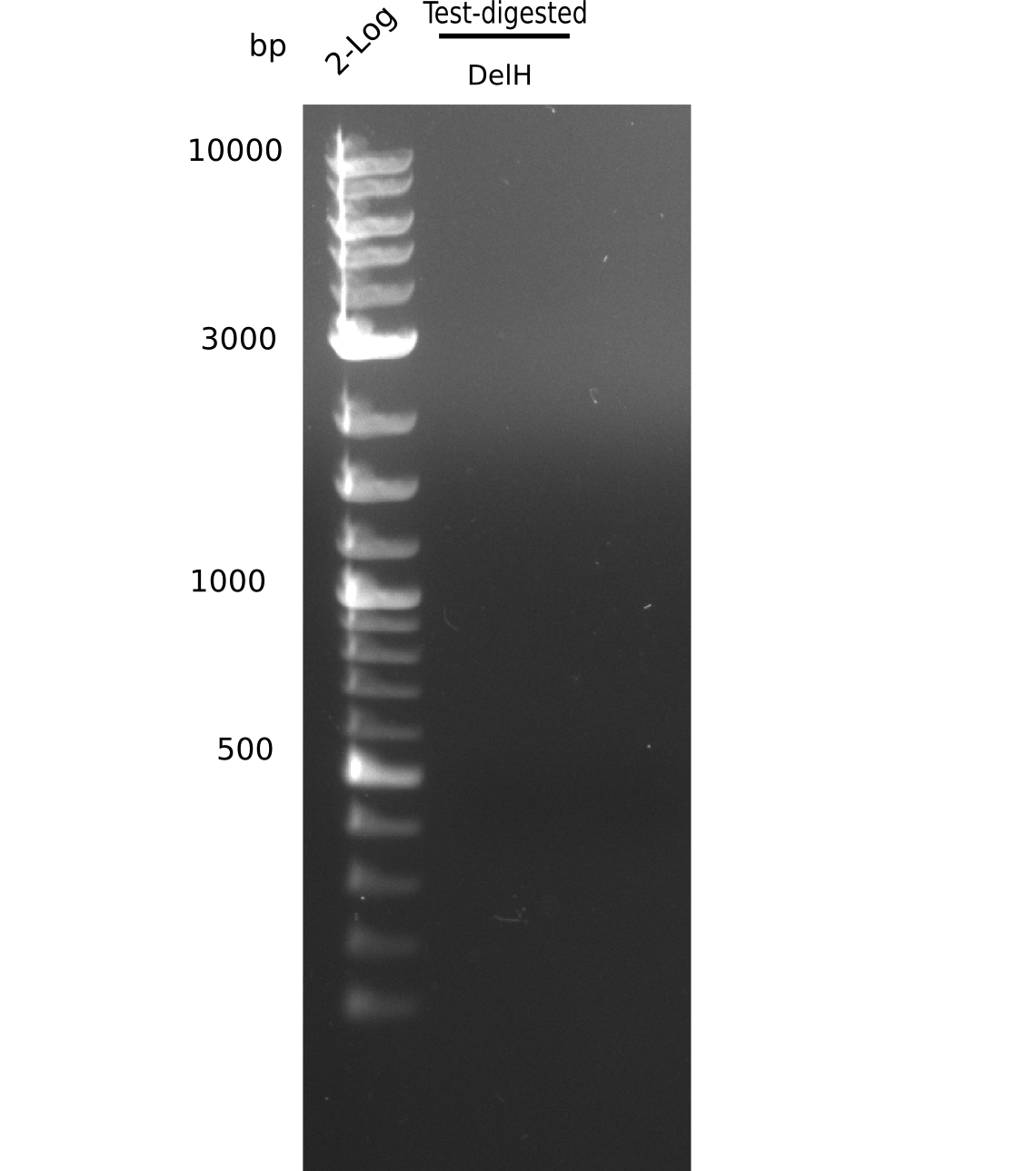
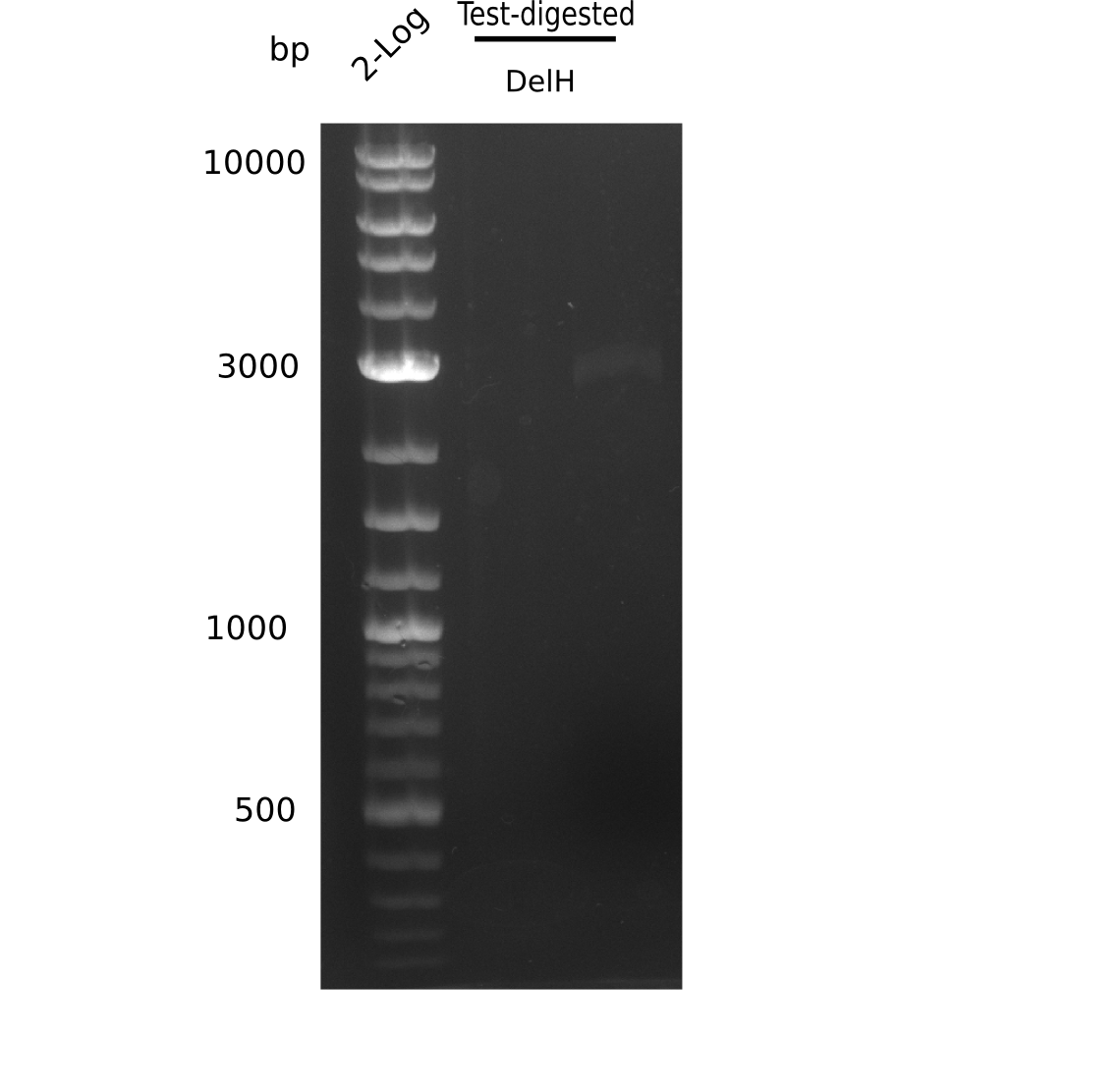
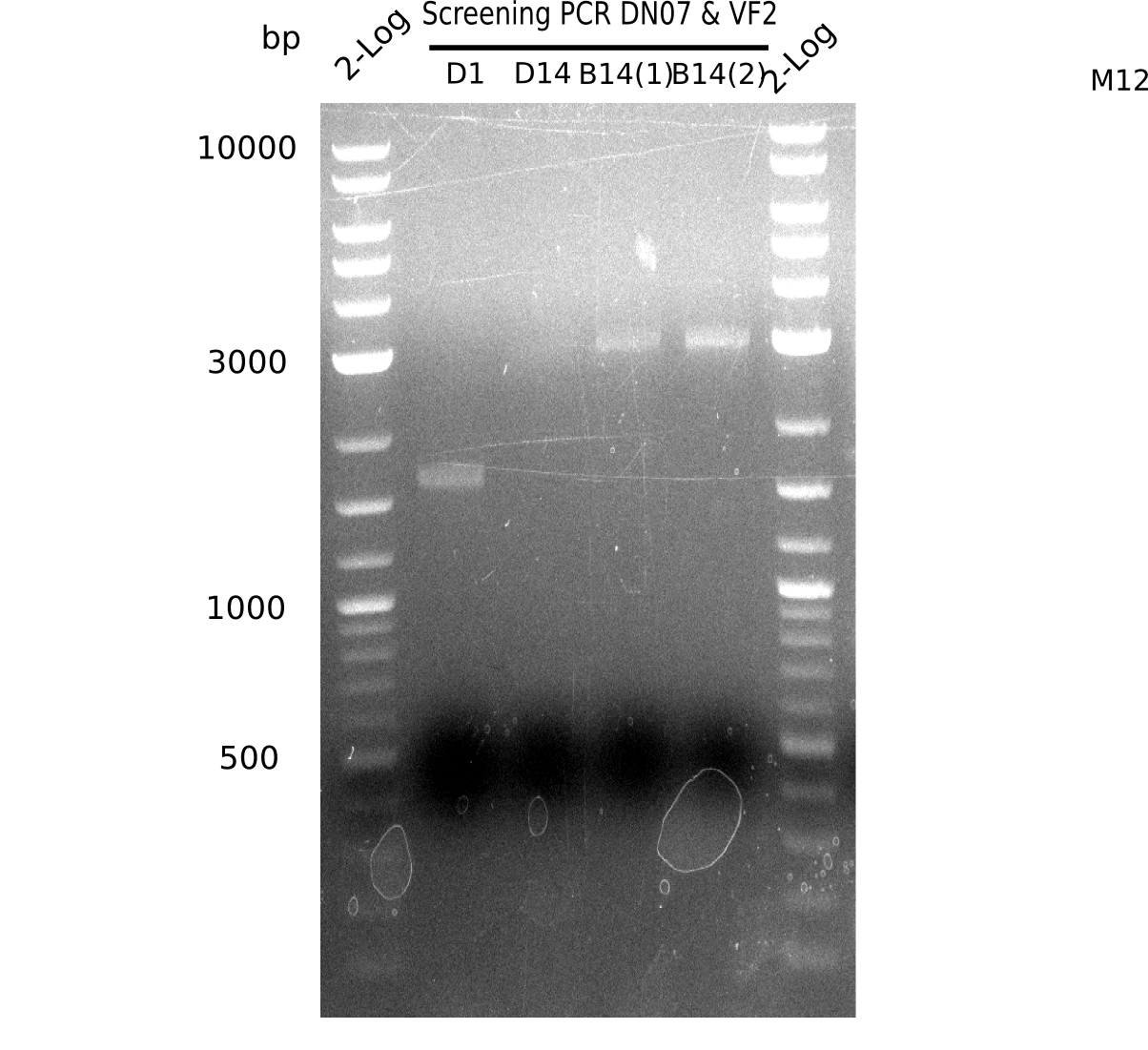
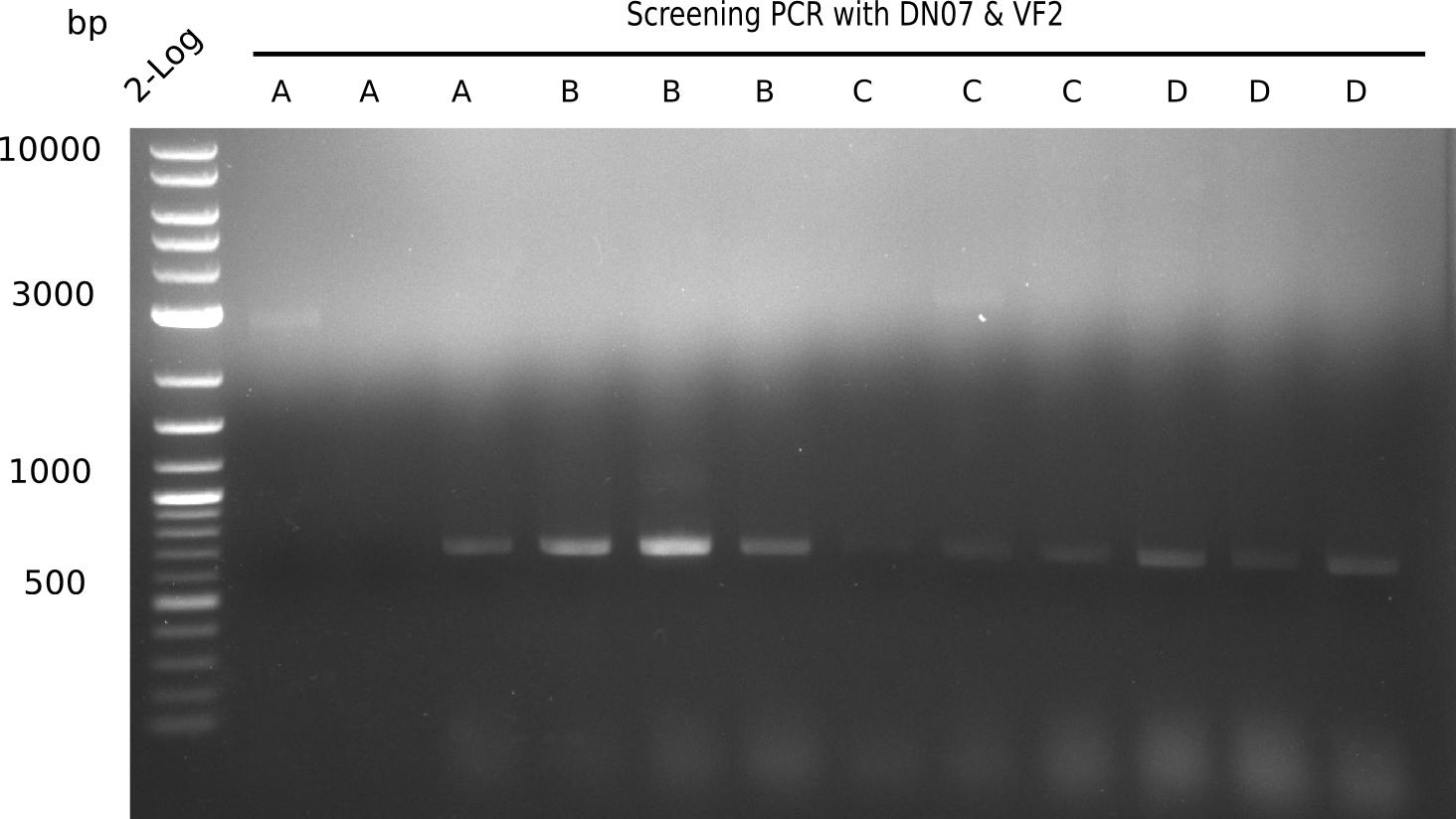
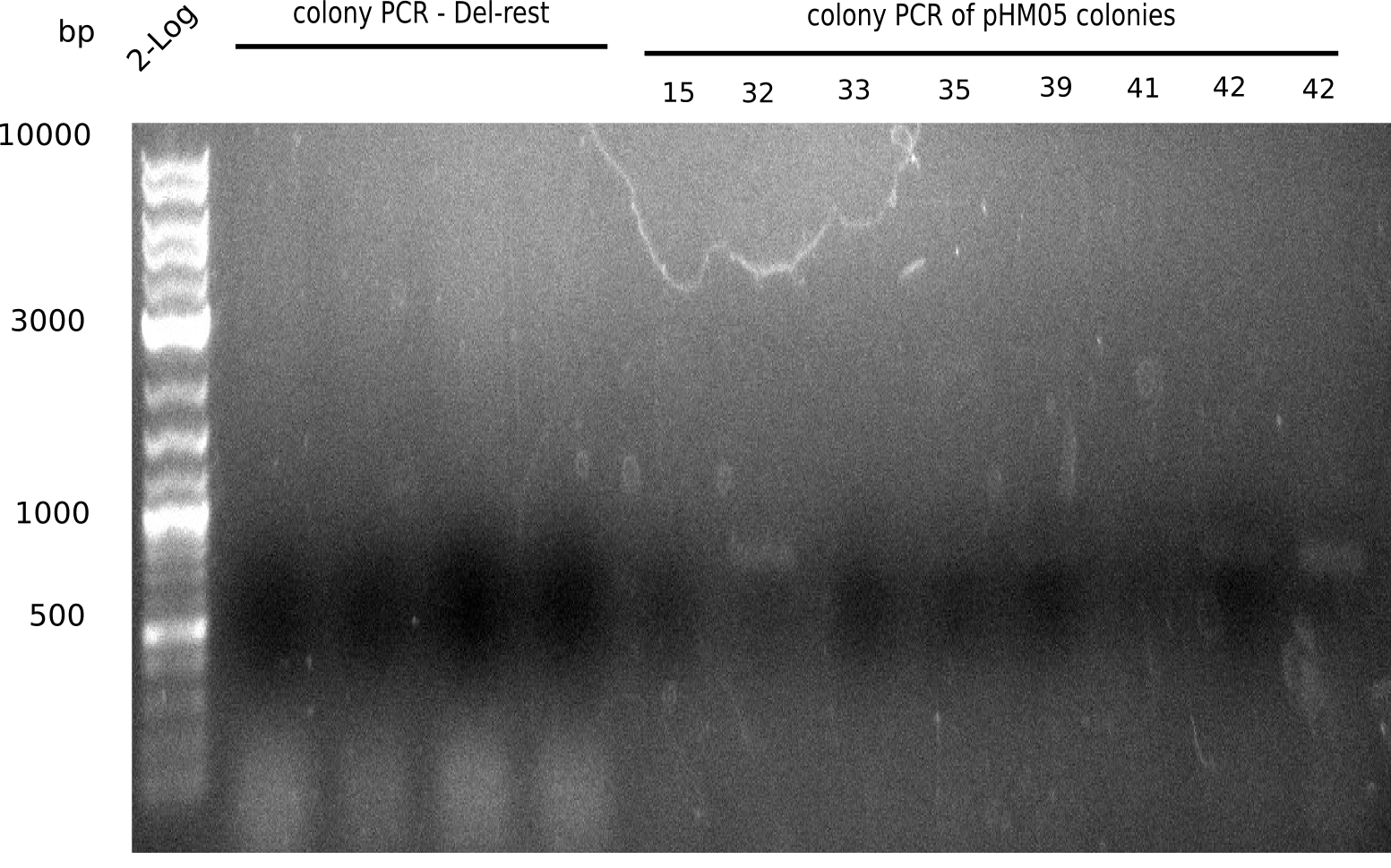
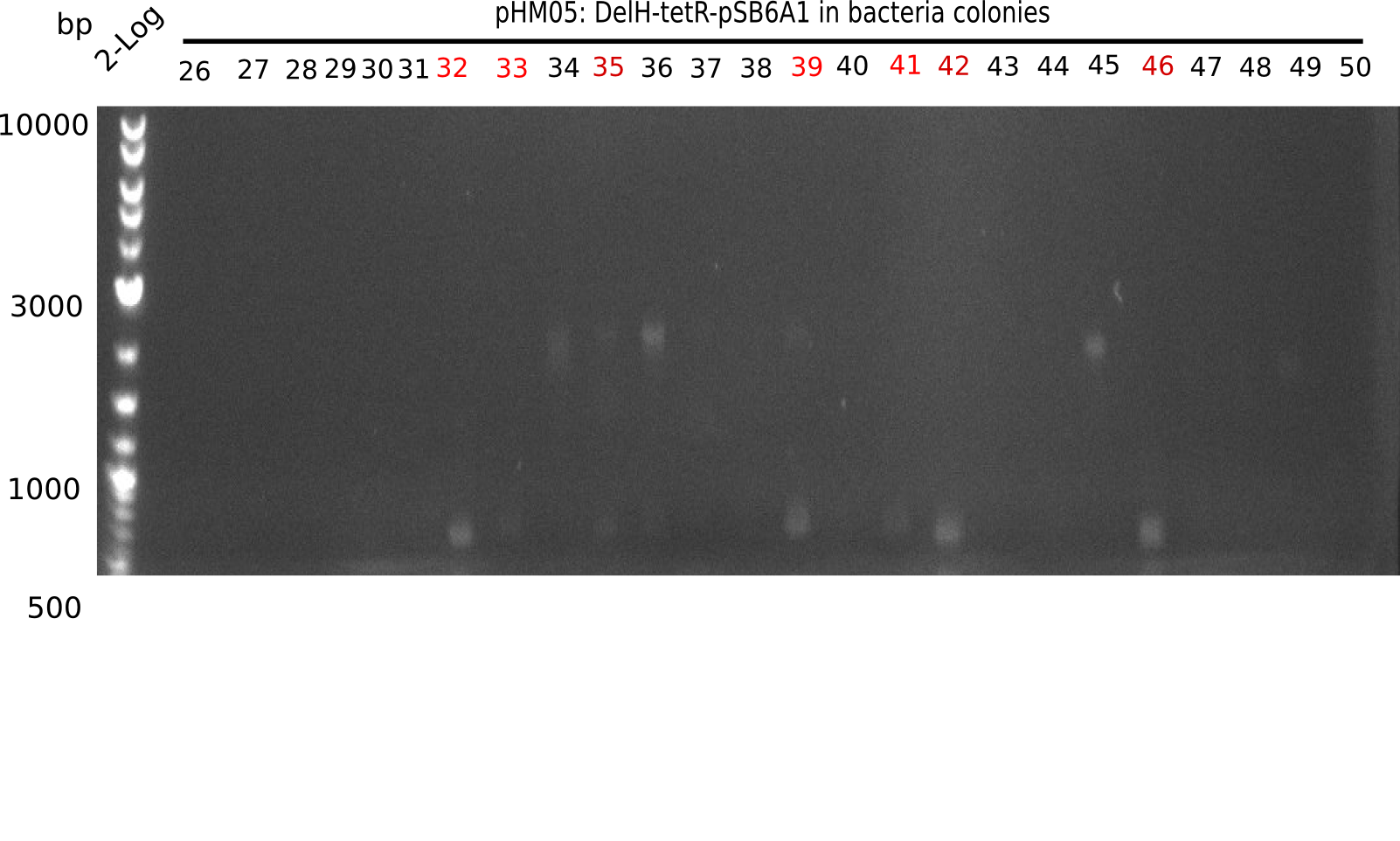
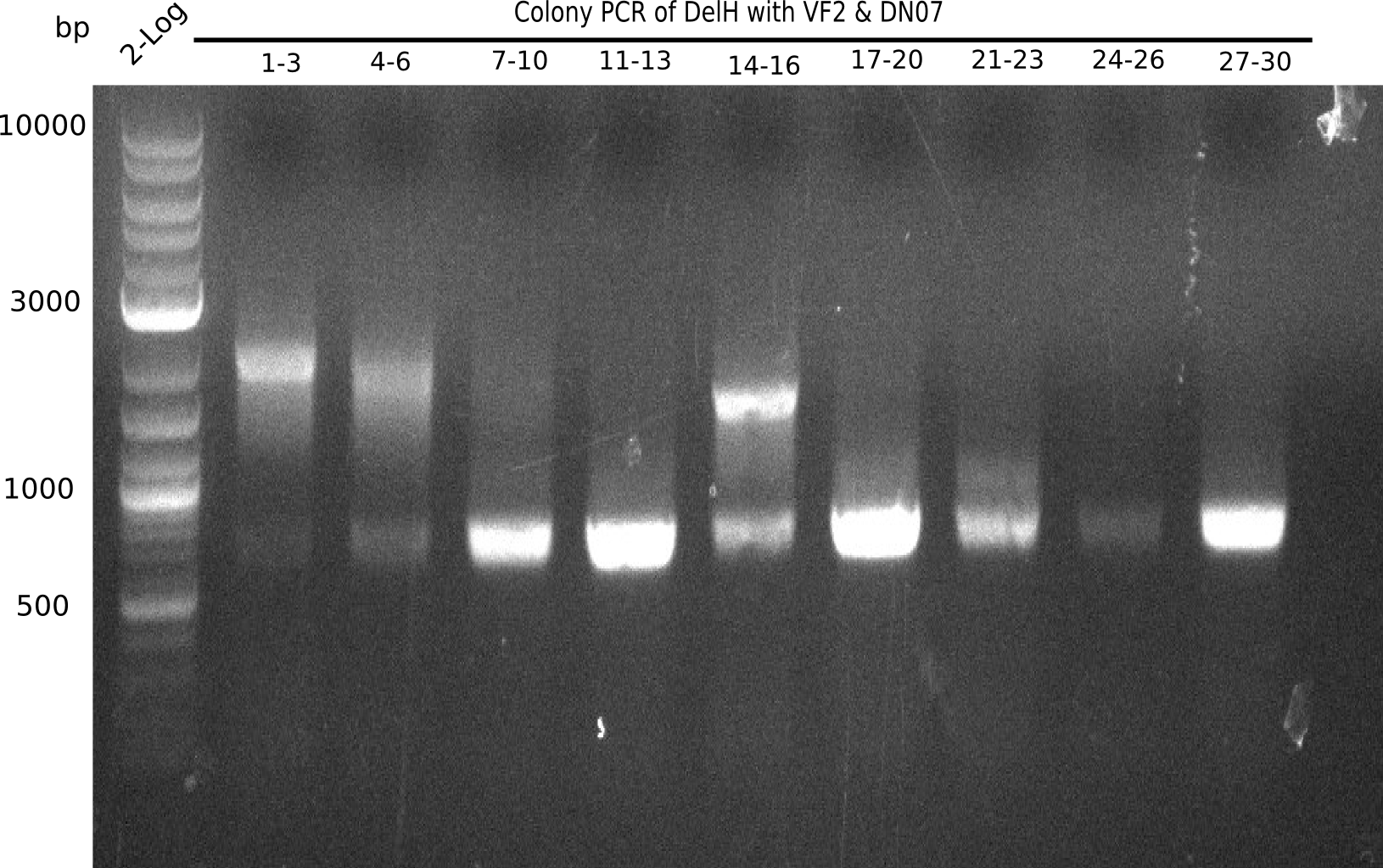
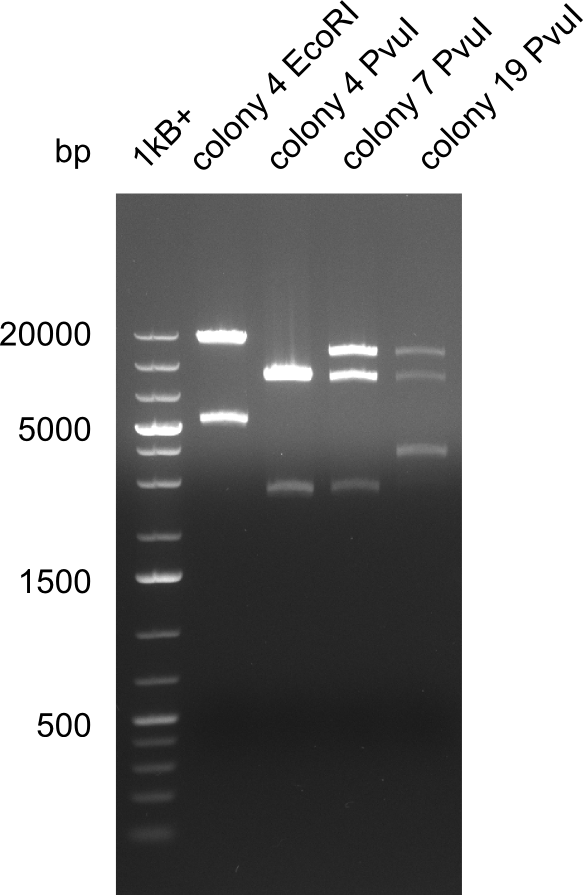
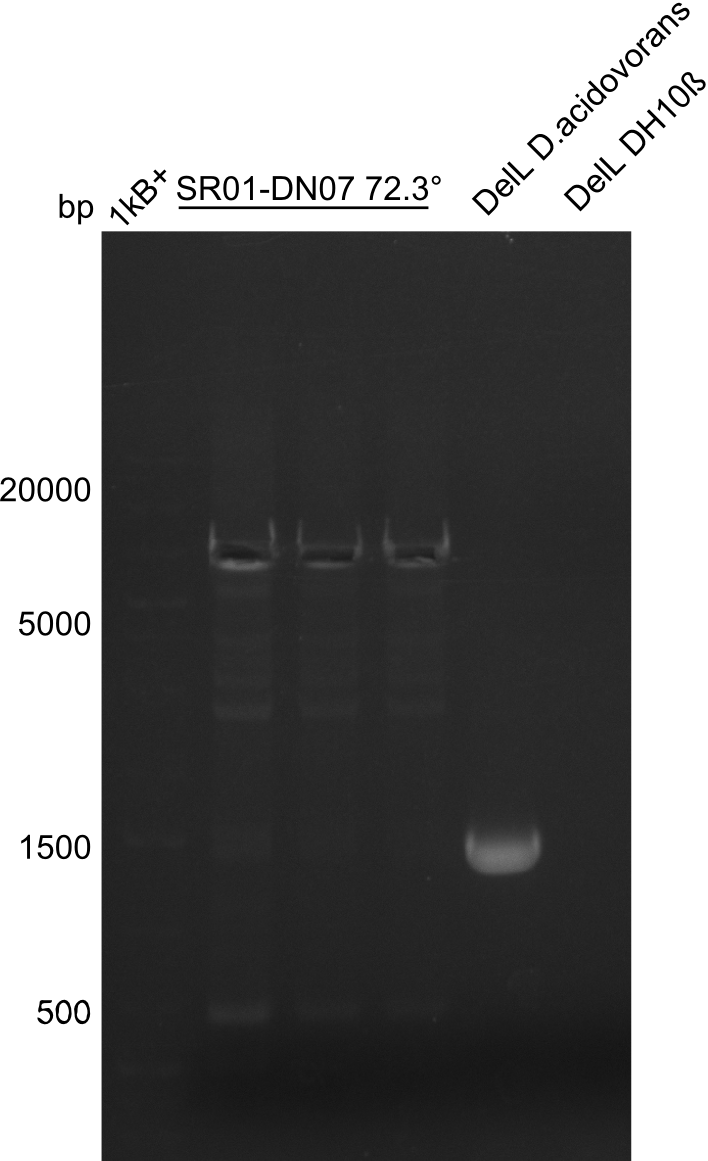
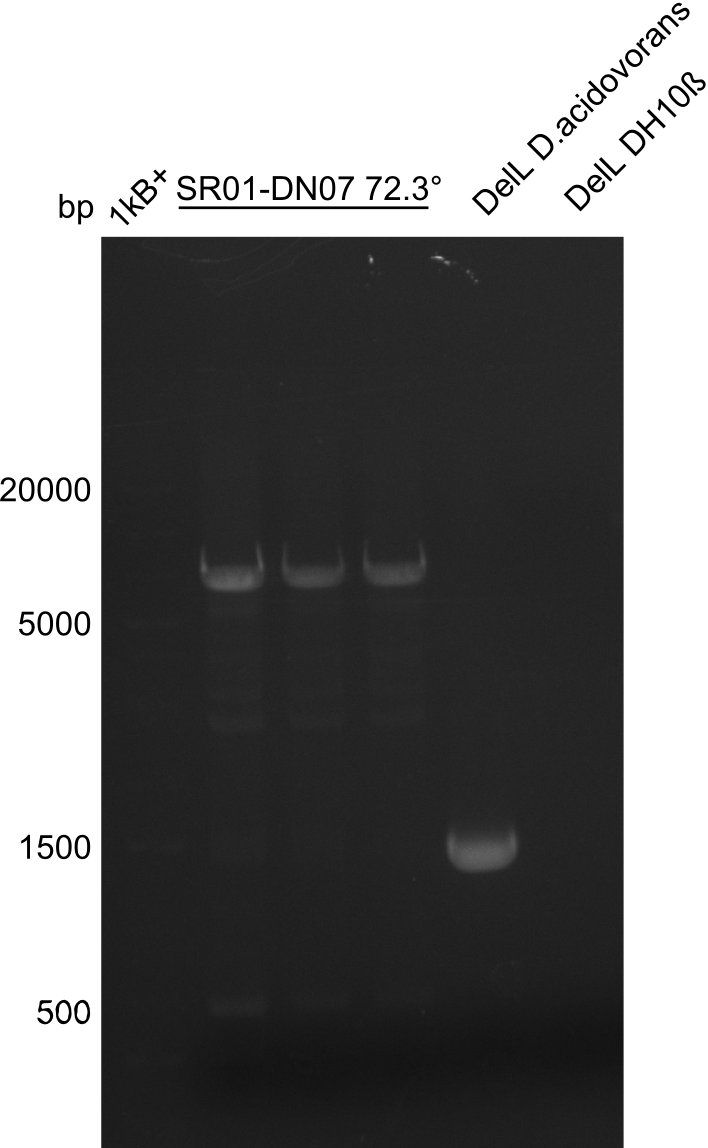
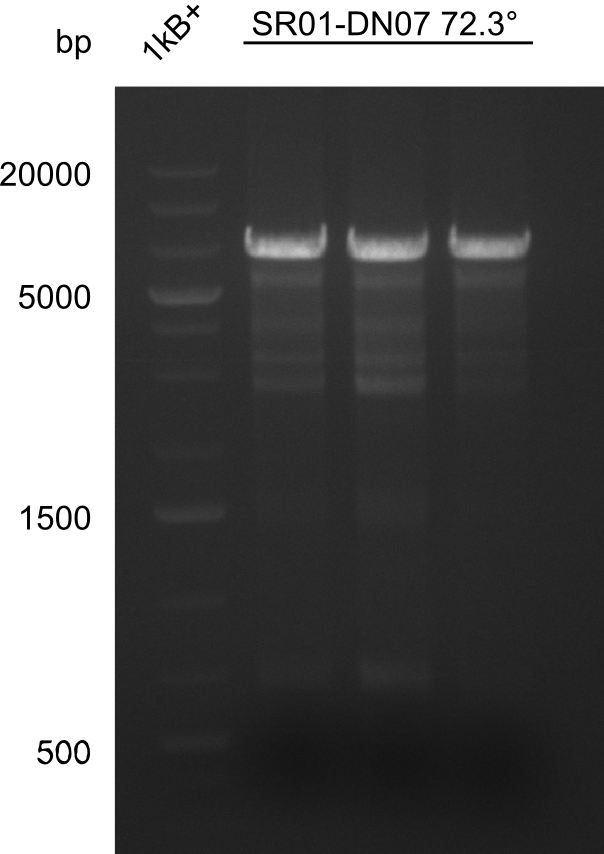
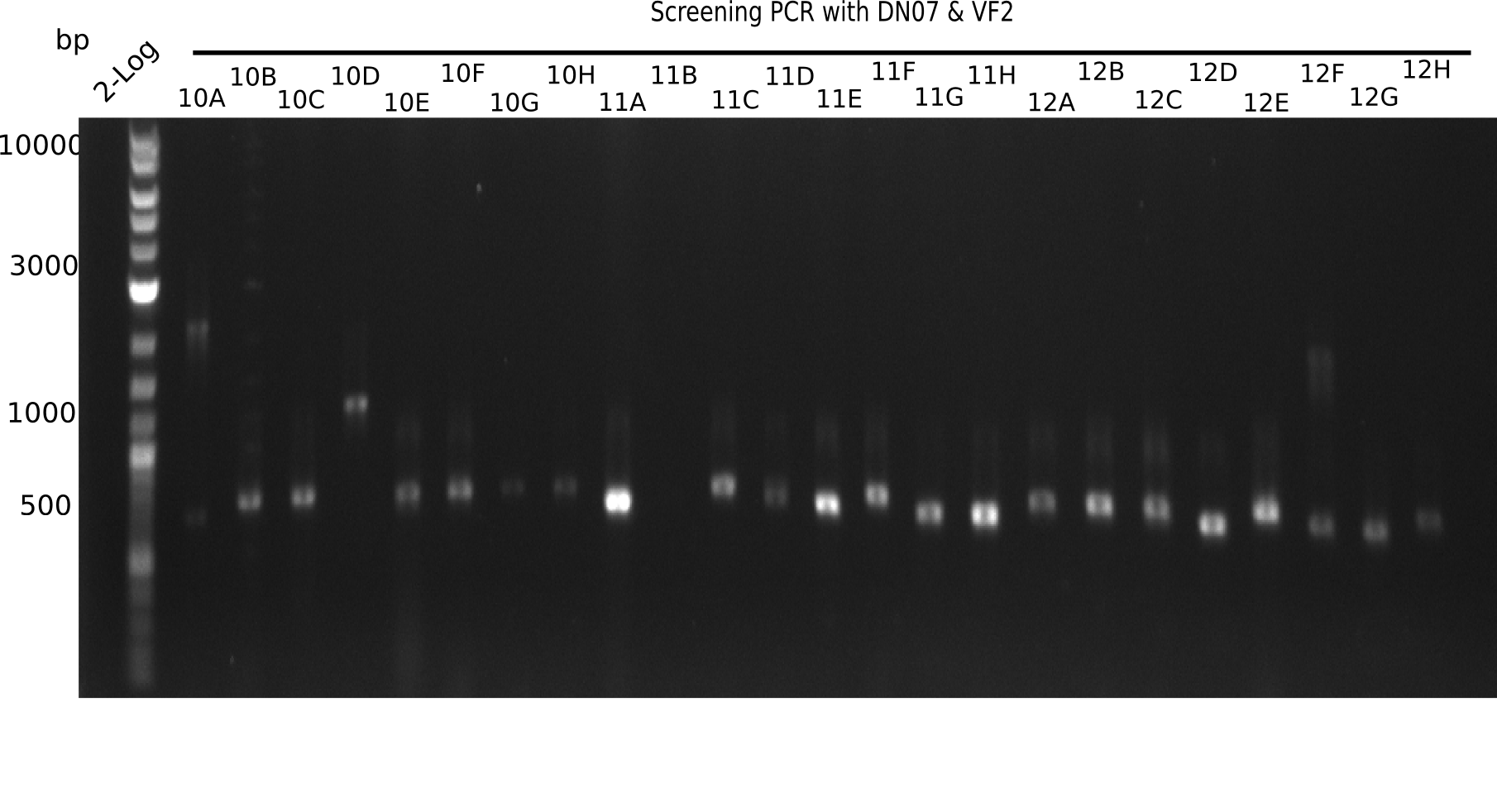
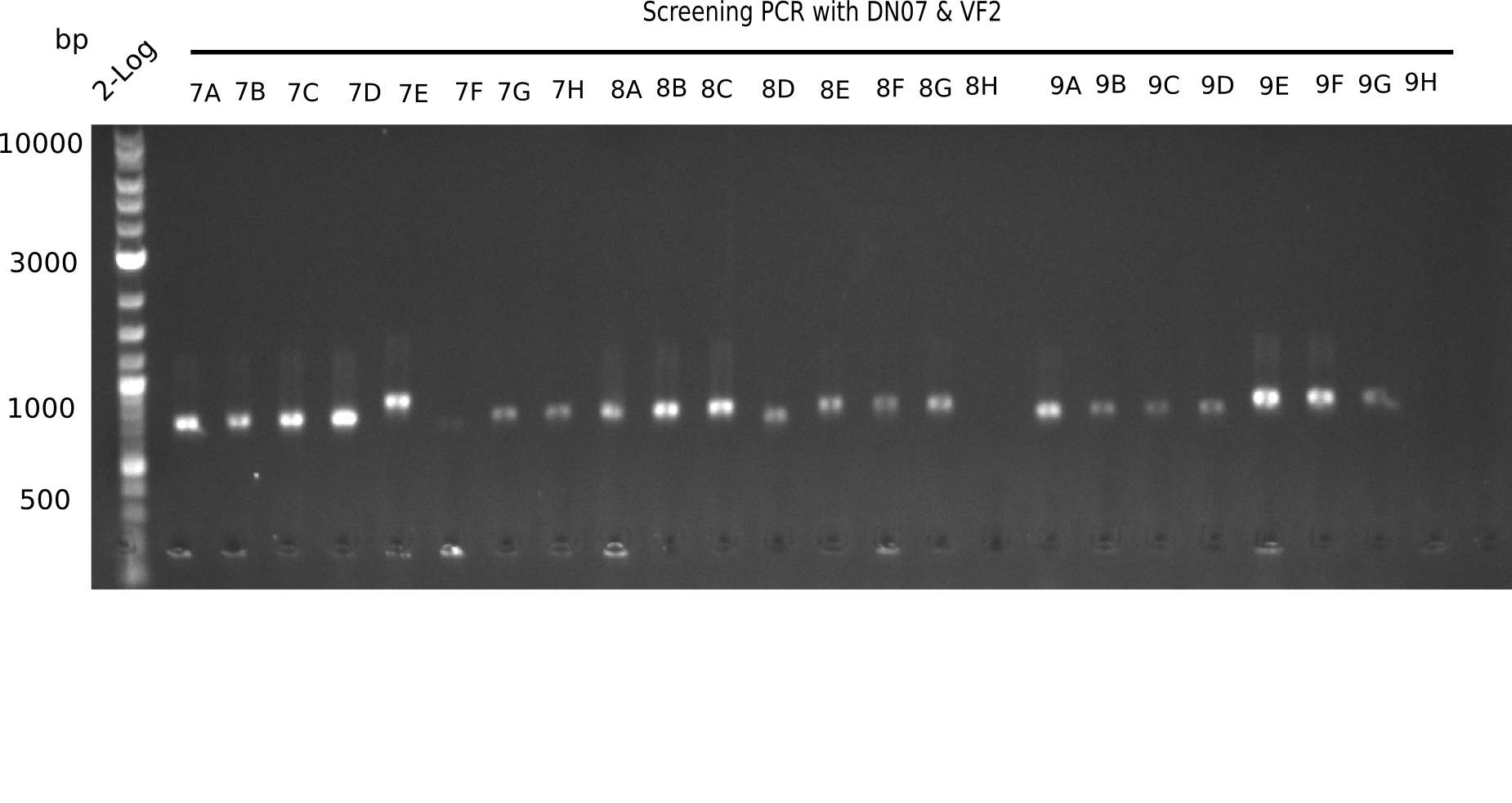
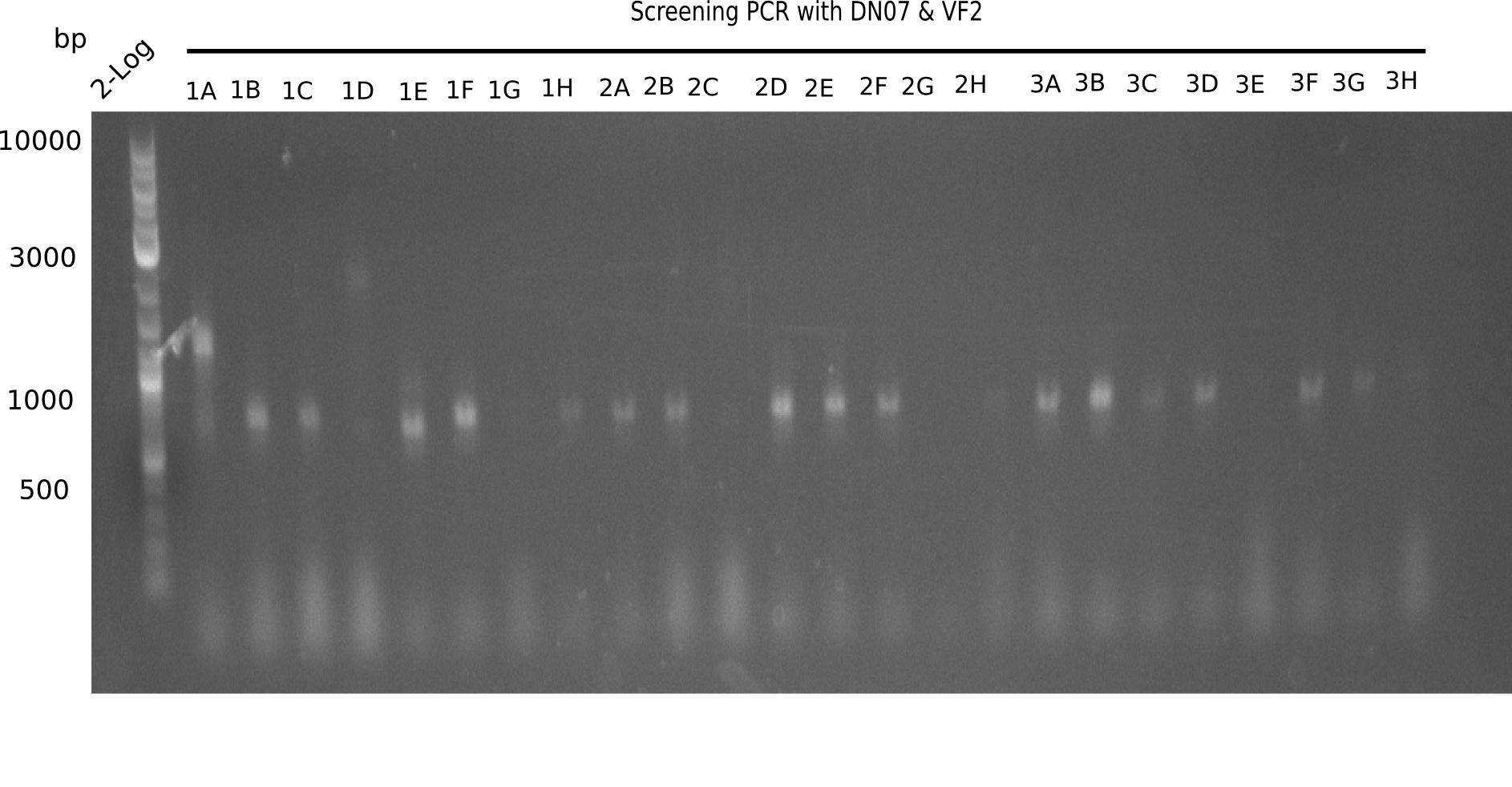
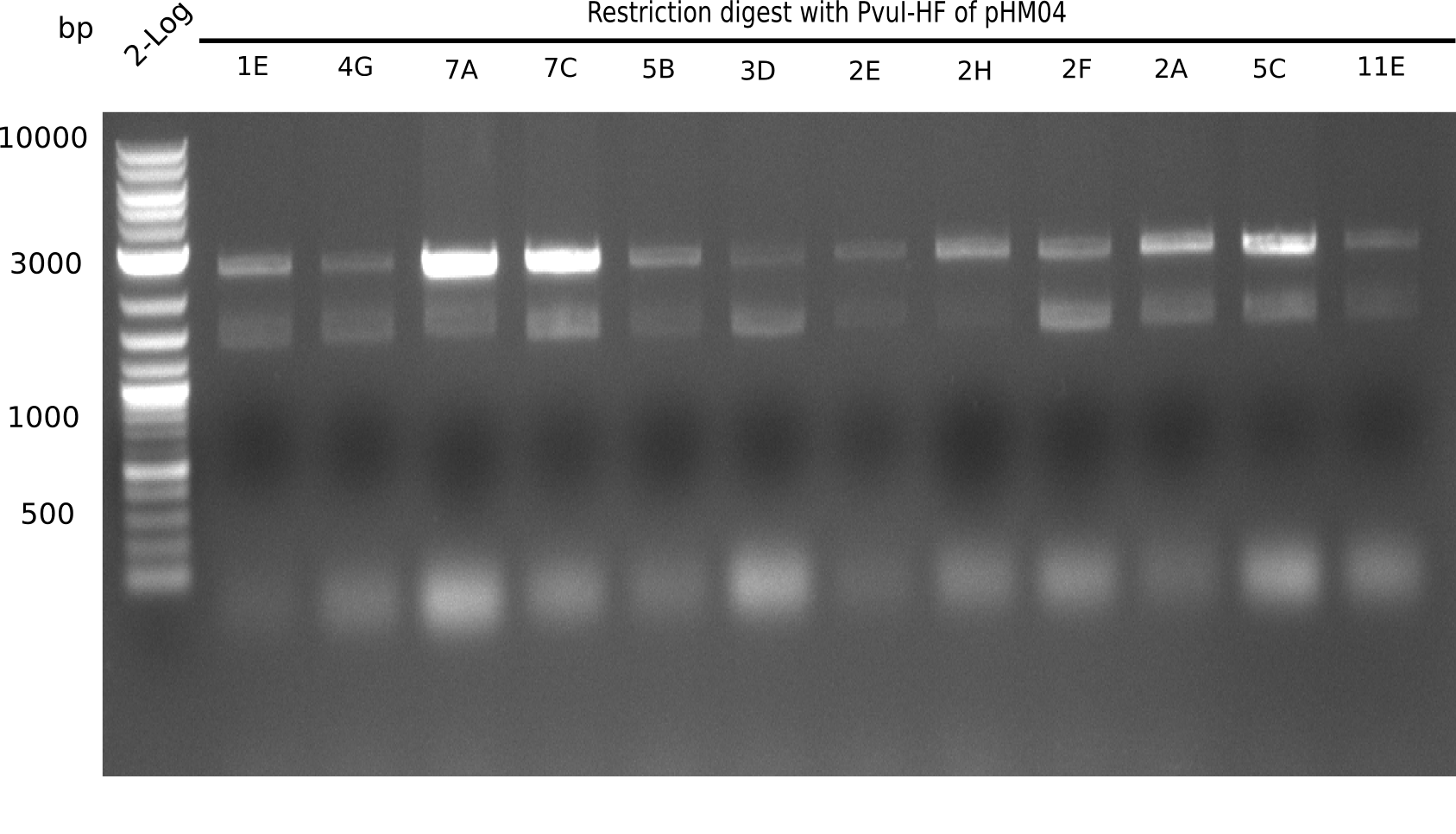
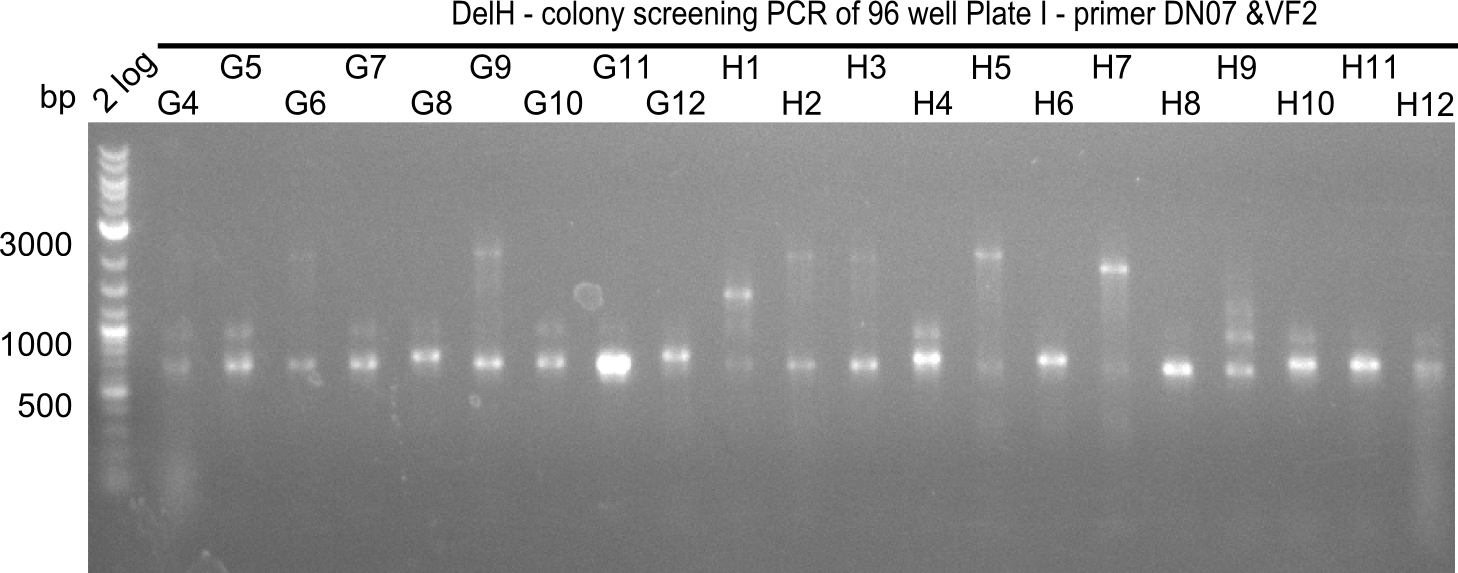
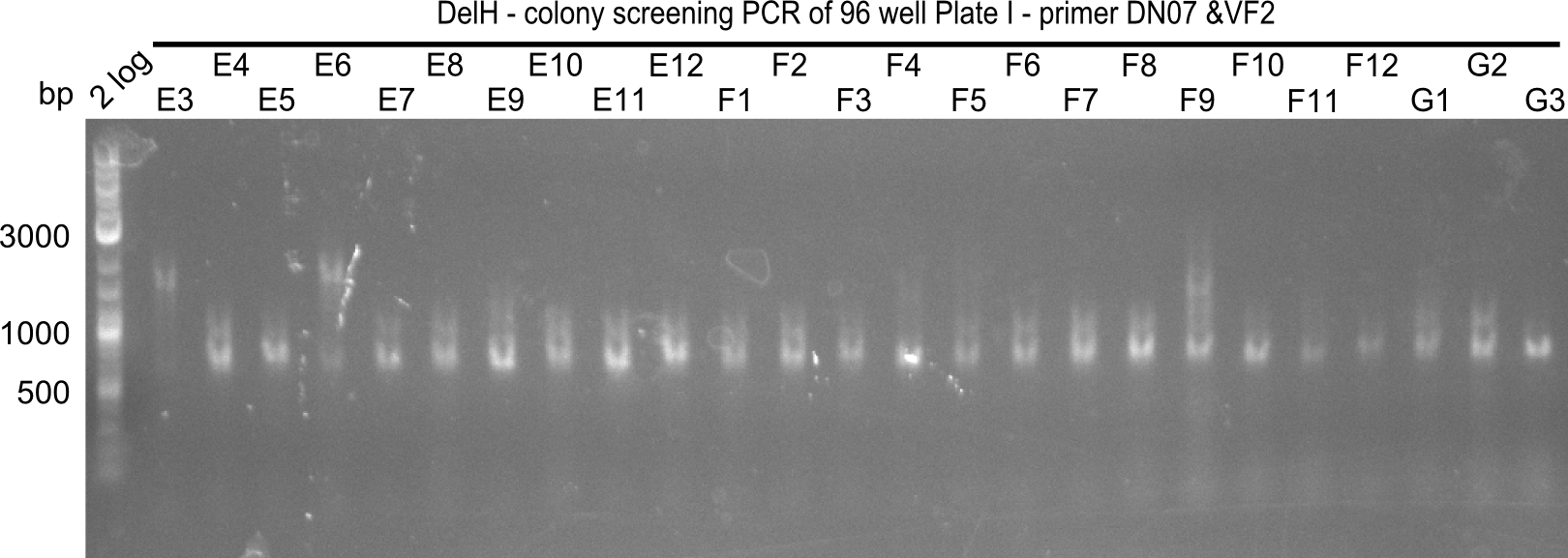
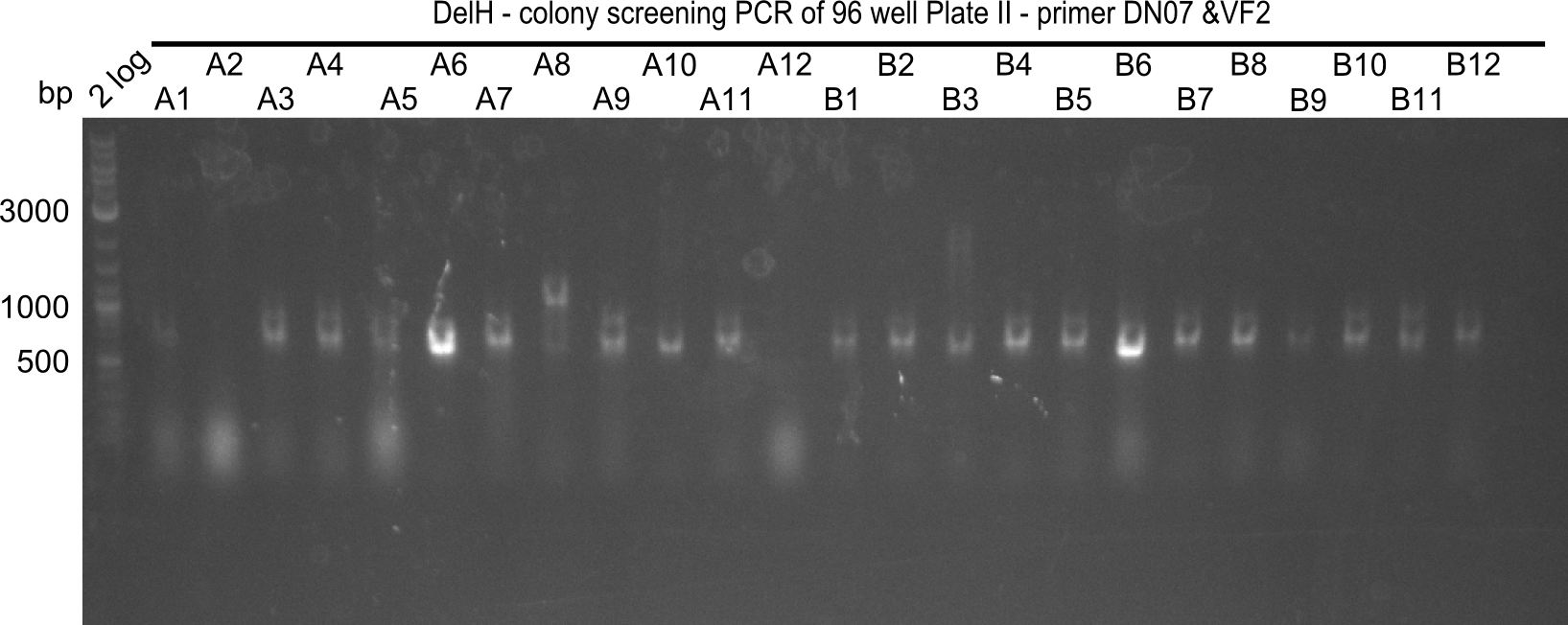
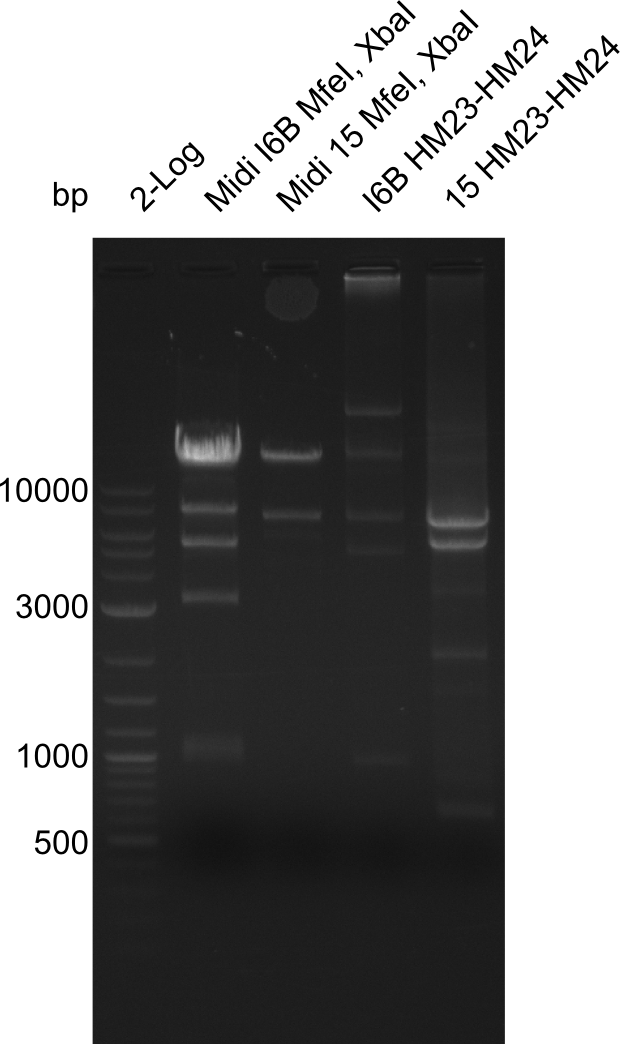
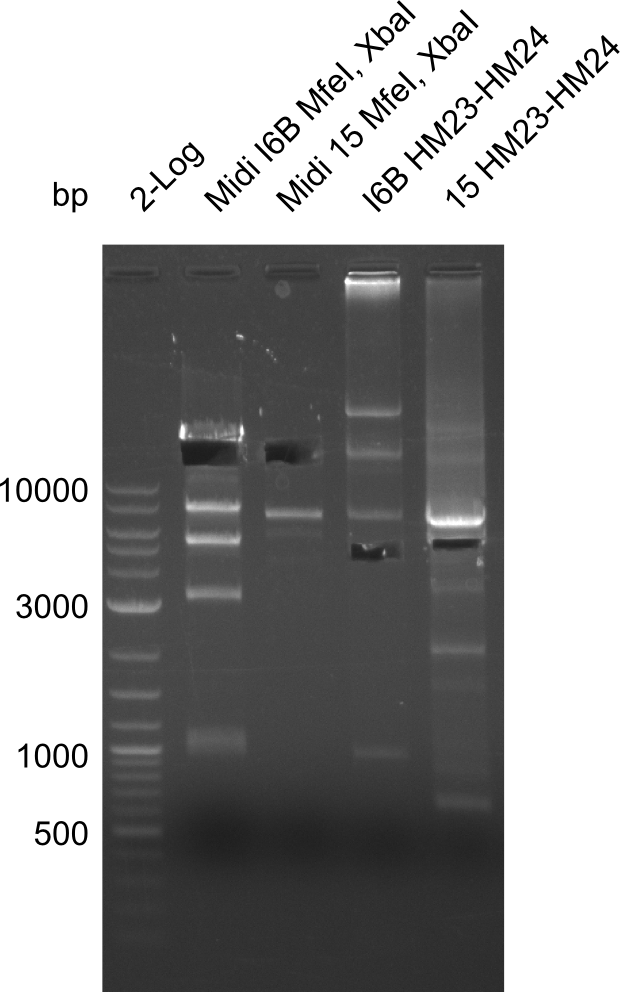
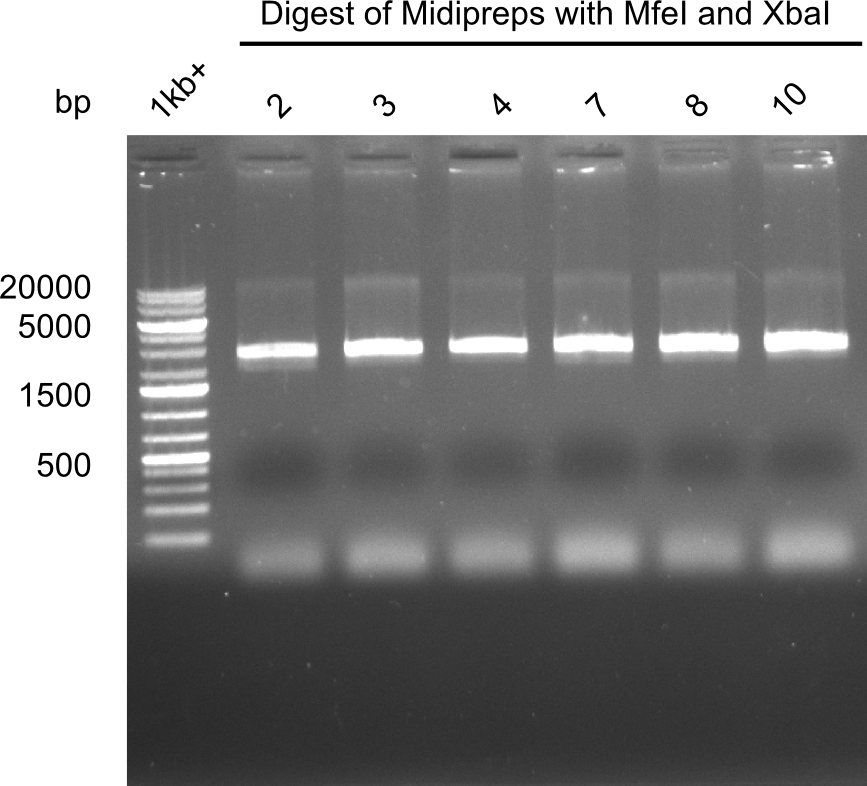
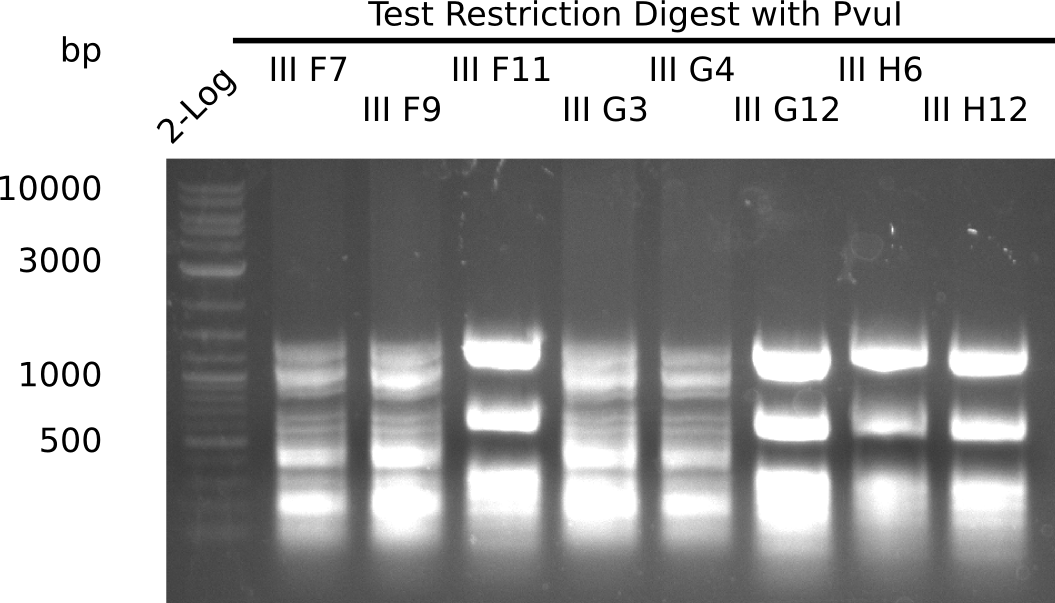
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After successful electroporation, we screened numerous clones by colony-PCR and test restriction digest (PvuI-HF). Midiprepped DNA dervied from clones positive for both methods were sent for sequencing. By sequencing of the transition sequence from the end of the pSB6A1 backbone without mRFP (pHM04) to the beginning of DelH, the assembly success of the Gibson assembly can be determined (Primer: reverse DN07 primer or VF2).Sequencing results showed truncating mutation at the beginning of DelH (in the primer region) for all clones. </p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 233: | Line 233: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 20</h1> | <h1>Week 20</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">As in week 19, the screening-PCRs showed colonies positive for the DelH contatining plasmid, whereas the restriction digest reveiled many of the screening results to be false positive. The remaining miniprepped colonies were sent in for sequencing. Sequencing results showed again truncating mutation at the beginning of DelH (in the primer region) for all clones. </p> |
</div> | </div> | ||
| Line 243: | Line 243: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 21</h1> | <h1>Week 21</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">None of the analyzed clones showed a correct sequence, which lead as to the assumptions following assumptions. First, we suspect DelH to be toxic for <i>E.coli</i>, thus only clones carrying the mutant DelH-plasmid survive. Second, we consider the low quality of the gibson primers as a possible explanation for the high number of mutations in the assemblies. To circumwent the latter problem, we will order HPLC purified primers. |
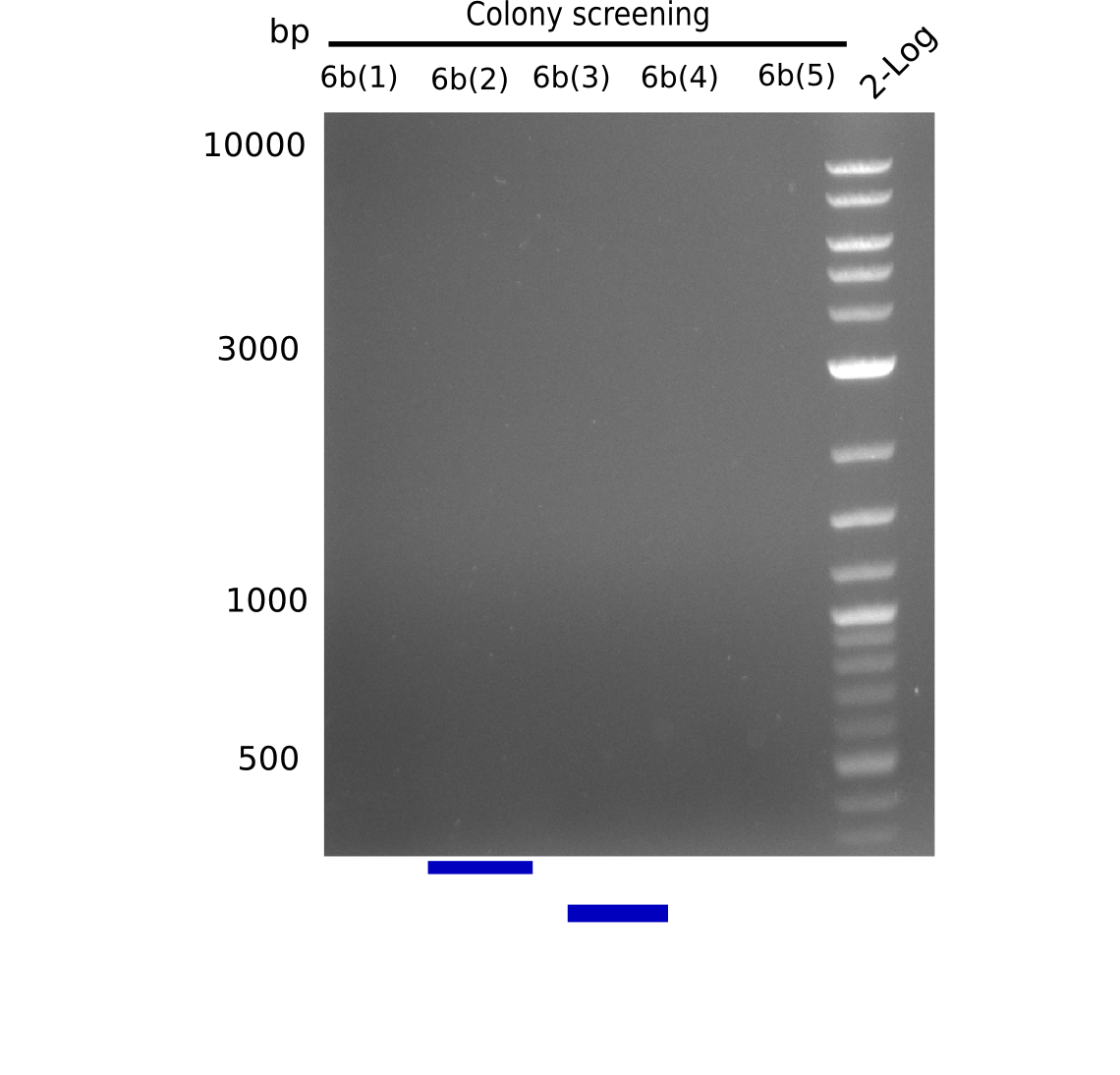
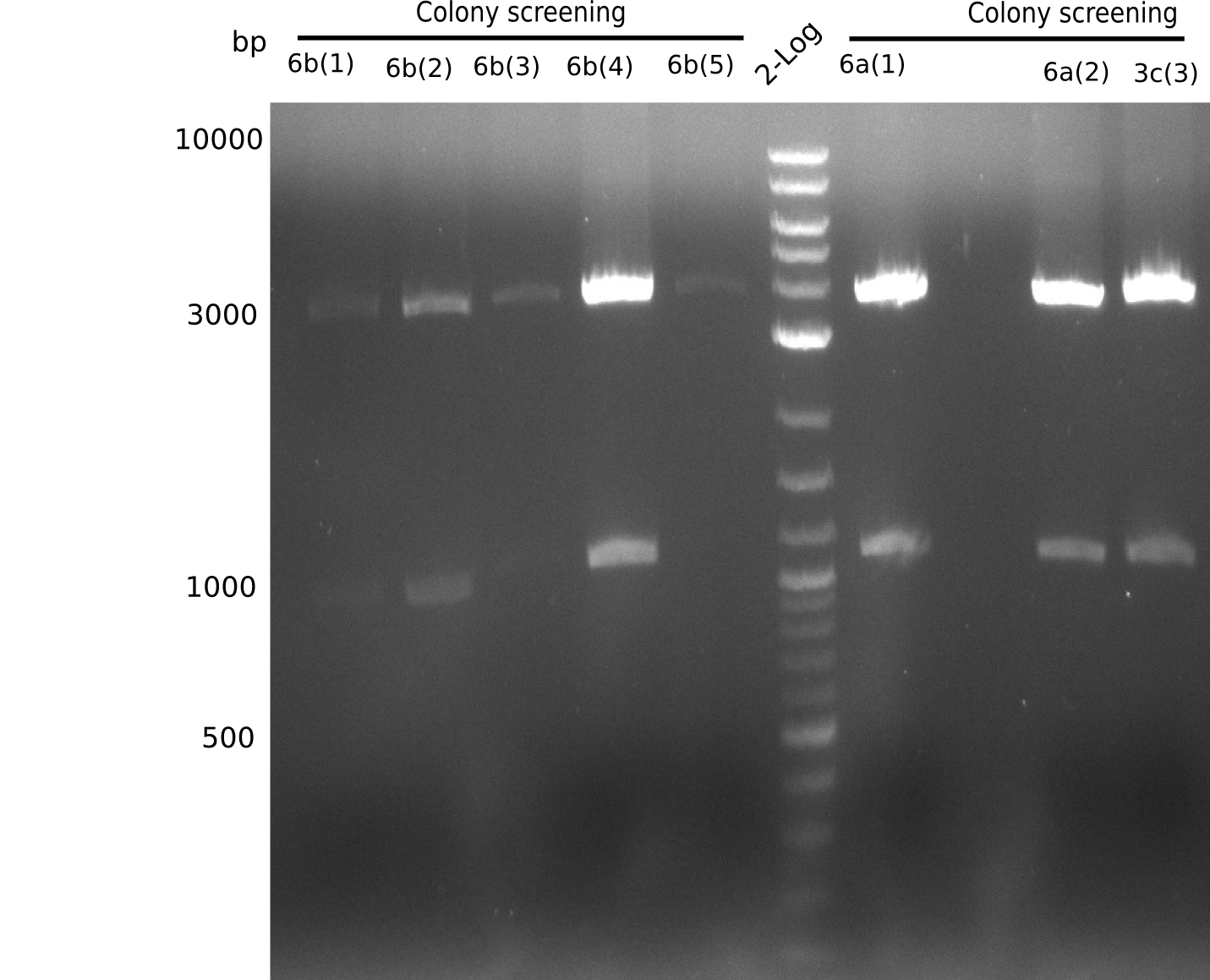
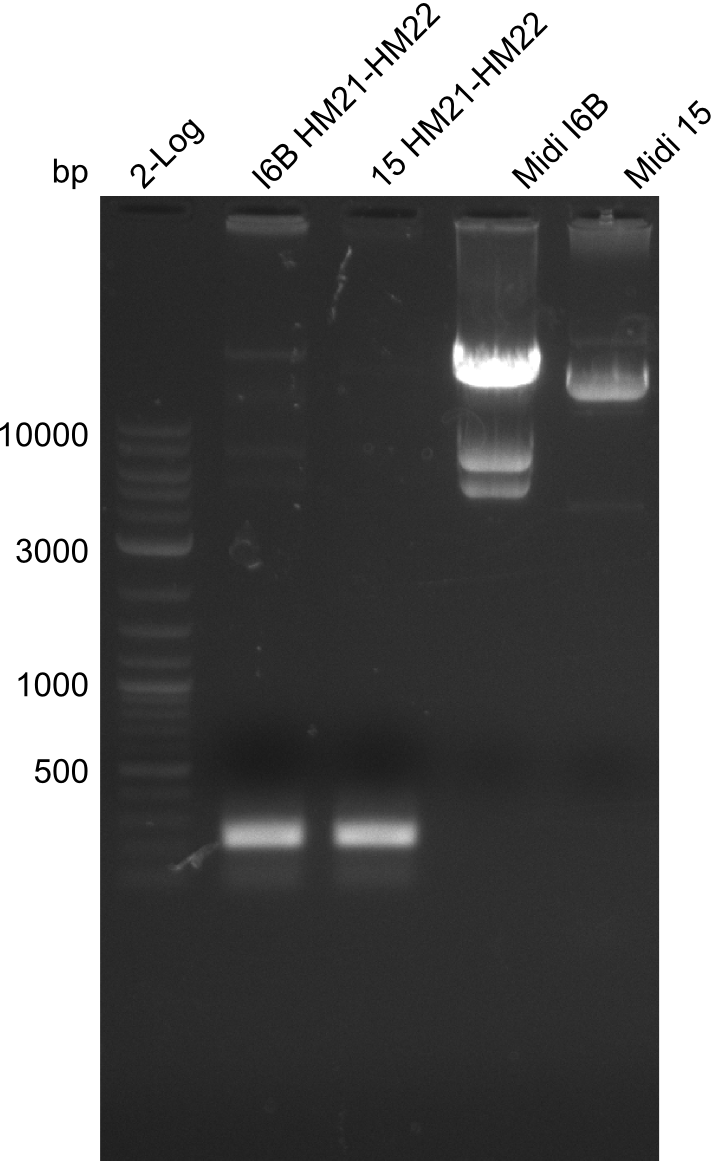
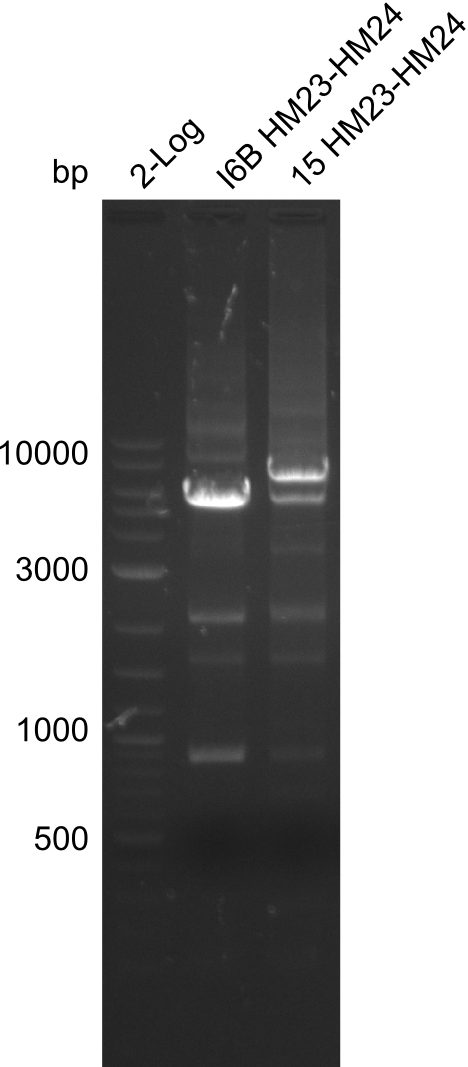
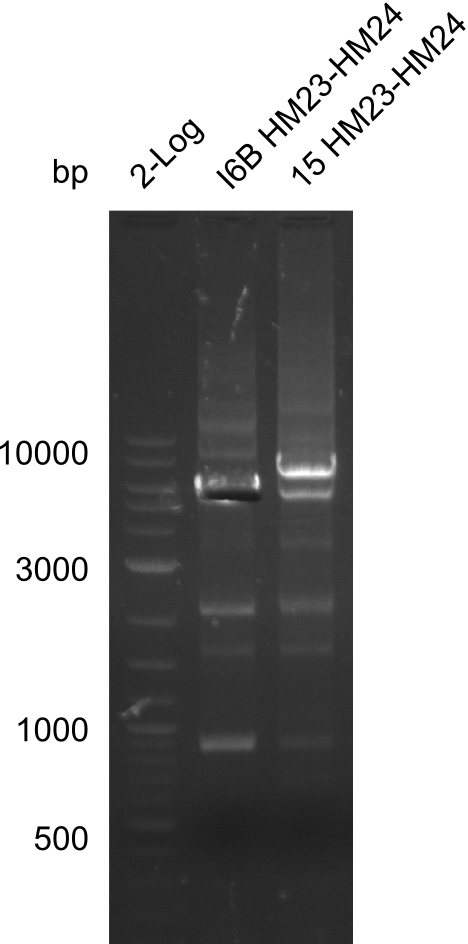
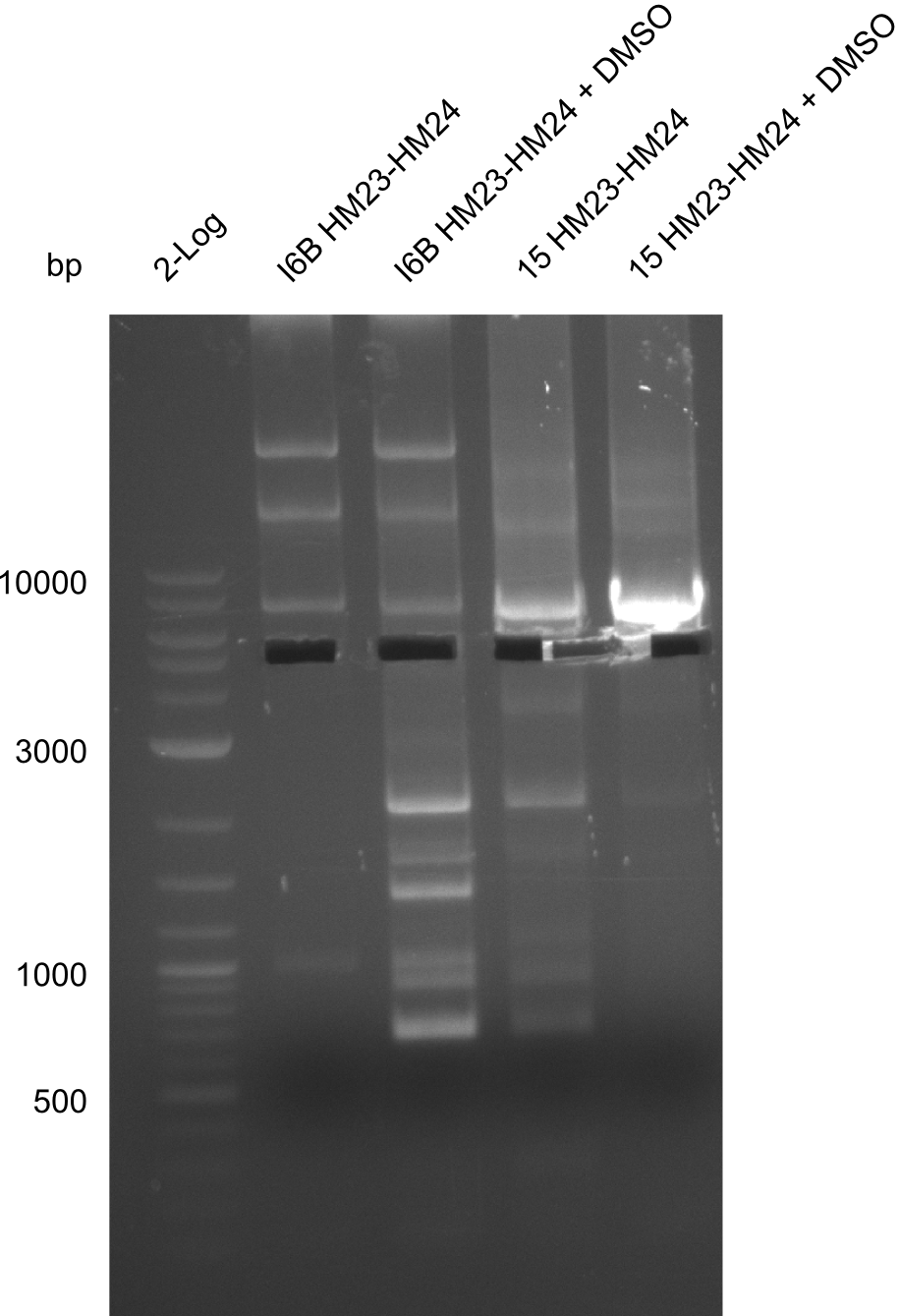
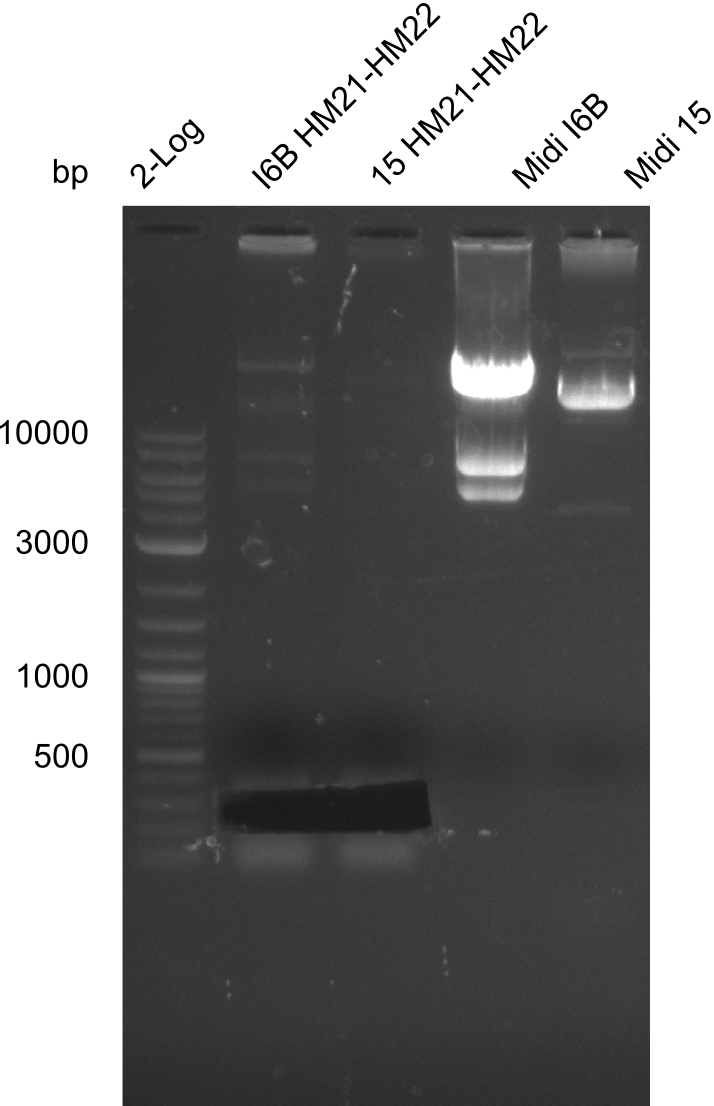
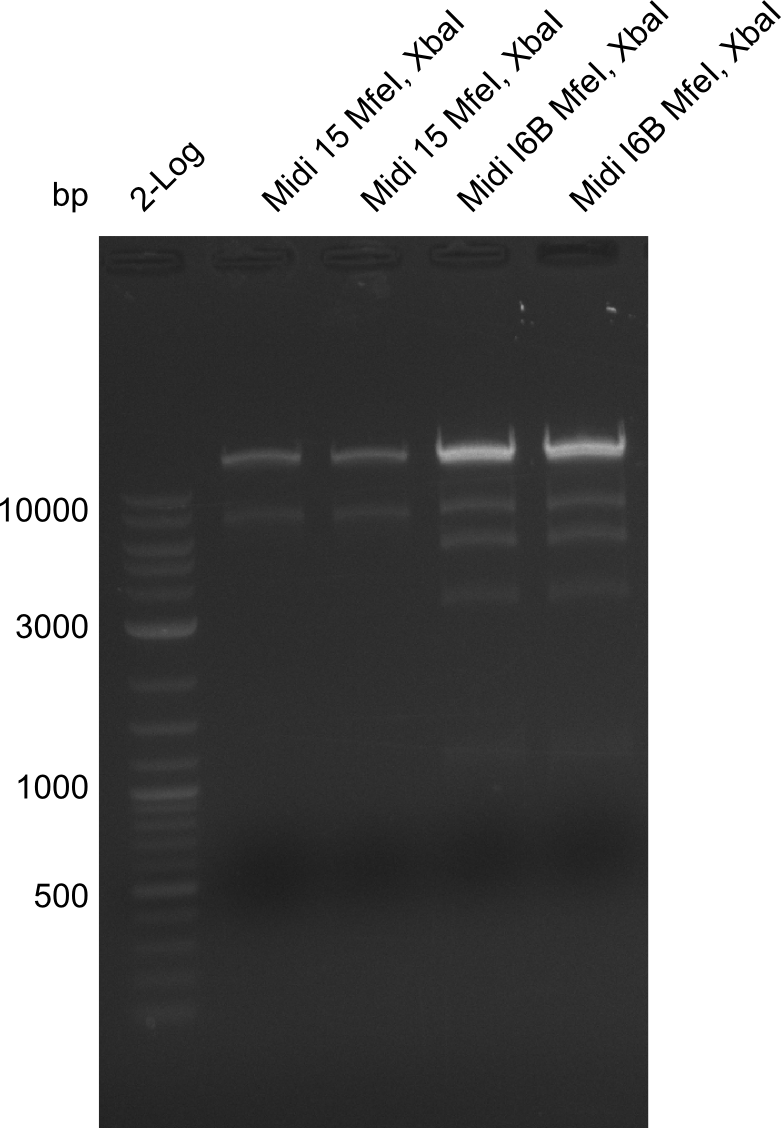
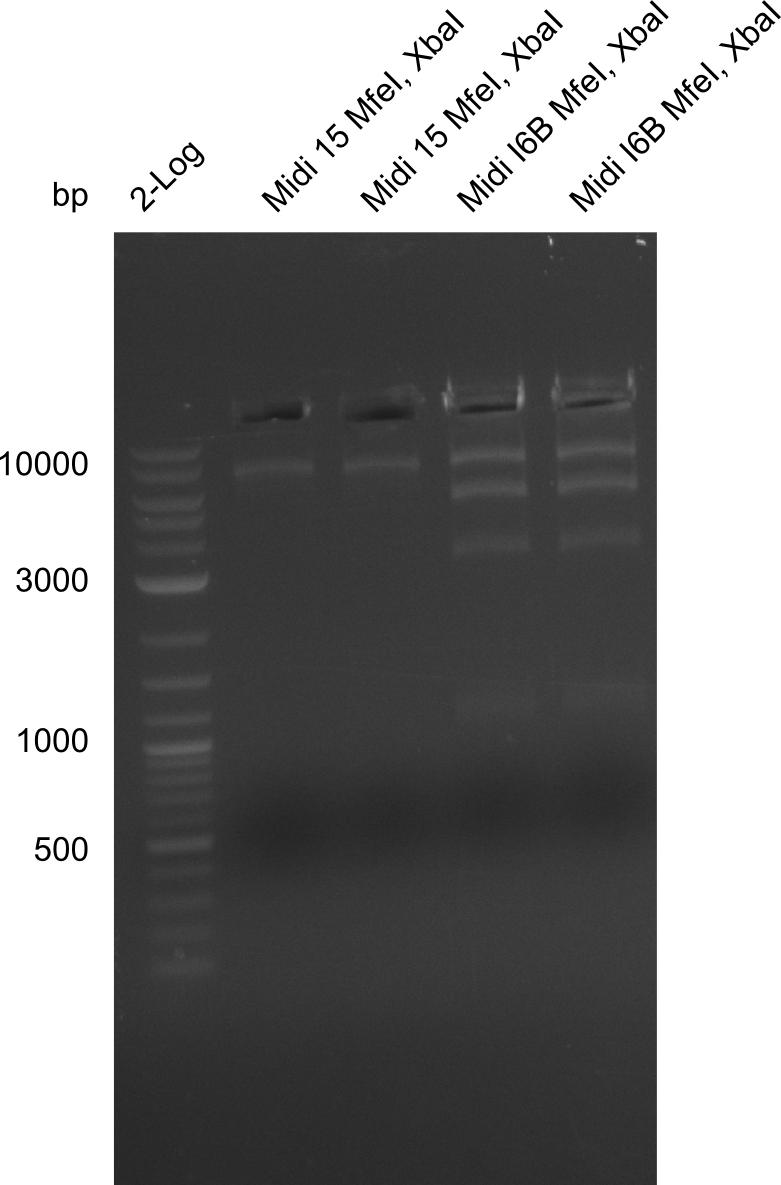
Additionally, we tried to eliminate the mutations in DelH clones I 6b and 15 by mutagenesis. Herefore, primers were ordered. </p> | Additionally, we tried to eliminate the mutations in DelH clones I 6b and 15 by mutagenesis. Herefore, primers were ordered. </p> | ||
</div> | </div> | ||
| Line 253: | Line 253: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 22</h1> | <h1>Week 22</h1> | ||
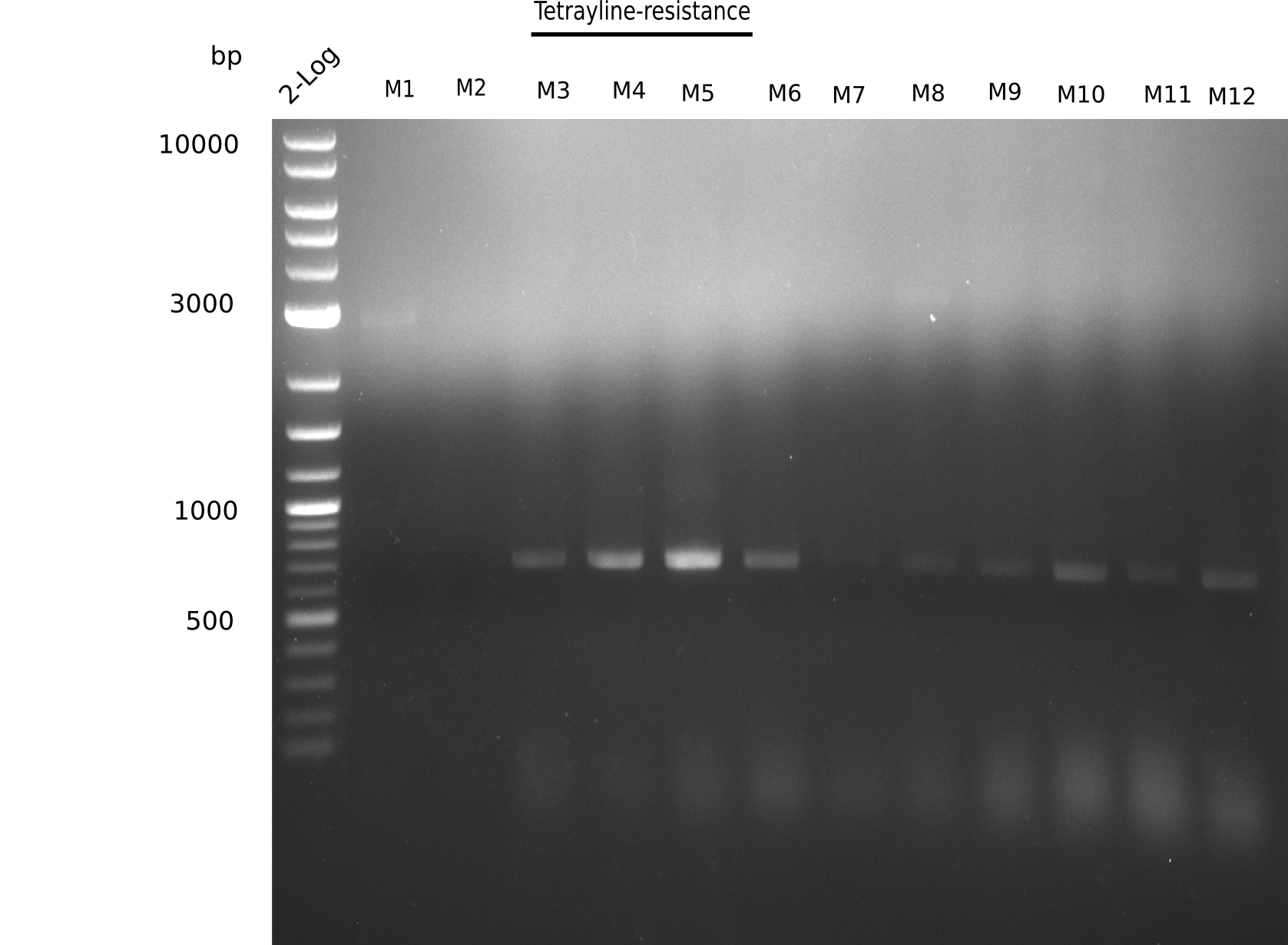
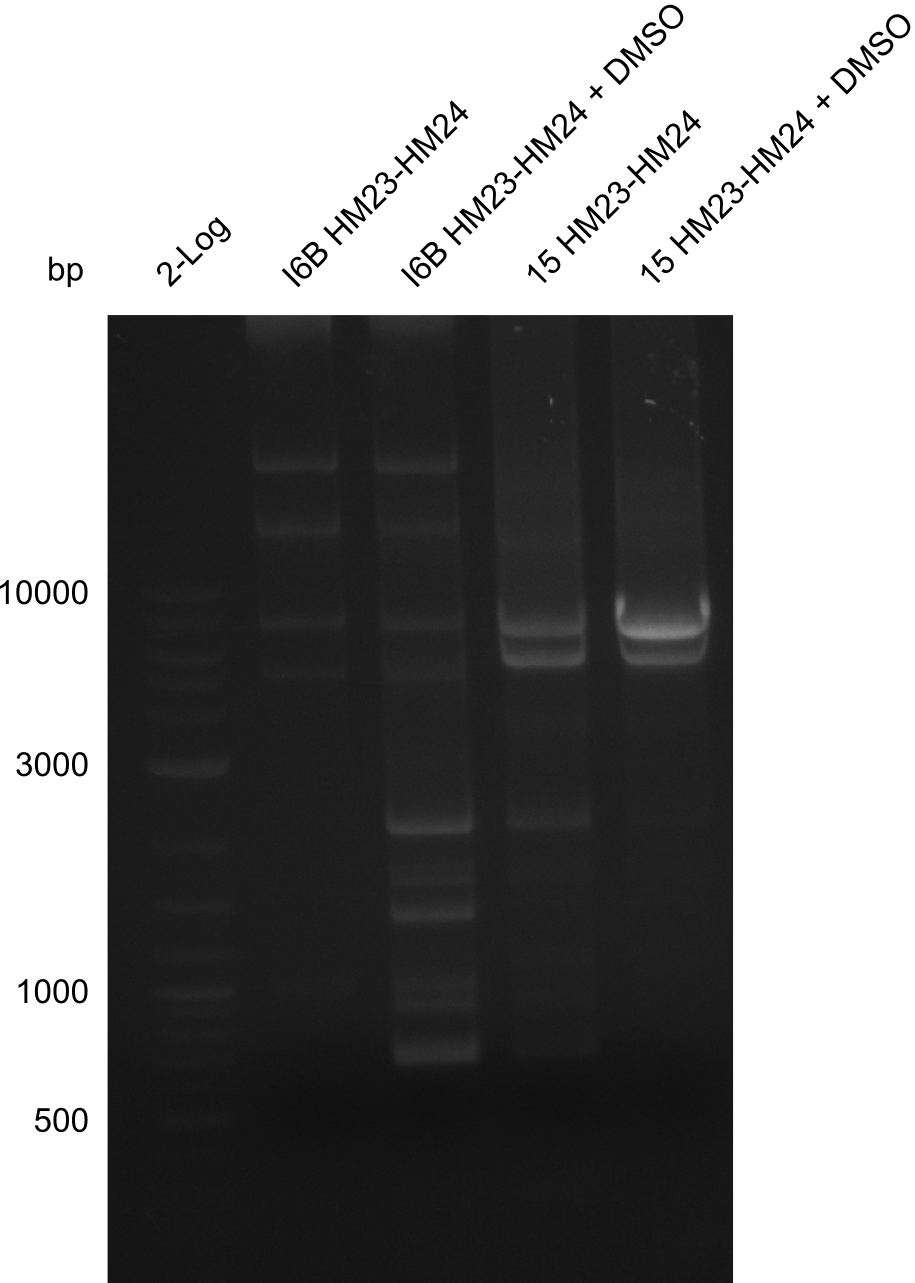
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">So far, we failed to | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">So far, we failed to obtain a single correct DelH clone. We suspect the DelH module to be toxic for <i>E. coli</i> when transformed without the other parts of the Del cluster, thus DelH-transformed cells would select for mutated plasmids. In order to reduce the selection pressure, we used <i>E. coli</i> BL21 DE3, known for increased expression of the lac repressor. This strategy also did not result in any correct clone. One reason might be, that the <i>E. coli</i> BL21 DE3 strain we obtained was actually a BL21 DE3 pLys strain, which itself is already Chloramphenicol resistant, thus not useful for screening and amplification of the DelH construct coded for on a chloramphenicol vector. |
| - | We | + | We obtained the correct <i>E. coli</i> BL21 DE3 strain as well as NEB <i>E. coli</i> turbo cells, which significantly overexpress the lac repressor. We found the lacZ-controlled expression of the latter to be very leaky when compared to <i>E. coli</i> BL21 DE3. |
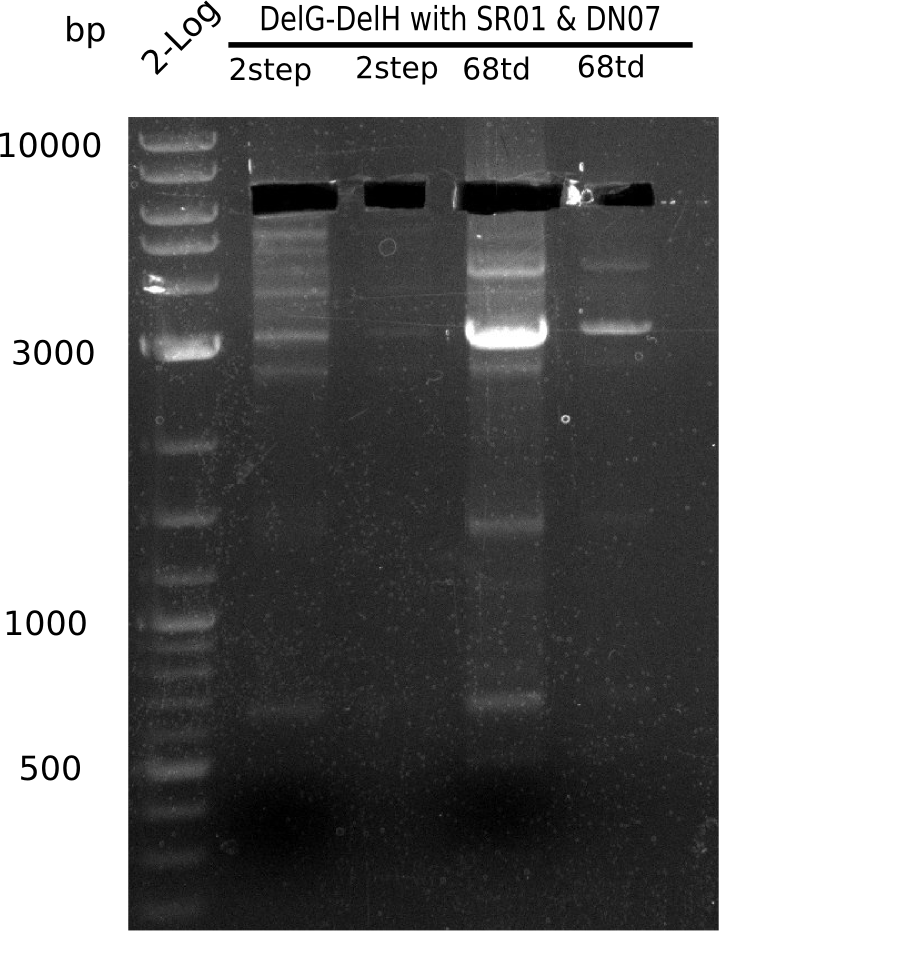
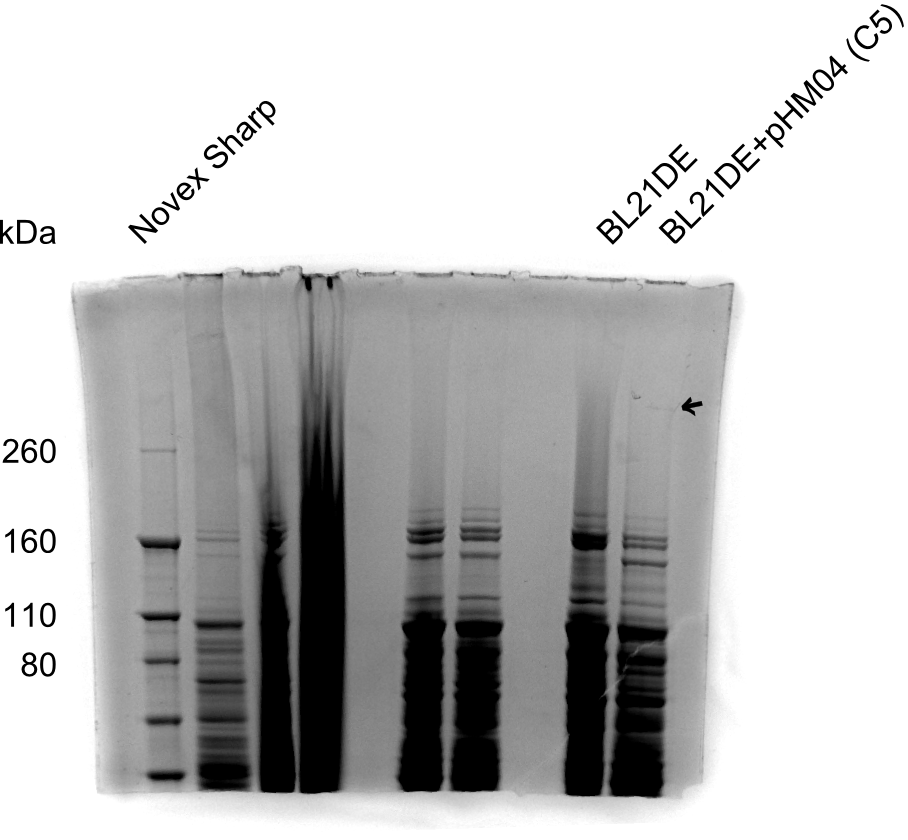
| - | Since the Gibson assembly of DelH using the HPLC purified primers also exclusively resulted in mutated clones, we developed two new cloning strategies to avoid expression of DelH. The first strategy uses a weak promoter and | + | Since the Gibson assembly of DelH using the HPLC purified primers also exclusively resulted in mutated clones, we developed two new cloning strategies to avoid expression of DelH. The first strategy uses a weak promoter and ribosomal binding site, the second introduces DelH in a ccdB helper construct. Lastly, we continued working on the mutagenesis approach to eliminate the mutation in clone C5. It was sent for sequencing... </p> |
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | <div class="item | + | <div class="item october first last"> |
<img src="data:image/png;base64,"/> | <img src="data:image/png;base64,"/> | ||
<div class="container"> | <div class="container"> | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | <h1>Week | + | <h1>Week 25</h1> |
| - | + | This week we tried to clone DelH into a plasmid with neither promotor nor ribosome binding site (pFS_03). We managed to obtain two clones which did not have the mutations usually observed at the beginning of DelH. This proves that DelH is in fact toxic. In parallel we constructed another plasmid (pFS_04) into which the correct DelH should then be ligated by common restriction enzyme based cloning. | |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 277: | Line 277: | ||
| - | <div class="col-sm-6"> | + | <div class="col-xs-12 col-sm-12 col-md-6"> |
| - | <div class="methods jumbotron" data-spy="scroll" data-target="#navbarExample" data-offset="0" | + | <div class="methods jumbotron" data-spy="scroll" data-target="#navbarExample" data-offset="0" style="height:364px"> |
| - | + | ||
| - | < | + | <div style="width:100%;"> |
| - | </ | + | </html>[[File:Heidelberg_ga_delf.png|450px]]<html> |
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
| - | |||
<div class="row"> | <div class="row"> | ||
| - | |||
| - | |||
<!--Start Weekly Labjournal--> | <!--Start Weekly Labjournal--> | ||
<!--Week1--> | <!--Week1--> | ||
| Line 301: | Line 298: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 334: | Line 329: | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 364: | Line 357: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 371: | Line 362: | ||
<div class="tab-pane active" id="a3"> | <div class="tab-pane active" id="a3"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | </html>{{:Team:Heidelberg/Templates/DelH | + | </html>{{:Team:Heidelberg/Templates/DelH overview3}}<html> |
</p> | </p> | ||
</div> | </div> | ||
| Line 396: | Line 387: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 403: | Line 392: | ||
| - | <div class="tab-pane active" id=" | + | <div class="tab-pane active" id="a4"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week4}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week4}}<html> | ||
| Line 419: | Line 408: | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | <li><a href="# | + | <li><a href="#a5" data-toggle="tab">Lab book</a></li> |
</ul> | </ul> | ||
| - | </div> | + | </div> |
| - | + | <div class="jumbotron"> | |
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="tab-content"> | <div class="tab-content"> | ||
| - | + | <div class="tab-pane active" id="a5"> | |
| - | + | ||
| - | <div class="tab-pane" id=" | + | |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week5}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week5}}<html> | ||
</p> | </p> | ||
</div> | </div> | ||
| - | |||
</div> | </div> | ||
</div> | </div> | ||
| Line 447: | Line 429: | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | <li><a href="# | + | <li><a href="#a6" data-toggle="tab">Lab book</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 459: | Line 439: | ||
| - | <div class="tab-pane" id=" | + | <div class="tab-pane active" id="a6"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week6}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week6}}<html> | ||
| Line 480: | Line 460: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 511: | Line 489: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0" |
| - | + | ||
| - | + | ||
| Line 519: | Line 495: | ||
| - | <div class="tab-pane" id="b8"> | + | <div class="tab-pane active" id="b8"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week8}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week8}}<html> | ||
| Line 528: | Line 504: | ||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
<!--Week9--> | <!--Week9--> | ||
| Line 535: | Line 510: | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | <li><a href="# | + | <li><a href="#a9" data-toggle="tab">Lab book</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 547: | Line 520: | ||
| - | <div class="tab-pane" id=" | + | <div class="tab-pane active" id="a9"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week9}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week9}}<html> | ||
| Line 568: | Line 541: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 575: | Line 546: | ||
| - | <div class="tab-pane" id="b10"> | + | <div class="tab-pane active" id="b10"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week10}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week10}}<html> | ||
| Line 596: | Line 567: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| - | |||
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 628: | Line 596: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 635: | Line 601: | ||
| - | <div class="tab-pane" id="b12"> | + | <div class="tab-pane active" id="b12"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week12}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week12}}<html> | ||
| Line 656: | Line 622: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 688: | Line 652: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
<div class="tab-content"> | <div class="tab-content"> | ||
| - | <div class="tab-pane" id="b14"> | + | <div class="tab-pane active" id="b14"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week14}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week14}}<html> | ||
| Line 715: | Line 677: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
<div class="tab-content"> | <div class="tab-content"> | ||
| - | <div class="tab-pane active | + | <div class="tab-pane active" id="b15"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week15}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week15}}<html> | ||
| Line 746: | Line 701: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 778: | Line 731: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 785: | Line 736: | ||
| - | <div class="tab-pane" id="b17"> | + | <div class="tab-pane active" id="b17"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/DelH week17}}<html> | </html>{{:Team:Heidelberg/Templates/DelH week17}}<html> | ||
| Line 806: | Line 757: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 838: | Line 787: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 870: | Line 817: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 902: | Line 847: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| Line 934: | Line 877: | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| - | |||
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 955: | Line 895: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| - | <!-- | + | <!-- Week 25 --> |
<div class="labjournal-weekly"> | <div class="labjournal-weekly"> | ||
<div> | <div> | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | <li class="active"><a href="# | + | <li class="active"><a href="#a25" data-toggle="tab">Overview</a></li> |
| - | <li><a href="# | + | <li><a href="#b25" data-toggle="tab">Lab book</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
<div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | |||
| - | |||
| - | |||
<div class="tab-content"> | <div class="tab-content"> | ||
| - | + | ||
| - | + | <div class="tab-pane active" id="a25"> | |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Templates/DelH_overview25}}<html> | ||
</p> | </p> | ||
</div> | </div> | ||
| - | + | <div class="tab-pane" id="b25"> | |
| - | <div class="tab-pane" id=" | + | |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/DelH_week25}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| Line 992: | Line 932: | ||
</div> | </div> | ||
</html> | </html> | ||
| + | {{:Team:Heidelberg/Templates/Footer-Nav}} | ||
{{:Team:Heidelberg/Templates/Footer-DelH}} | {{:Team:Heidelberg/Templates/Footer-DelH}} | ||
Latest revision as of 03:33, 29 October 2013


 "
"