Team:Heidelberg/Delftibactin/DelRest
From 2013.igem.org
Nils.kurzawa (Talk | contribs) |
|||
| (55 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:Heidelberg/Templates/Navigation}} | {{:Team:Heidelberg/Templates/Navigation}} | ||
| + | {{:Team:Heidelberg/Templates/scrollbar-css}} | ||
| + | |||
<html> | <html> | ||
<style type="text/css"> | <style type="text/css"> | ||
| Line 11: | Line 9: | ||
p { | p { | ||
font-size:14px; | font-size:14px; | ||
| + | text-align:justify; | ||
| + | } | ||
| + | .carousel-inner { | ||
| + | margin-top:20%; | ||
| + | } | ||
| + | .tab-content .wikitable { | ||
| + | font-size:9pt; | ||
} | } | ||
</style> | </style> | ||
<div class="container"> | <div class="container"> | ||
<!--Project Description--> | <!--Project Description--> | ||
| - | <div | + | <div> |
| - | <h1><span style="font-size:180%;color:#FFCC00;">Del Rest.</span><span class="text-muted" style="font-size:120%"> Creating a 32 | + | <h1><span style="font-size:180%;color:#FFCC00;">Del Rest.</span><span class="text-muted" style="font-size:120%"> Creating a 32 kb plasmid.</span></h1> |
| - | + | ||
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col-sm- | + | <div class="col-sm-12 col-md-6"> |
<!--Months--> | <!--Months--> | ||
| - | <ul class="pagination" style="margin-bottom:2%; margin-left: | + | <ul class="pagination" style="margin-bottom:2%; margin-left:17%;"> |
<!--<button type="button" class="btn btn-default">May</button> | <!--<button type="button" class="btn btn-default">May</button> | ||
| Line 65: | Line 70: | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | This week the project DelRest was launched | + | This week the project "DelRest" was launched, which aims at creating a single, 32 kb plasmid enabling the expression of most genes from the <i>D. Acidovorans </i> Del cluster, namely DelA-G and DelL-P (note: the 18 kbp gene encoding DelH is cloned onto a seperate plasmid). Due to the shere size and complexity of the DelRest construct, we decided to use Gibson cloning. |
| - | + | This weeks goal was amplify the pSB4K5 backbone from the partsregistry with primers giving the intended overlaps as well as the desired genes from our host organism <i>D. Acidovorans </i>. | |
| - | + | Therefore, Gibson primers for the amplification of the target backbone pSB4K5 were designed, which also introduce a Gibson-overlap to DelA, the first gene present in the Del cluster. Furthermore Gibson primers for amplifying the fragments DelA-G and DelO-P were ordered. The last Gibson primer pair used to amplify DelL consequently carries the required overlap to the beginning of the mRFP reporter present the pSB4K5 insert part BBa_J04450. Check out our vectormap if you are curious about the detailed cloning strategy and primer design. | |
</p> | </p> | ||
| Line 78: | Line 83: | ||
<h1>Week 11</h1> | <h1>Week 11</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | During the past week we managed to amplify | + | During the past week we managed to amplify a fragment of 11 kbp encoding the genes DelA-F from genomic DNA of <i>D. Acidovorans</i>. We were further able to amplify the backbone fragment from pSB4K5 carrying the desired overlaps to the corresponding fragments to be amplified from the Del cluster. We also succeeded in the amplification of the very last gene in our construct, DelL. Unfortunately, the amplification of DelF-G and DelO-P turned out to be more difficult than expected. Since our initial plan, which required the amplification of DelO-P using a Gibson primer which introduces an overlap to DelL and furthermore introduces an artificial ribosom binding site did not work out, we changed our strategy. We orderd shorter versions of all our Gibson primers. Using these shorter primers not bearing any overlaps to other fragments, we will try to amplify the desired genes in order to obtain a specific template for the reamplification with the primers carrying the needed overlaps. Furthermore, we will optimize the PCR conditions for the amplification of the DelF-G fragment. |
</p> | </p> | ||
| Line 90: | Line 95: | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | By the end of last week we | + | By the end of last week we found out, that the short primers we had ordered did not improve amplification of the missing fragments from <i>D. Acidovorans. </i> Additionly, further analysis of the Delftibactin cluster, led to the discovery of a predicted promoter present in front of DelO-P potentially essential for DelO-P expression. Therefore, we not only decided to design new primers for the amplyfication of DelF-G, but also modifie the entire strategy concerning the DelO-P fragment. To ensure the expression of DelO-P in our target organism <i> E.coli </i> we decided to amplify DelO-P together with its putative promoter. In consequence, we ordered new primers for DelO-P and also for the last DelA-G fragment in order to introduce the required new overhang to the new DelO-P fragment. The correlating primers can be found in our new vector map. |
</p> | </p> | ||
| Line 101: | Line 106: | ||
<h1>Week 13</h1> | <h1>Week 13</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | In the previous week several of the | + | In the previous week, several of the newly ordered primer pairs improved our PCRs for DelO-P and DelF-G. However, results were not convincing enough to use these amplicons for Gibson assembly. Therefore, we spend this week optimizing the PCRs for the abovementioned fragments. In addition we validated the amplicon of DelG with restriction digest. Validation of the other PCR products did not succeed, mostly due to very low amount of DNA. |
</p> | </p> | ||
</div> | </div> | ||
| Line 111: | Line 116: | ||
<h1>Week 14</h1> | <h1>Week 14</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | + | We were still struggeling with getting the correct amplicons for the fragments encoding DelF-G as well as DelO-P. Furthermore, the restriction digests of the already successfully amplified fragments needed to be repeated using higher amounts of DNA, as results of the previous test digests were rather inconclusive. Furthermore, samples of these fragments were send for sequencing. | |
</p> | </p> | ||
</div> | </div> | ||
| Line 122: | Line 127: | ||
<h1>Week 15</h1> | <h1>Week 15</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| + | Single read sequencing of the PCR amplified fragments DelA-E, DelL as well as the backbone pSB4K5 | ||
| + | was carried out by GATC. We blasted the obtained seqeuences against the reference sequence based on the <i>D. acidovorans SPH-1</i> strain available on NCBI. Although our sequence matched to the reference sequence, we found a significant number of missmatches, which was too diverse and high to be simply explained by random mutations introduced by our polymerase during PCR (note: we used a high-fidelity proofreading polymerase suitable for GC-rich templates and long amplicons). We thus hypothesized that the SPH-1 strain based on which our cloning strategy was designed might have a significant number of single-nucleotide polymorphisms in the Del cluster compared to the <i>D. acidovorans</i> DSM-39 strain, which we used as template for all PCRs (note: there is no complete genomic sequence available for the DSM-39 strain on NCBI). We further hypothesized, that this difference in sequence between the DSM-39 strain used as PCR template and the SPH-1 strain based on which our primers were designed could explain our troubles with the PCR amplifications of DelF-G and DelO-P. In consequence, we ordered the SPH-1 strain from the DSMZ in order to obtain a suitable template for our PCRs. This solved all our PCR problems right away and we were able to get all amplicons required for cloning the DelRest construct within this week. Furthermore, we successfully validated our amplicons by restriction digest and sequencing. | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 131: | Line 138: | ||
<h1>Week 16</h1> | <h1>Week 16</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| + | Last week we successfully amplified all the fragments needed to complete the DelRest cloning. Therefore, we went ahaed and performed Gibson assembly in order to assembel the final pFSN plasmid carrying bearing the DelRest genes. The assembly mix was transformed into <i>E. coli DH10beta</i> electrocompetent cells. Screening PCRs showed that the assembly was successful, calling for futher validation of the clones. | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 141: | Line 149: | ||
<h1>Week 17</h1> | <h1>Week 17</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| + | From colonies which were positive for screening last week, we rescued 3 plasmids. Test restriction digest were conducted and showed our clones to be correct. | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 151: | Line 160: | ||
<h1>Week 18</h1> | <h1>Week 18</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| + | We send one of our DelRest constructs that showed the right restriction pattern in the test digest for sequencing. As it would be quite costly to sequence the whole 32 kb plasmid, we focused on the ligation sites between the different assembly fragments, which are most prone to insertion of errors. Although all insert fragments sequenced, including the ligation sites between DelA and DelF, DelO-P and DelL, were 100 % correct, we detected a mutation within the mRFP cds (note: we wanted to use mRFP in order to confirm expression from our DelRest plasmid). FACS analysis of <i> E. coli </i> bearing the DelRest construnct confirmed that mRFP expression was impaired, likely due to the corresponding mutation. However, as mRFP was only meant to be a general expression control on our construct, we did not start correcting out construct by mutagenesis. Instead, we started preparing samples for an SDS page in order to directly proof the expression of the Del genes by Coomassie staining. | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 160: | Line 170: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 19</h1> | <h1>Week 19</h1> | ||
| + | The Coomassie staining we carried out last week did not display all expected bands clearly. This occured most likely due to the low amount of protein loaded onto the corresponding SDS page. Therefore the SDS page was repeated. | ||
| + | This expression analysis confirmed the results indicated by the previous SDS-page. We were able to show that DelE and DelG are expressed at a detectable level. | ||
| + | As we could confirm the successful cloning and functioning of the DelRest expression plasmid named pFSN (for Florian-Sophie-Nils :)) the DelRest subproject was succssfully completed and the personal resources of the DelRest group were shifted to the DelH group as well as the wiki. | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
</p> | </p> | ||
| Line 171: | Line 184: | ||
</div> | </div> | ||
</div><!-- /.carousel --> | </div><!-- /.carousel --> | ||
| + | </div> | ||
| + | <div class="col-sm-12 col-md-6" style="margin-top:7%"> | ||
| + | <div class="jumbotron"> | ||
| + | |||
| + | <div style="width:100%;"> | ||
| + | <a class="fancybox fancyGraphical" rel="group" href="https://static.igem.org/mediawiki/2013/e/e3/Heidelberg_ga_delf.png"> | ||
| - | + | <img style="width:100%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 4px; border-color: grey;" src="https://static.igem.org/mediawiki/2013/e/e3/Heidelberg_ga_delf.png"></img> | |
| - | + | ||
| - | + | </a> | |
| - | + | ||
</div> | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| - | <div class="col-sm- | + | <div class="container"> |
| + | <div class="row"> | ||
| + | <div class="col-sm-12"> | ||
<!--Start Weekly Labjournal--> | <!--Start Weekly Labjournal--> | ||
| Line 198: | Line 223: | ||
</div> | </div> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 204: | Line 229: | ||
<div class="tab-pane active" id="a10"> | <div class="tab-pane active" id="a10"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week10_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 285: | Line 310: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a11"> | <div class="tab-pane active" id="a11"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Templates/Del_week11_overview}}<html> | ||
</p> | </p> | ||
| Line 370: | Line 396: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 376: | Line 402: | ||
<div class="tab-pane active" id="a12"> | <div class="tab-pane active" id="a12"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week12_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 438: | Line 464: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a13"> | <div class="tab-pane active" id="a13"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week13_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 516: | Line 542: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 522: | Line 548: | ||
<div class="tab-pane active" id="a14"> | <div class="tab-pane active" id="a14"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week14_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 585: | Line 611: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a15"> | <div class="tab-pane active" id="a15"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week15_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 672: | Line 698: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a16"> | <div class="tab-pane active" id="a16"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week16_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 743: | Line 769: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
<div class="tab-pane active" id="a17"> | <div class="tab-pane active" id="a17"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Del_week17_overview}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 771: | Line 797: | ||
</div> | </div> | ||
| + | <!--Week 18--> | ||
| + | <div class="labjournal-weekly"> | ||
| + | <div> | ||
| + | <ul class="nav nav-tabs"> | ||
| + | <li class="active"><a href="#a18" data-toggle="tab">Overview</a></li> | ||
| + | <li><a href="#z18" data-toggle="tab">Gibson assembly and validation</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="jumbotron weekly"> | ||
| + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| + | |||
| + | <div class="tab-content"> | ||
| + | <div class="tab-pane active" id="a18"> | ||
| + | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Templates/Del_week18_overview}}<html> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <div class="tab-pane" id="z18"> | ||
| + | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Template/Del_week18_Gibson}}<html> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <!--Week 19--> | ||
| + | <div class="labjournal-weekly"> | ||
| + | <div> | ||
| + | <ul class="nav nav-tabs"> | ||
| + | <li class="active"><a href="#a19" data-toggle="tab">Overview</a></li> | ||
| + | <li><a href="#z19" data-toggle="tab">Gibson assembly and validation</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="jumbotron weekly"> | ||
| + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| + | |||
| + | <div class="tab-content"> | ||
| + | <div class="tab-pane active" id="a19"> | ||
| + | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Templates/Del_week19_overview}}<html> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <div class="tab-pane" id="z19"> | ||
| + | <p style="font-size:12pt; text-align:justify;"> | ||
| + | </html>{{:Team:Heidelberg/Template/Del_week19_Gibson}}<html> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
| + | {{:Team:Heidelberg/Templates/Footer-Nav}} | ||
{{:Team:Heidelberg/Templates/Footer-DelRest}} | {{:Team:Heidelberg/Templates/Footer-DelRest}} | ||
Latest revision as of 01:37, 29 October 2013


Del Rest. Creating a 32 kb plasmid.
Week 10
This week the project "DelRest" was launched, which aims at creating a single, 32 kb plasmid enabling the expression of most genes from the D. Acidovorans Del cluster, namely DelA-G and DelL-P (note: the 18 kbp gene encoding DelH is cloned onto a seperate plasmid). Due to the shere size and complexity of the DelRest construct, we decided to use Gibson cloning. This weeks goal was amplify the pSB4K5 backbone from the partsregistry with primers giving the intended overlaps as well as the desired genes from our host organism D. Acidovorans . Therefore, Gibson primers for the amplification of the target backbone pSB4K5 were designed, which also introduce a Gibson-overlap to DelA, the first gene present in the Del cluster. Furthermore Gibson primers for amplifying the fragments DelA-G and DelO-P were ordered. The last Gibson primer pair used to amplify DelL consequently carries the required overlap to the beginning of the mRFP reporter present the pSB4K5 insert part BBa_J04450. Check out our vectormap if you are curious about the detailed cloning strategy and primer design.
Week 11
During the past week we managed to amplify a fragment of 11 kbp encoding the genes DelA-F from genomic DNA of D. Acidovorans. We were further able to amplify the backbone fragment from pSB4K5 carrying the desired overlaps to the corresponding fragments to be amplified from the Del cluster. We also succeeded in the amplification of the very last gene in our construct, DelL. Unfortunately, the amplification of DelF-G and DelO-P turned out to be more difficult than expected. Since our initial plan, which required the amplification of DelO-P using a Gibson primer which introduces an overlap to DelL and furthermore introduces an artificial ribosom binding site did not work out, we changed our strategy. We orderd shorter versions of all our Gibson primers. Using these shorter primers not bearing any overlaps to other fragments, we will try to amplify the desired genes in order to obtain a specific template for the reamplification with the primers carrying the needed overlaps. Furthermore, we will optimize the PCR conditions for the amplification of the DelF-G fragment.
Week 12
By the end of last week we found out, that the short primers we had ordered did not improve amplification of the missing fragments from D. Acidovorans. Additionly, further analysis of the Delftibactin cluster, led to the discovery of a predicted promoter present in front of DelO-P potentially essential for DelO-P expression. Therefore, we not only decided to design new primers for the amplyfication of DelF-G, but also modifie the entire strategy concerning the DelO-P fragment. To ensure the expression of DelO-P in our target organism E.coli we decided to amplify DelO-P together with its putative promoter. In consequence, we ordered new primers for DelO-P and also for the last DelA-G fragment in order to introduce the required new overhang to the new DelO-P fragment. The correlating primers can be found in our new vector map.
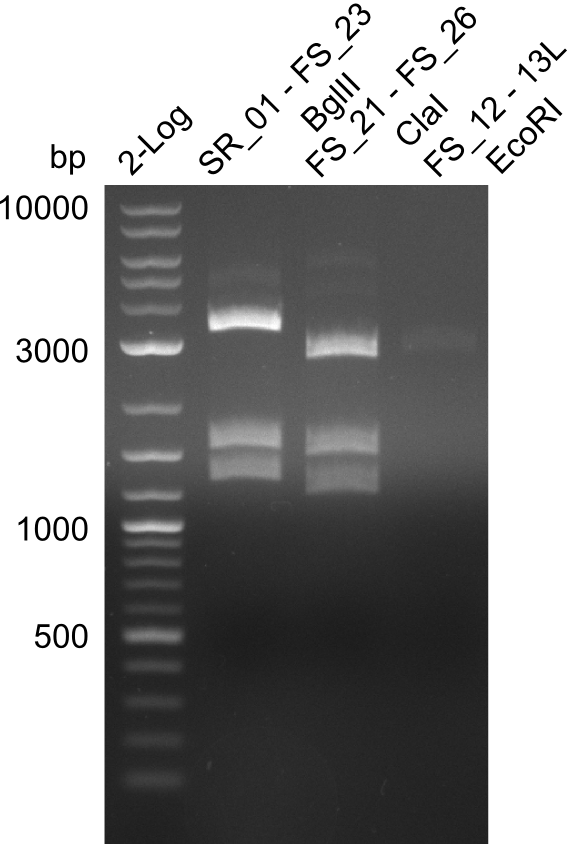
Week 13
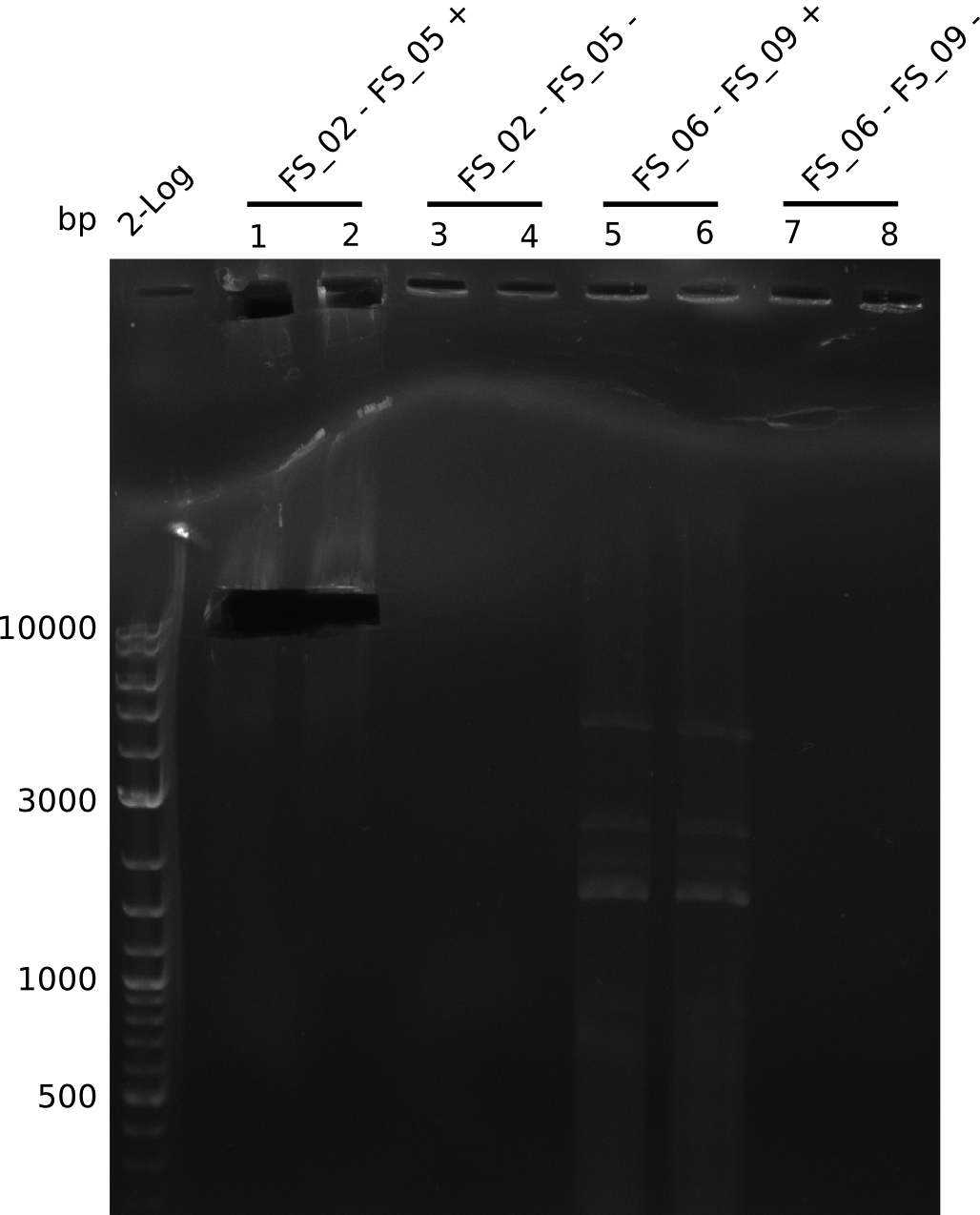
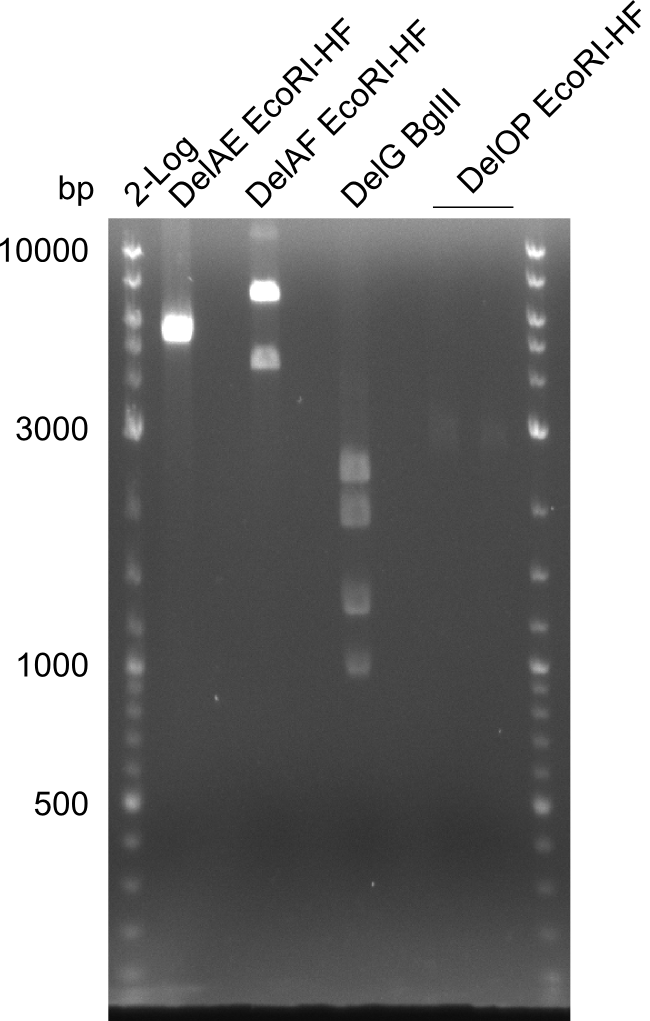
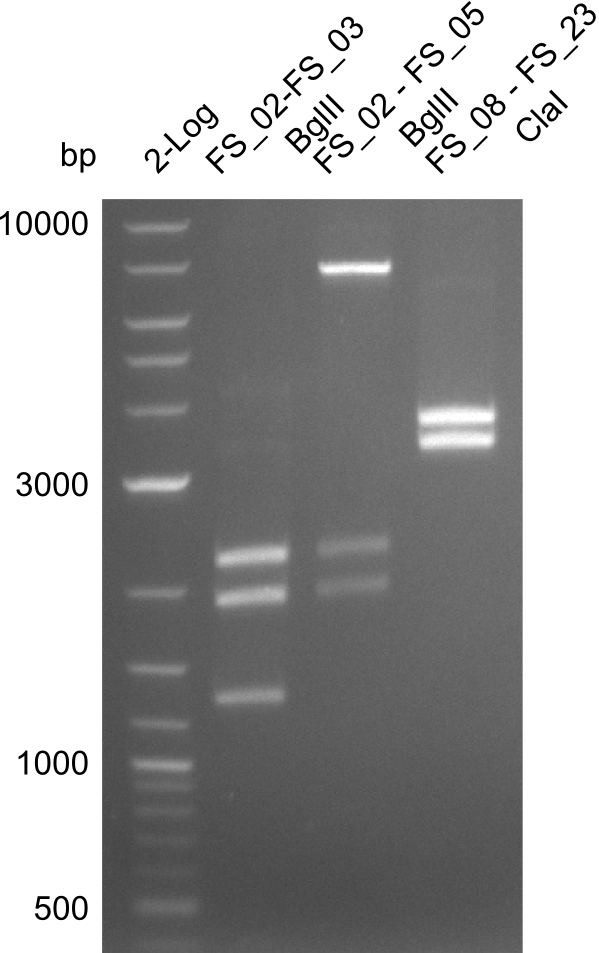
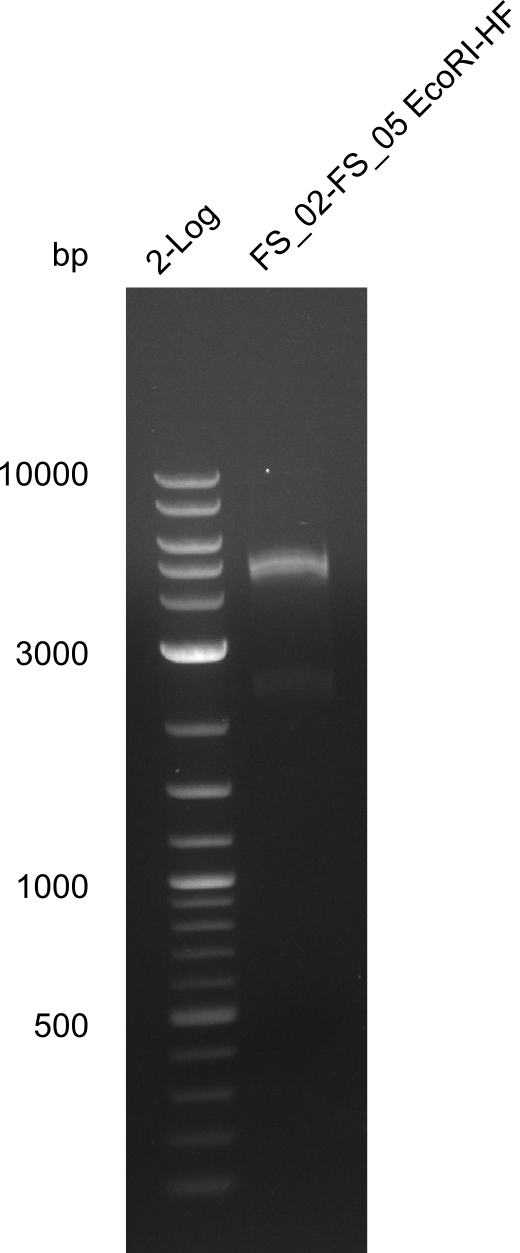
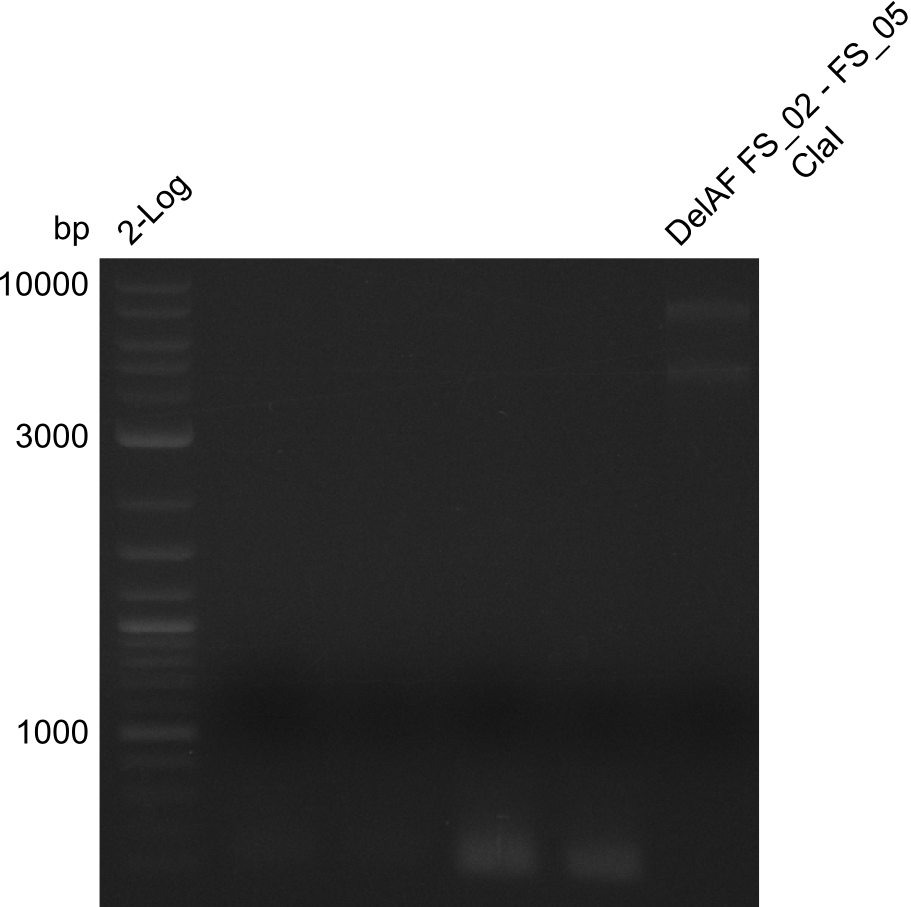
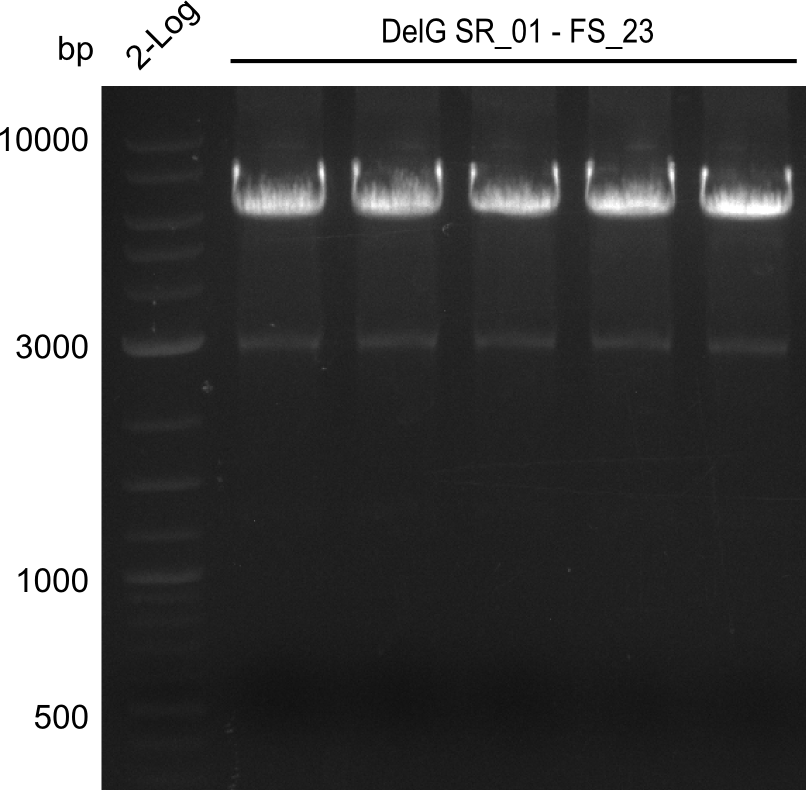
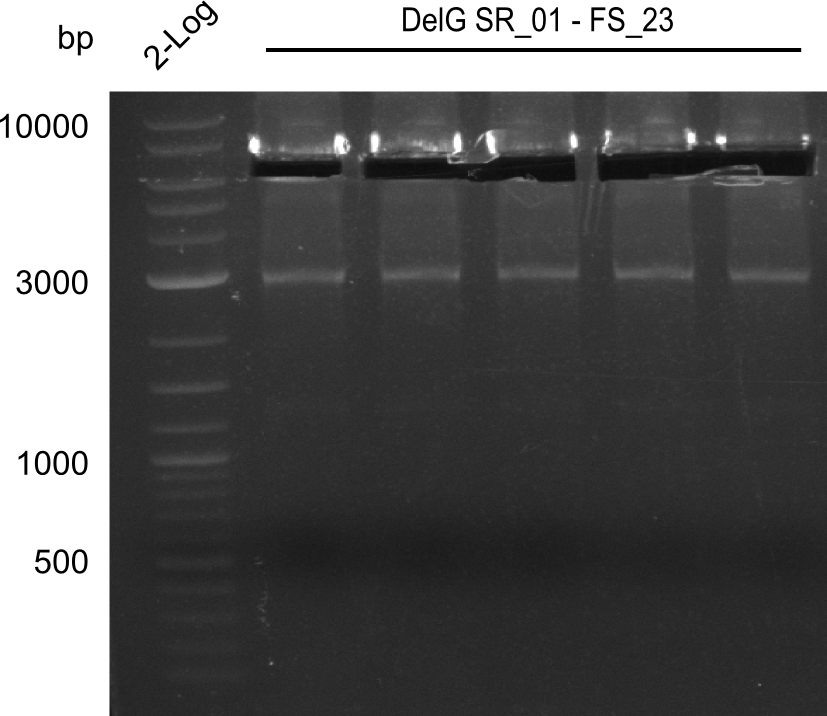
In the previous week, several of the newly ordered primer pairs improved our PCRs for DelO-P and DelF-G. However, results were not convincing enough to use these amplicons for Gibson assembly. Therefore, we spend this week optimizing the PCRs for the abovementioned fragments. In addition we validated the amplicon of DelG with restriction digest. Validation of the other PCR products did not succeed, mostly due to very low amount of DNA.
Week 14
We were still struggeling with getting the correct amplicons for the fragments encoding DelF-G as well as DelO-P. Furthermore, the restriction digests of the already successfully amplified fragments needed to be repeated using higher amounts of DNA, as results of the previous test digests were rather inconclusive. Furthermore, samples of these fragments were send for sequencing.
Week 15
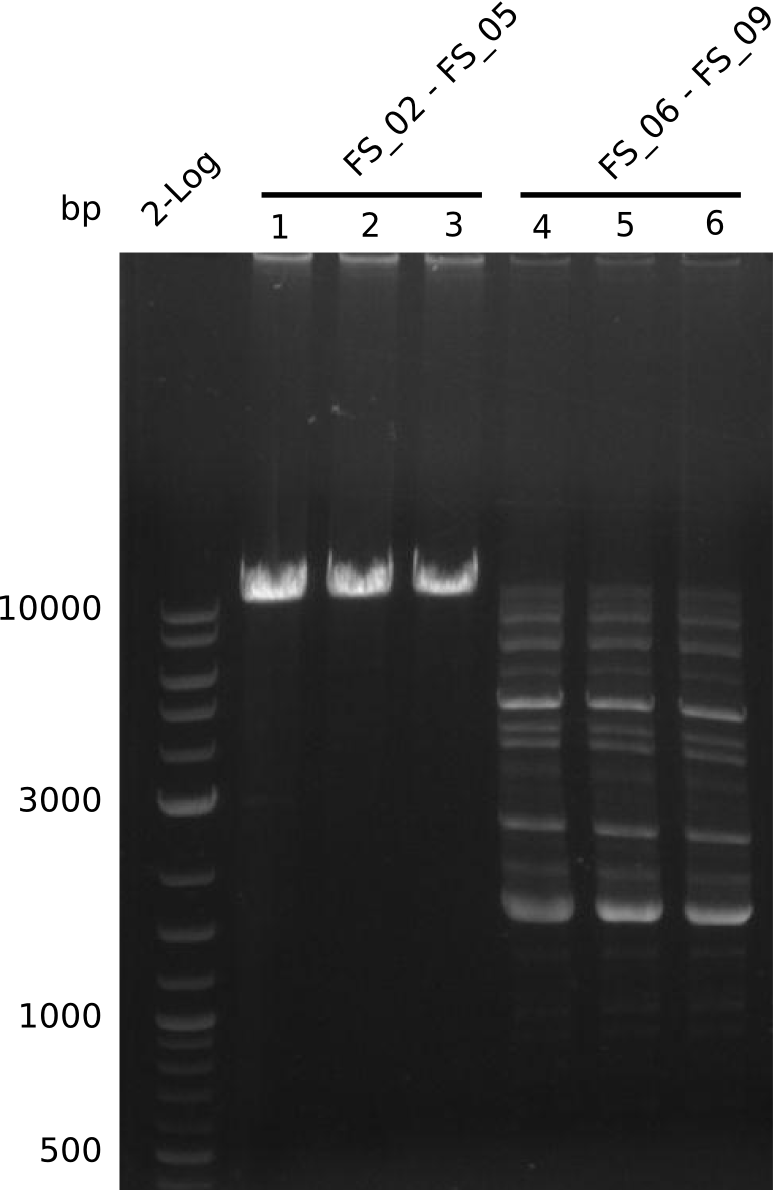
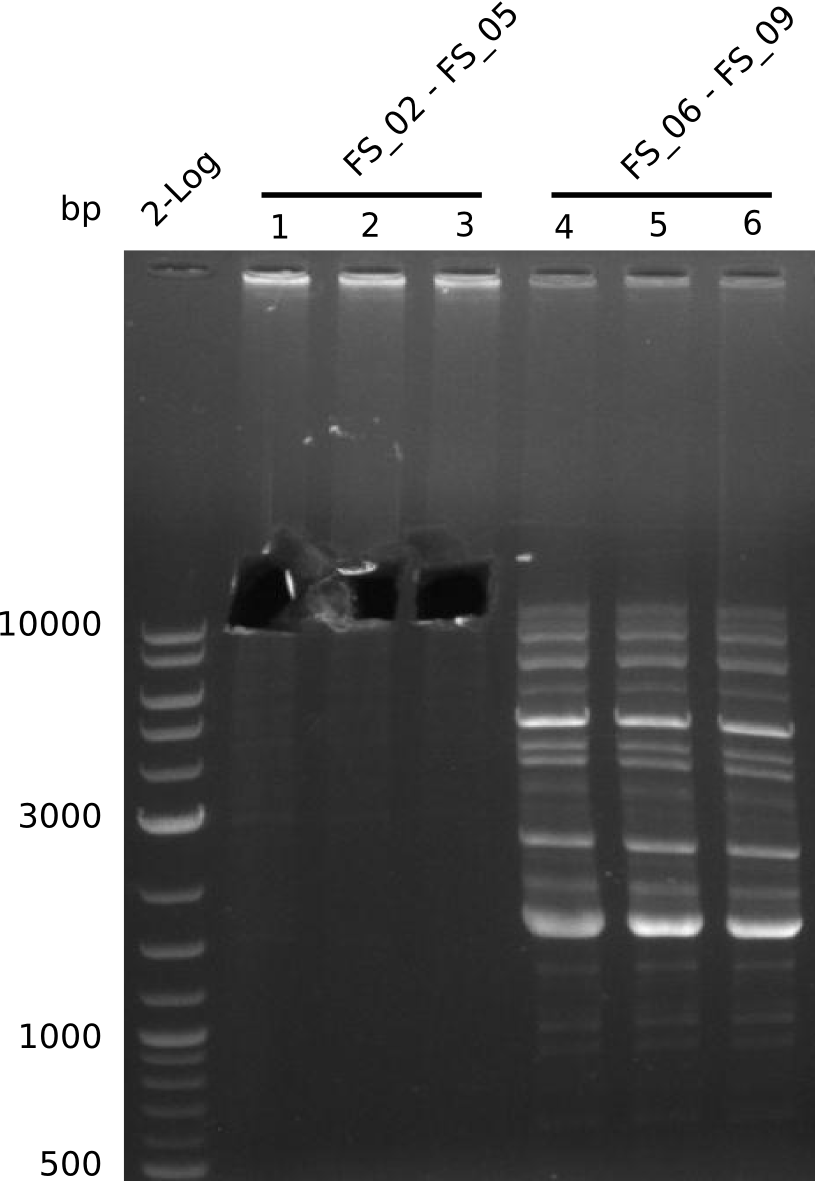
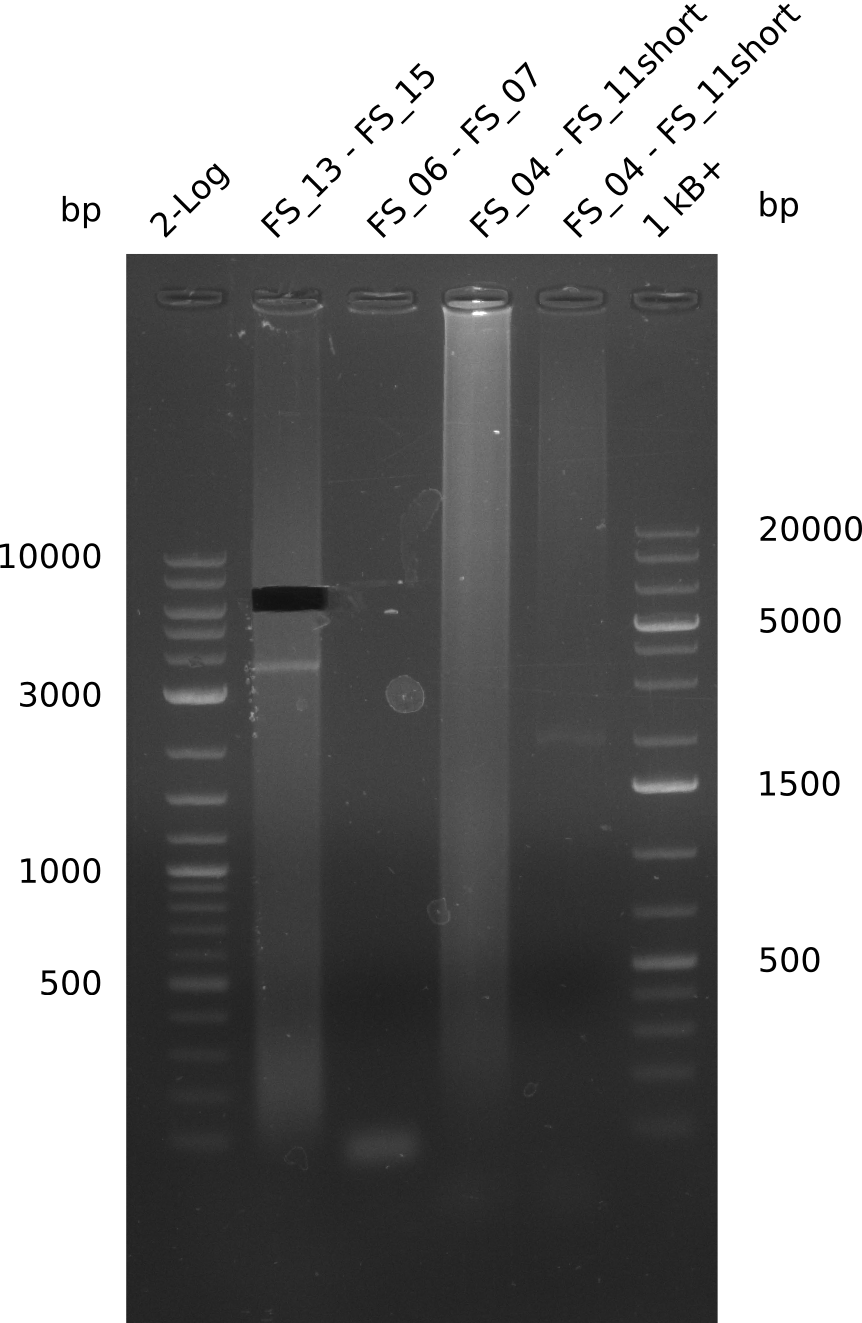
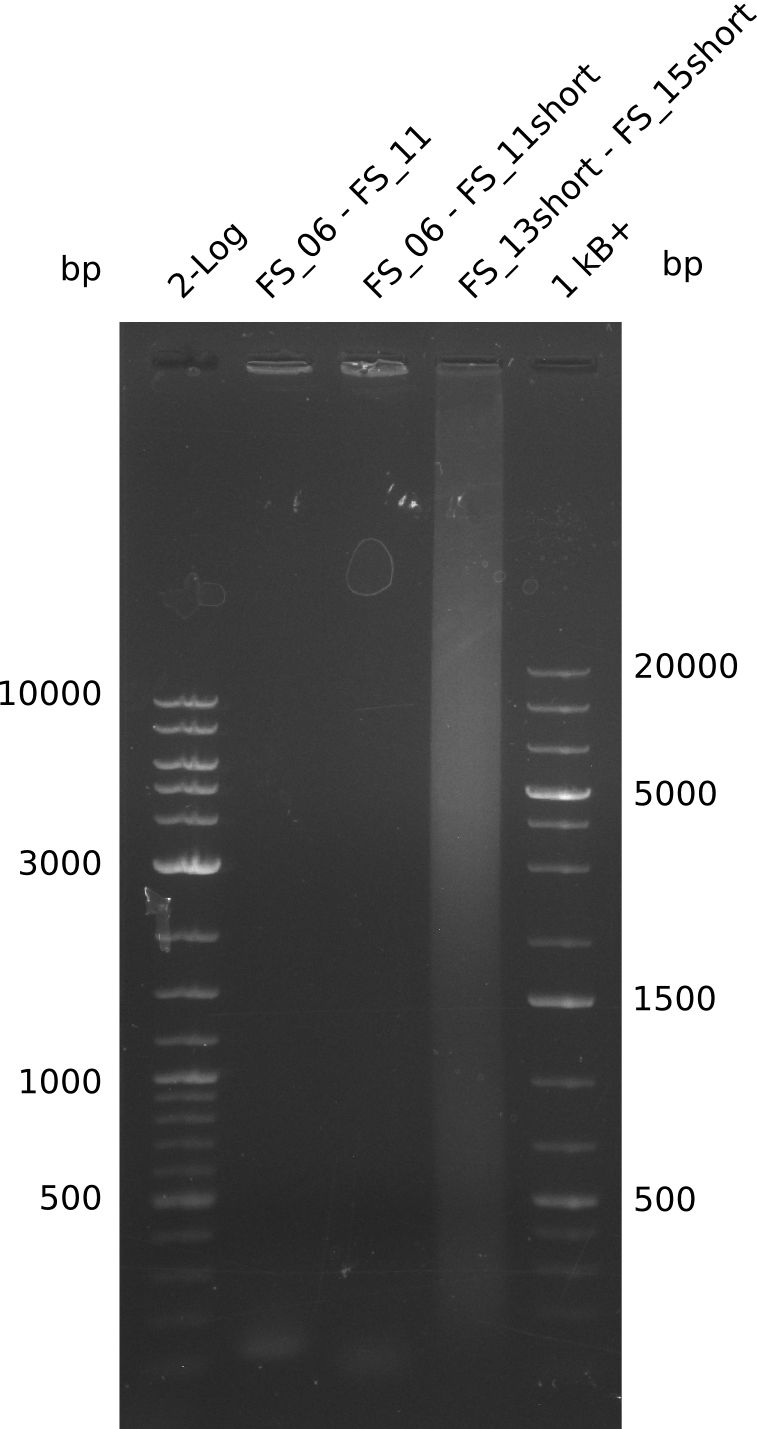
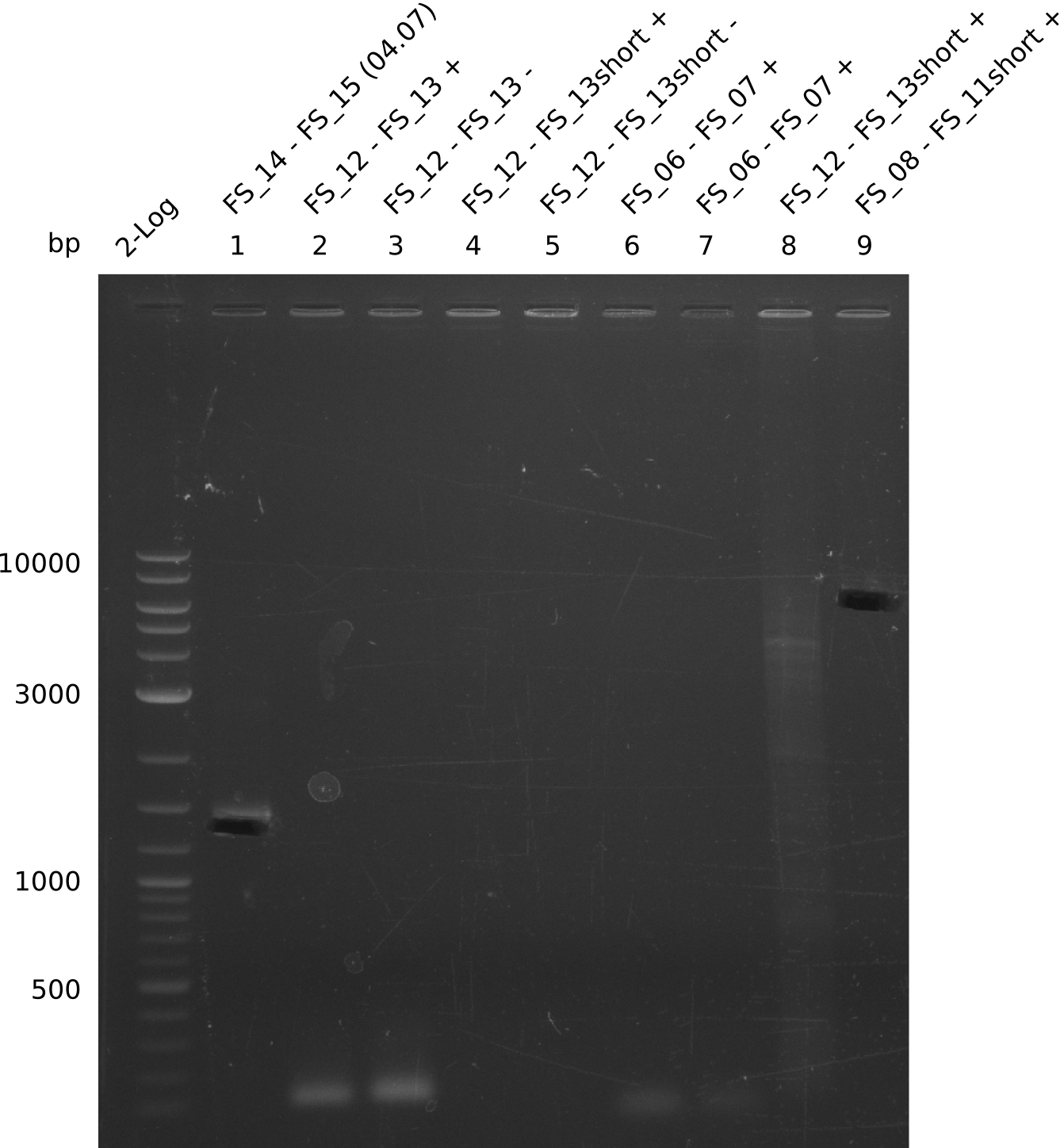
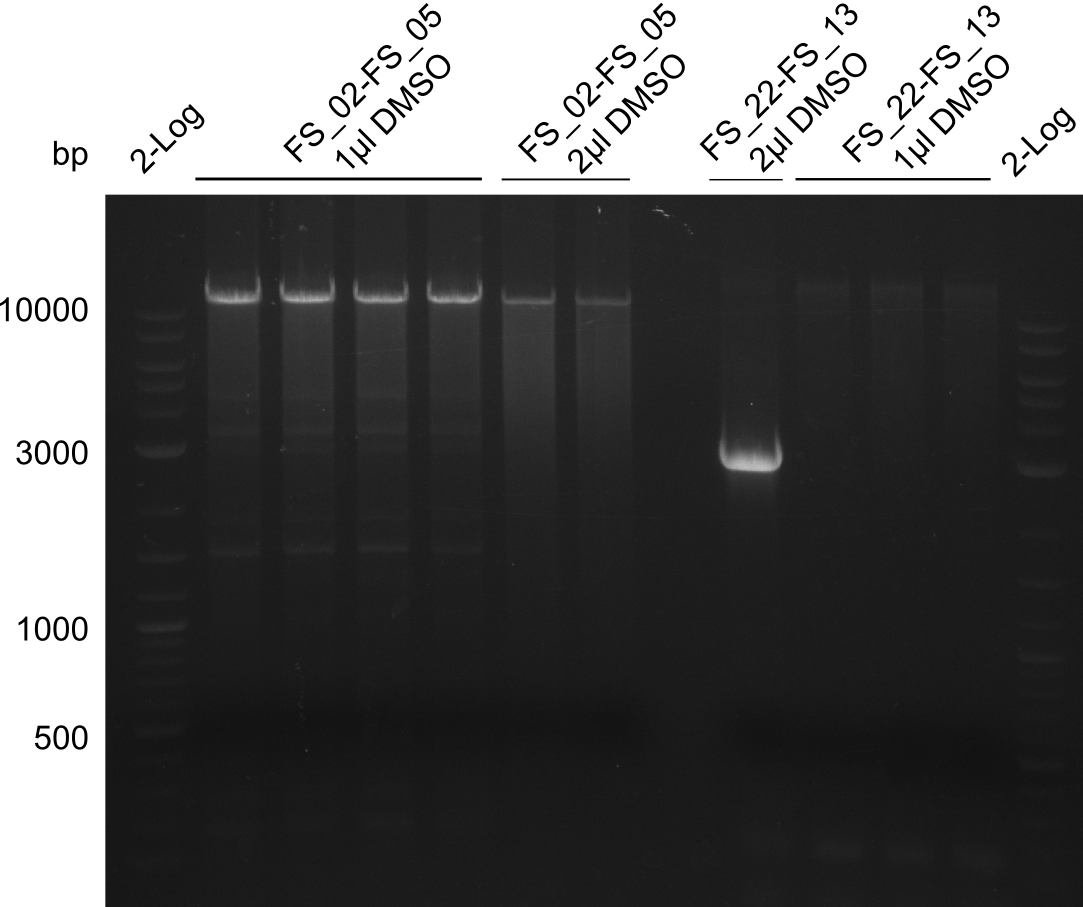
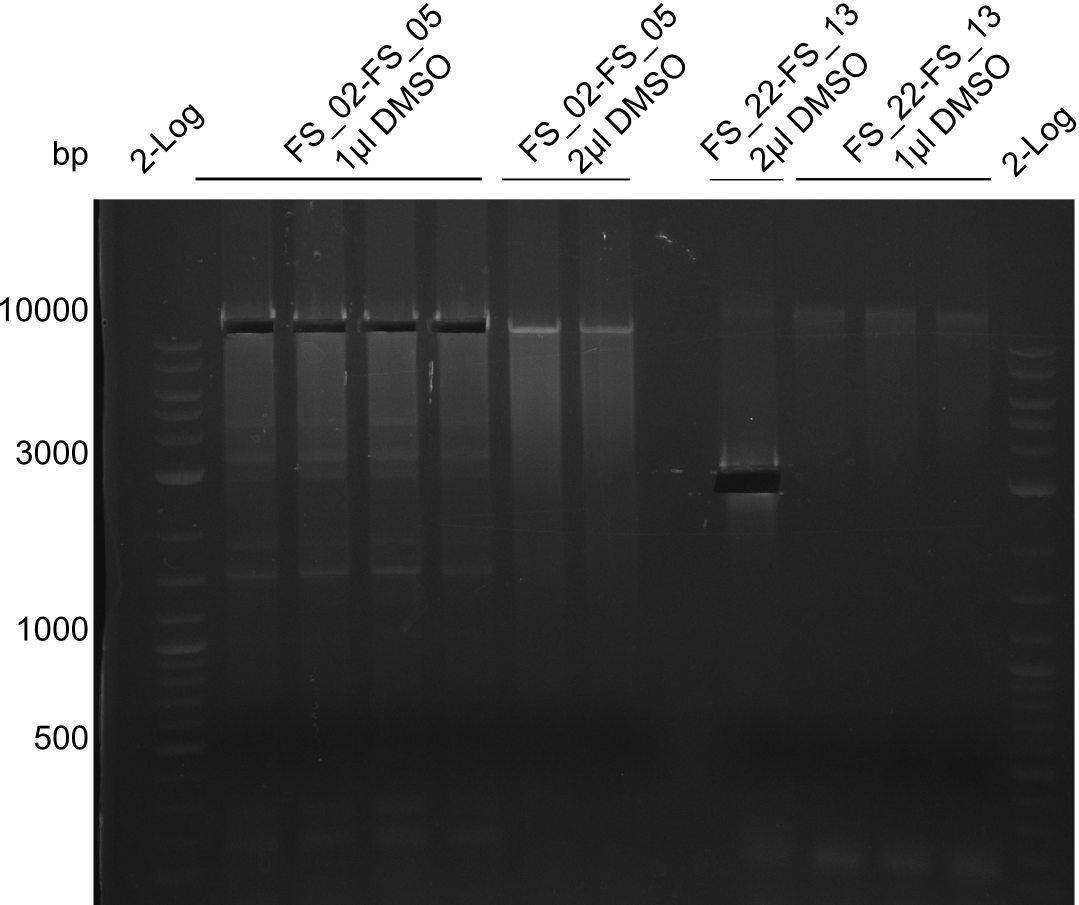
Single read sequencing of the PCR amplified fragments DelA-E, DelL as well as the backbone pSB4K5 was carried out by GATC. We blasted the obtained seqeuences against the reference sequence based on the D. acidovorans SPH-1 strain available on NCBI. Although our sequence matched to the reference sequence, we found a significant number of missmatches, which was too diverse and high to be simply explained by random mutations introduced by our polymerase during PCR (note: we used a high-fidelity proofreading polymerase suitable for GC-rich templates and long amplicons). We thus hypothesized that the SPH-1 strain based on which our cloning strategy was designed might have a significant number of single-nucleotide polymorphisms in the Del cluster compared to the D. acidovorans DSM-39 strain, which we used as template for all PCRs (note: there is no complete genomic sequence available for the DSM-39 strain on NCBI). We further hypothesized, that this difference in sequence between the DSM-39 strain used as PCR template and the SPH-1 strain based on which our primers were designed could explain our troubles with the PCR amplifications of DelF-G and DelO-P. In consequence, we ordered the SPH-1 strain from the DSMZ in order to obtain a suitable template for our PCRs. This solved all our PCR problems right away and we were able to get all amplicons required for cloning the DelRest construct within this week. Furthermore, we successfully validated our amplicons by restriction digest and sequencing.
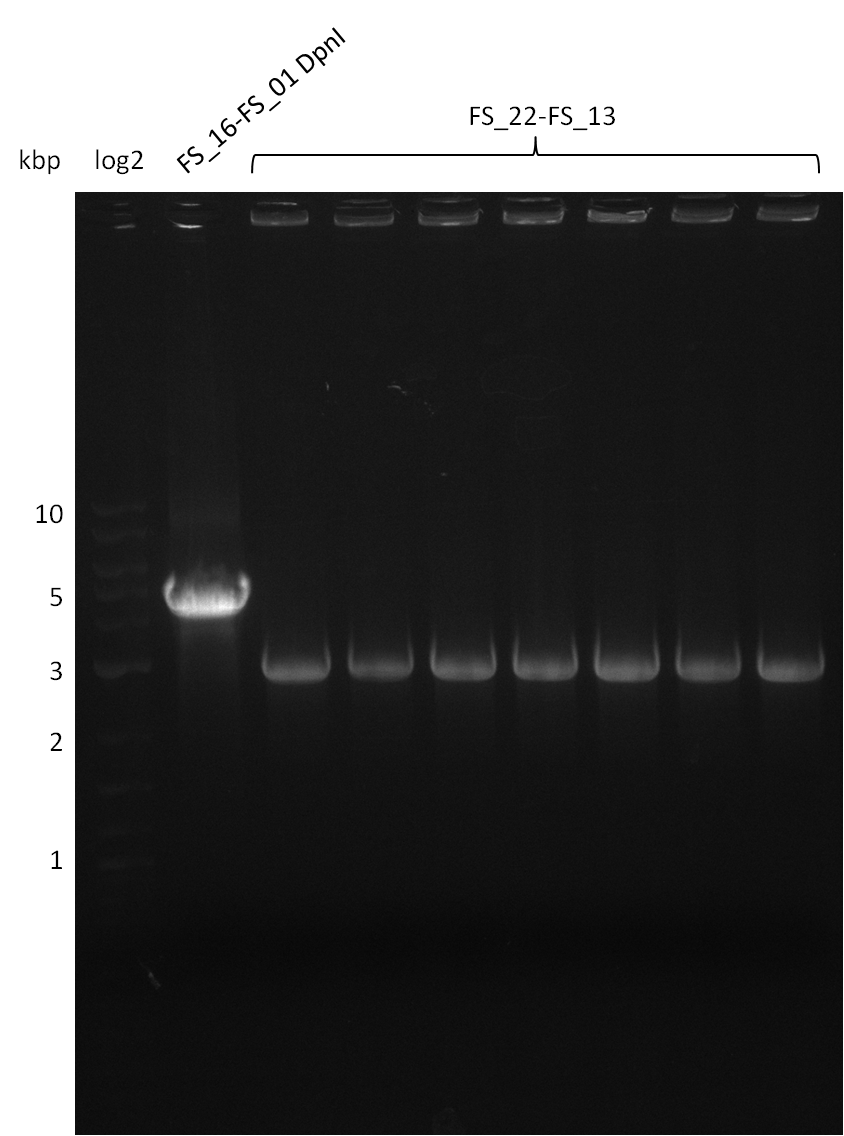
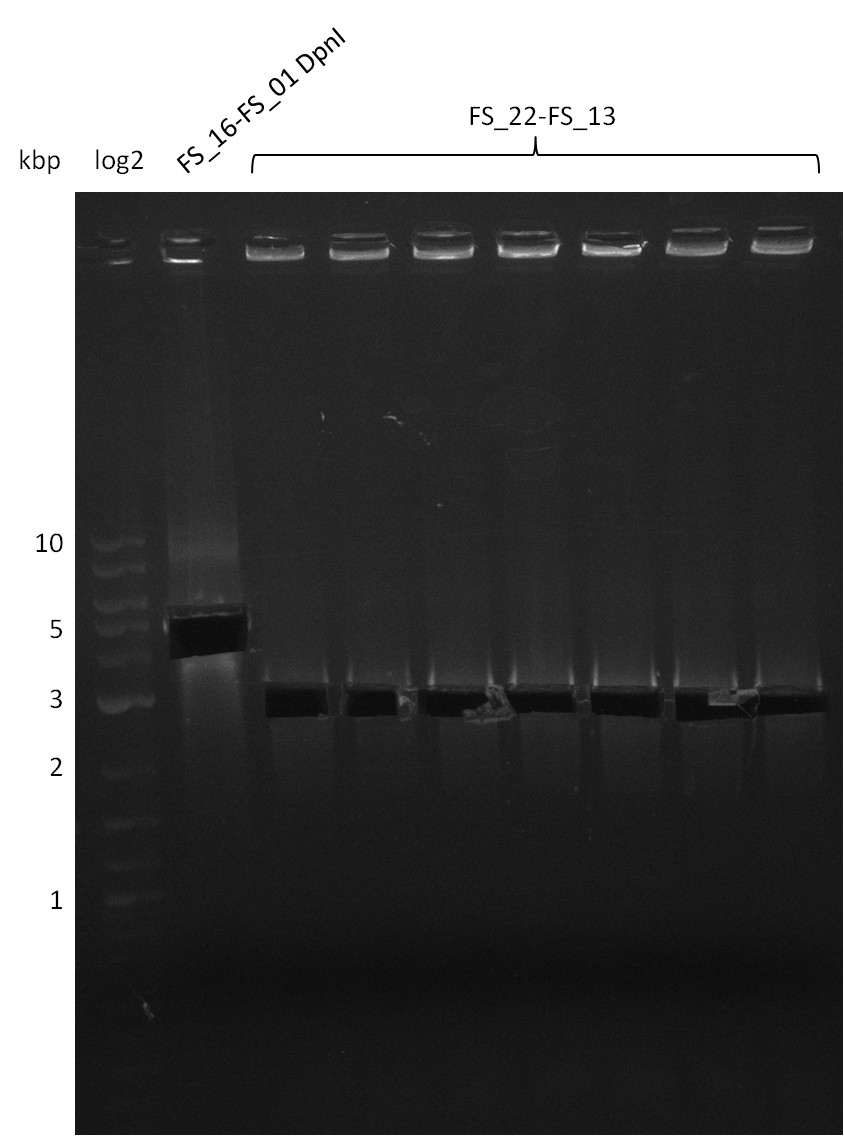
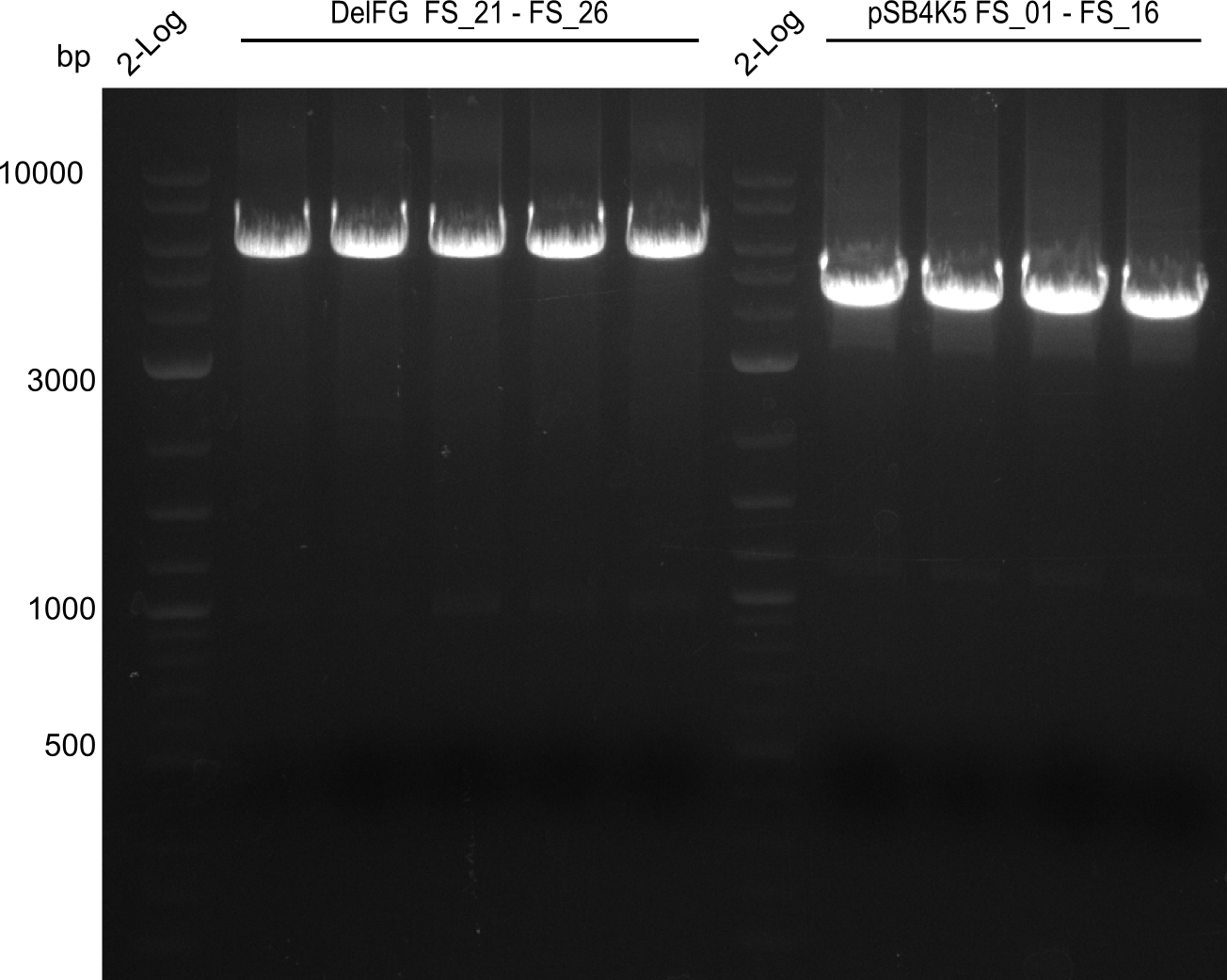
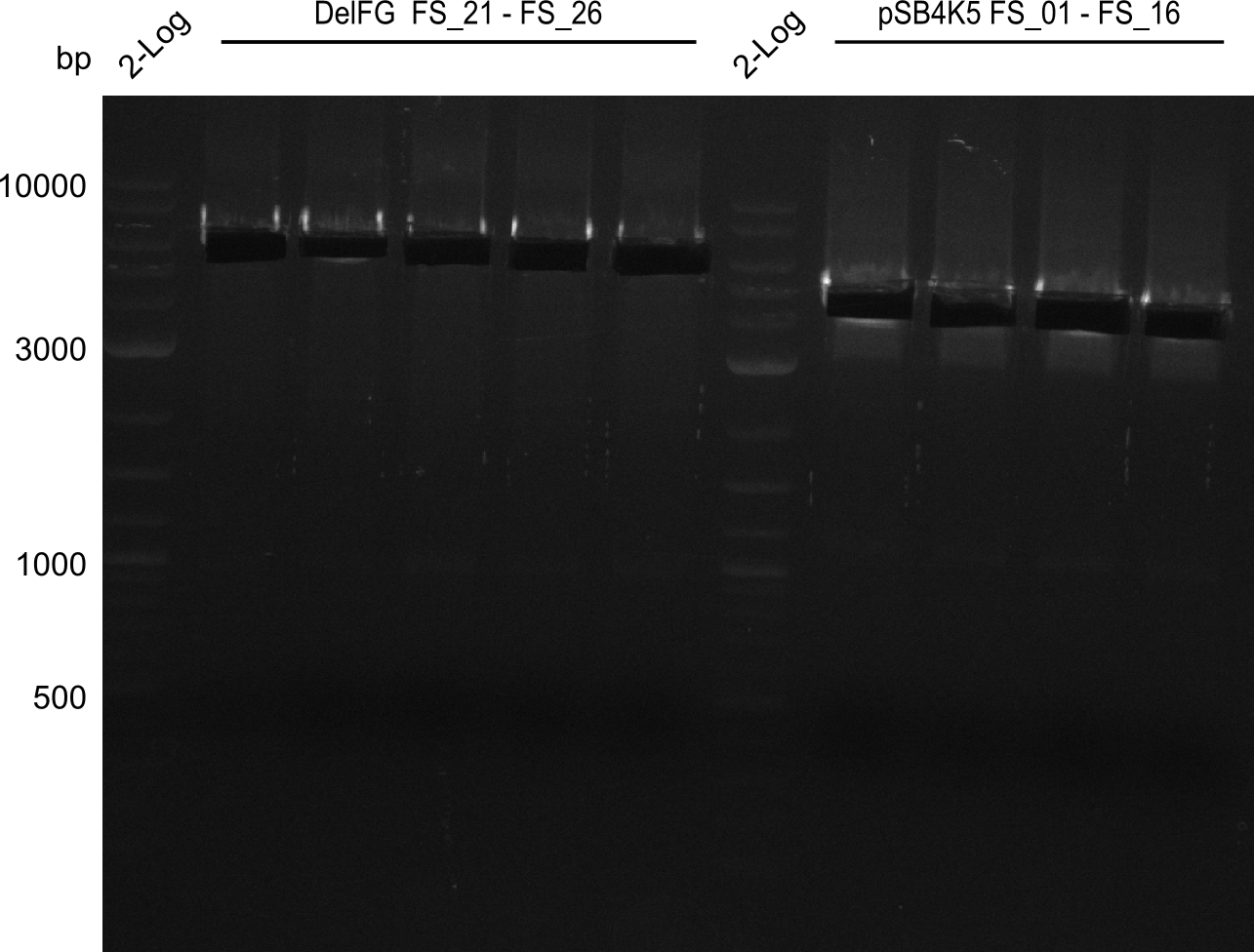
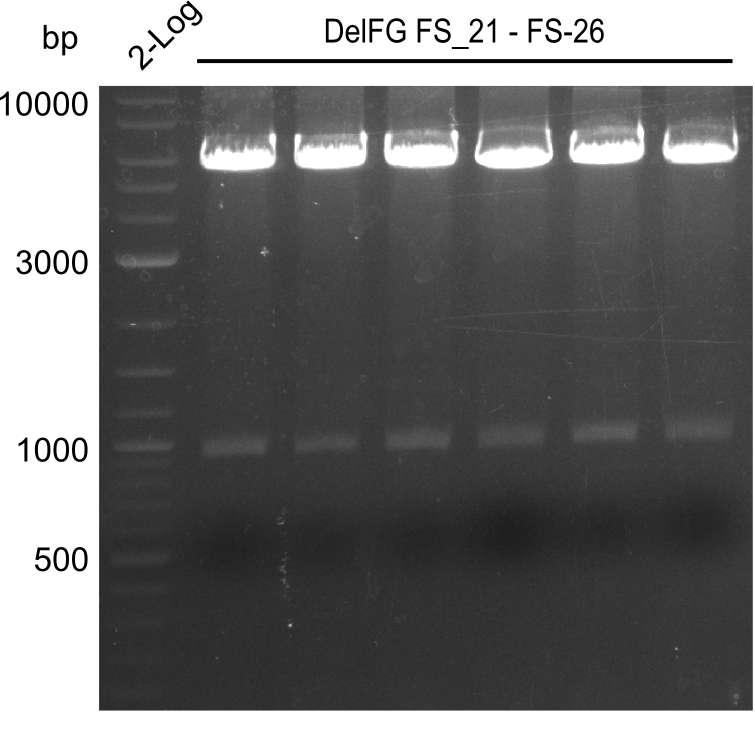
Week 16
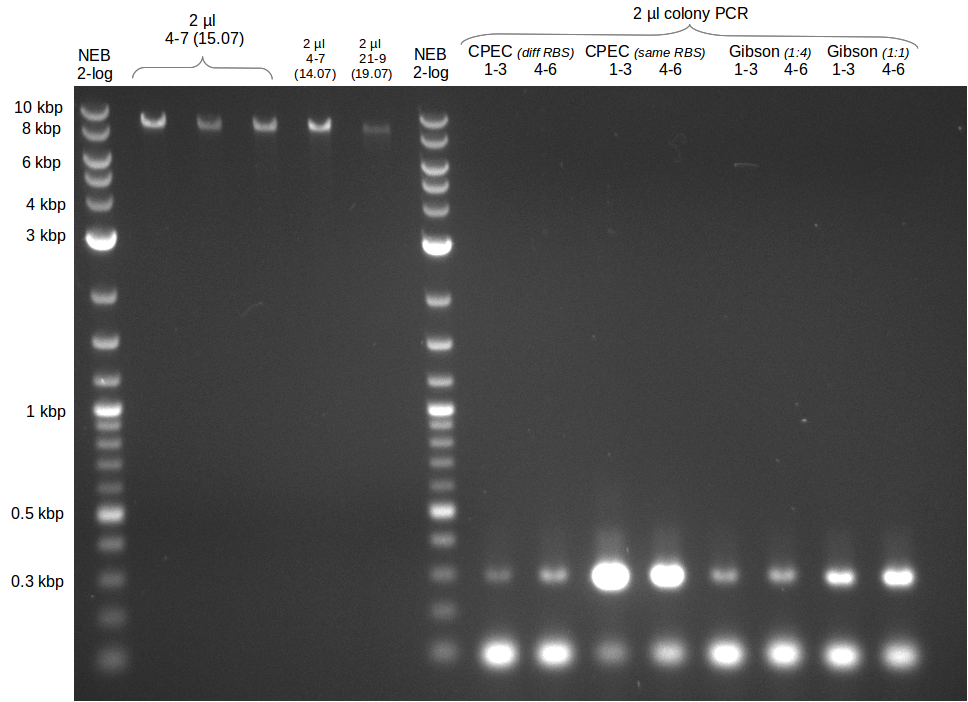
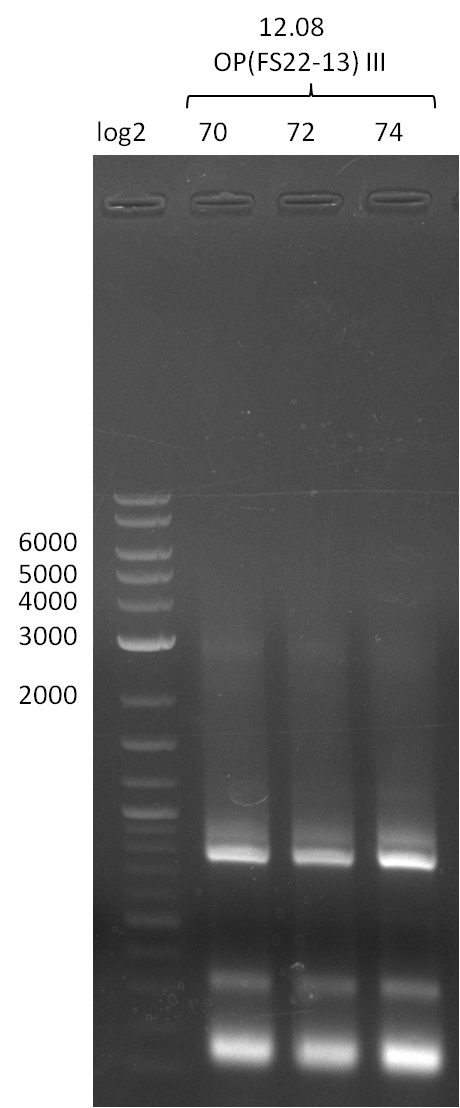
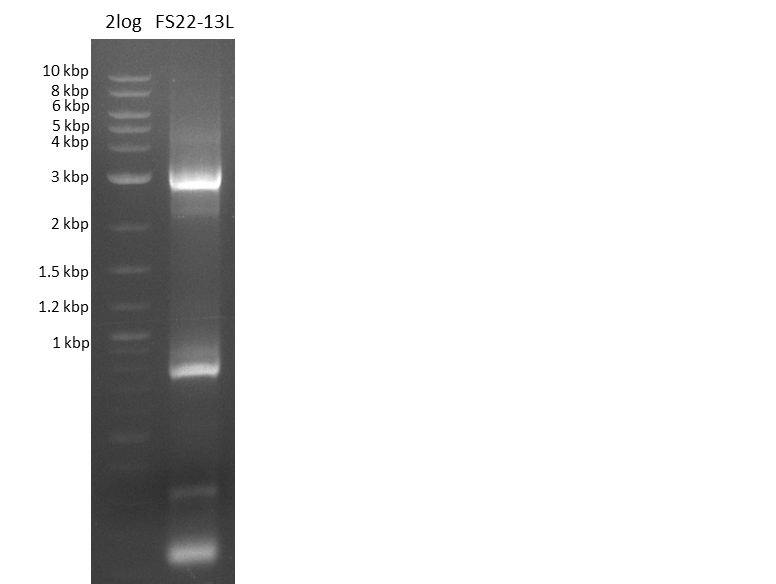
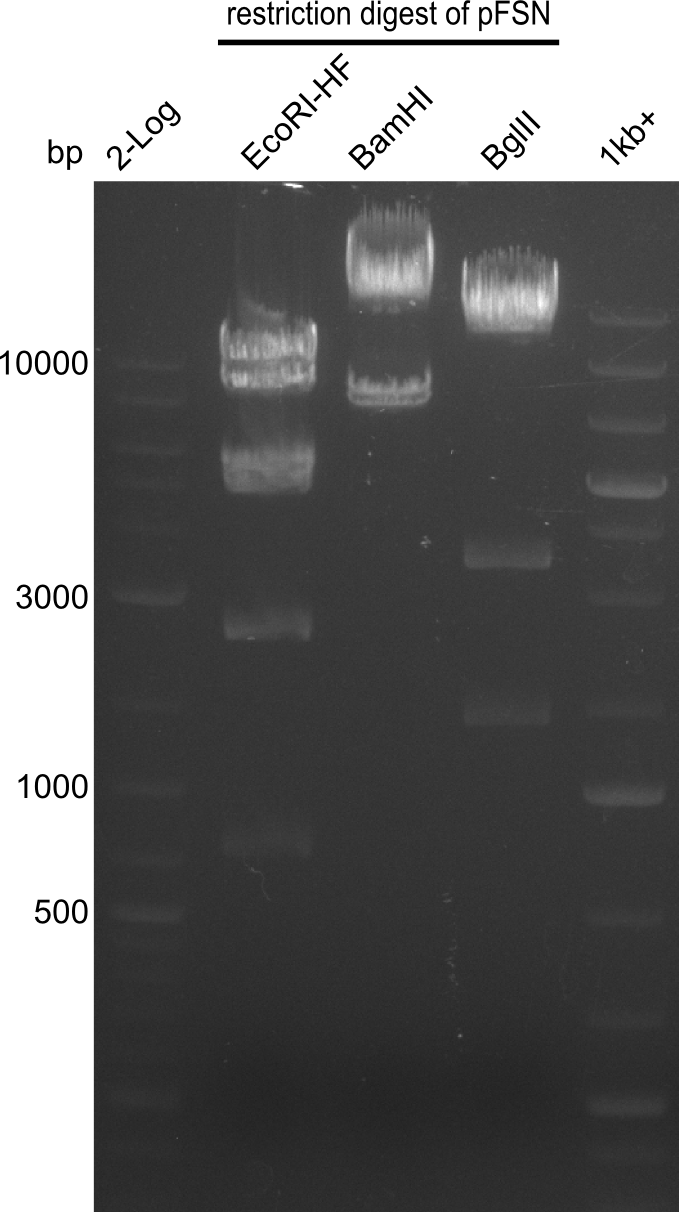
Last week we successfully amplified all the fragments needed to complete the DelRest cloning. Therefore, we went ahaed and performed Gibson assembly in order to assembel the final pFSN plasmid carrying bearing the DelRest genes. The assembly mix was transformed into E. coli DH10beta electrocompetent cells. Screening PCRs showed that the assembly was successful, calling for futher validation of the clones.
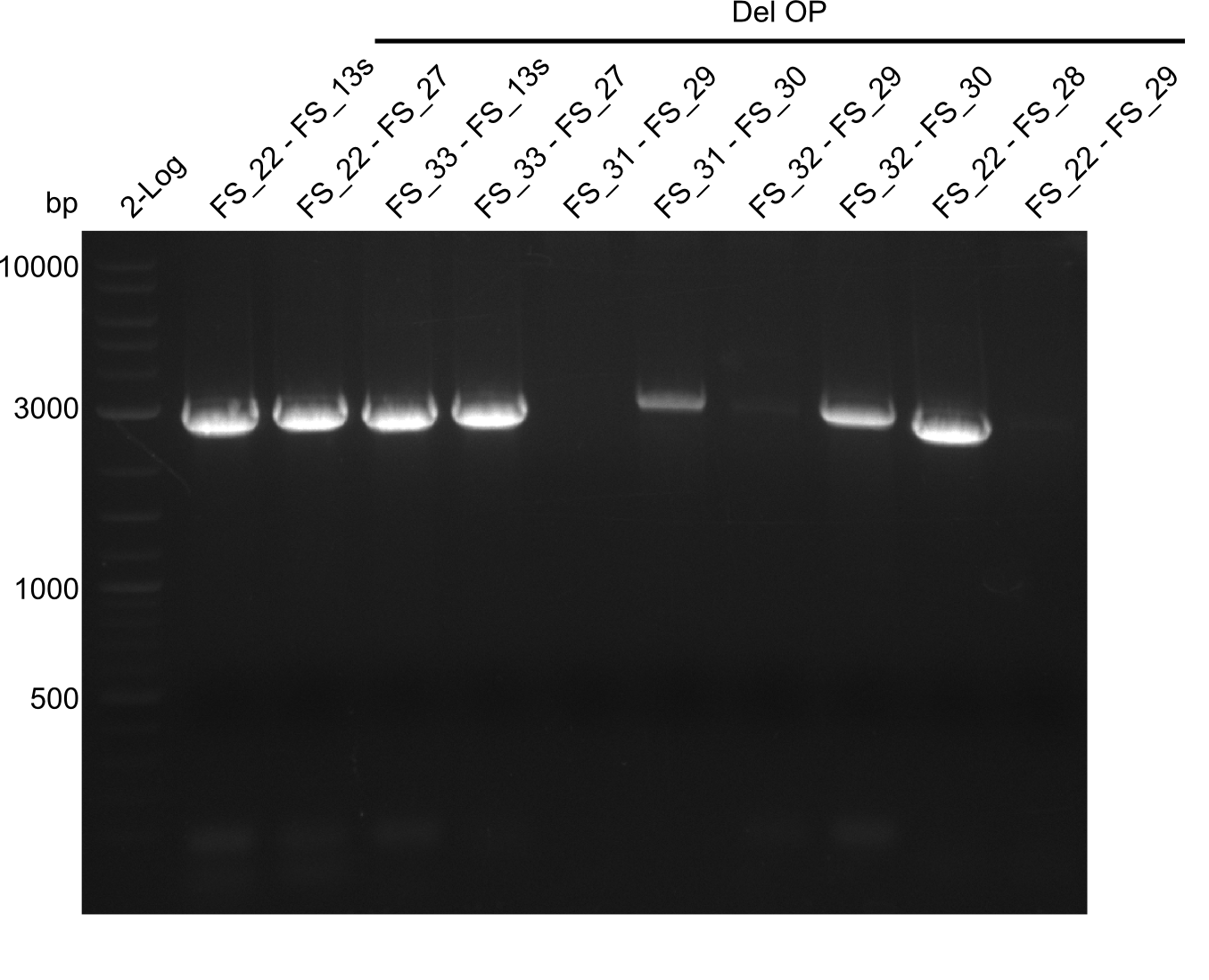
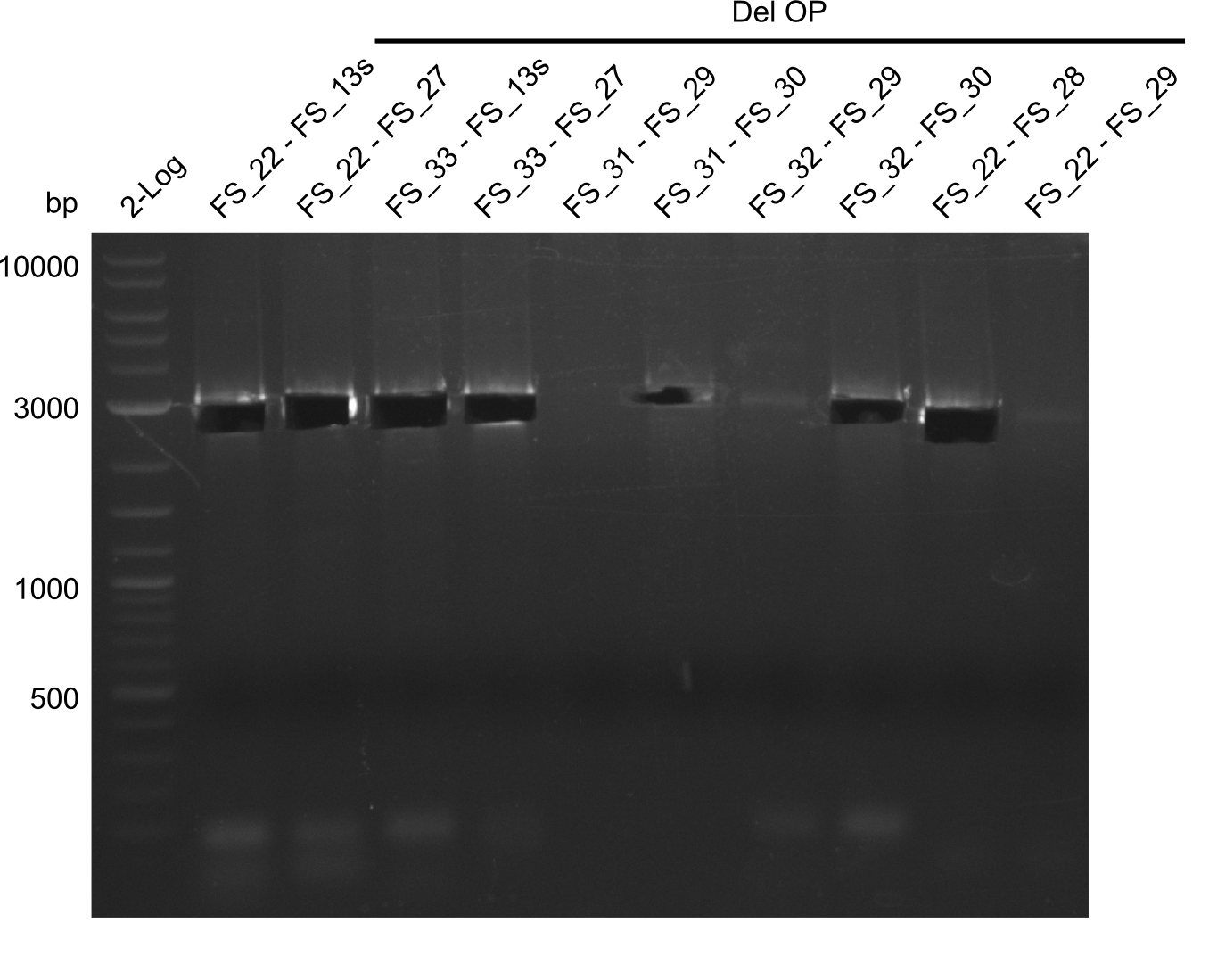
Week 17
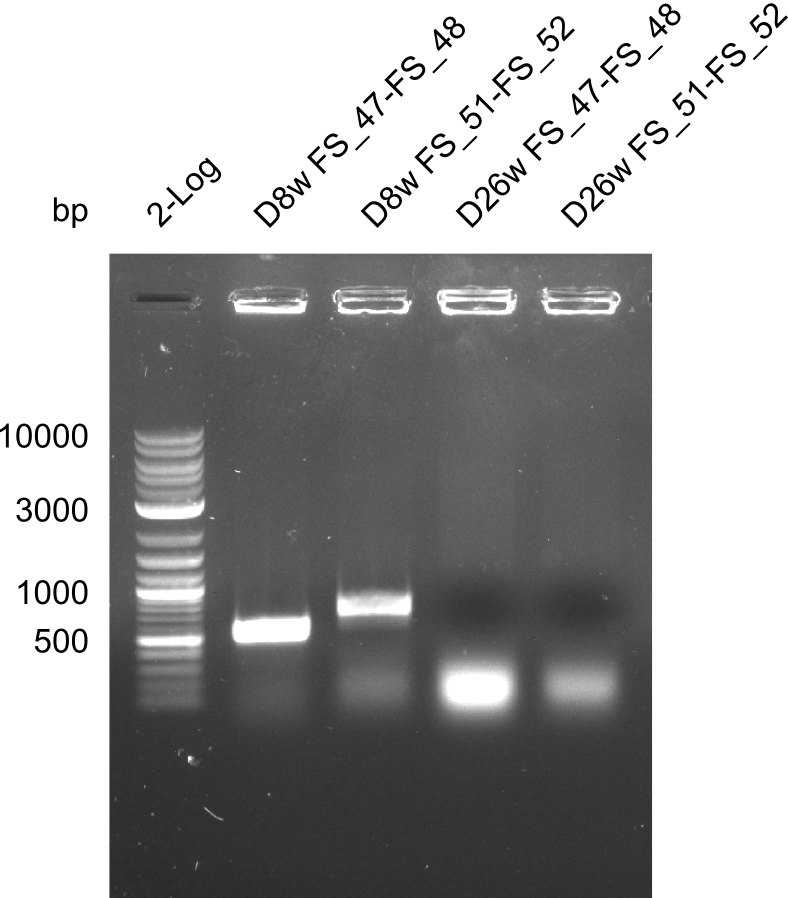
From colonies which were positive for screening last week, we rescued 3 plasmids. Test restriction digest were conducted and showed our clones to be correct.
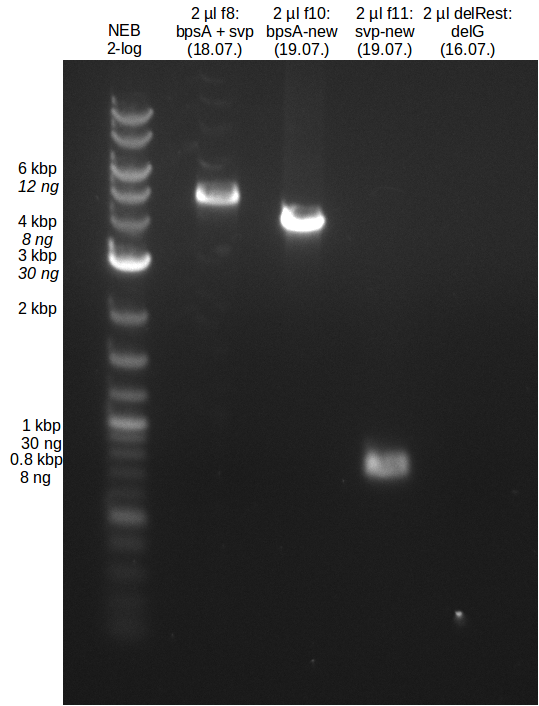
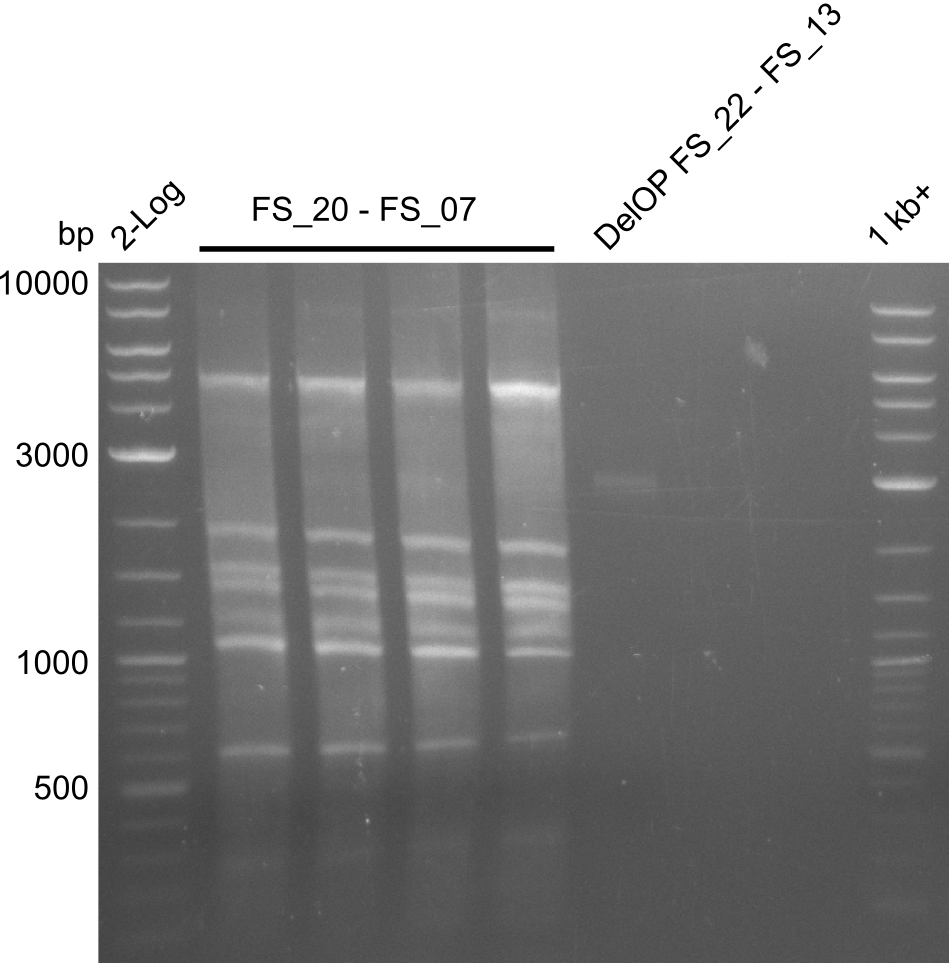
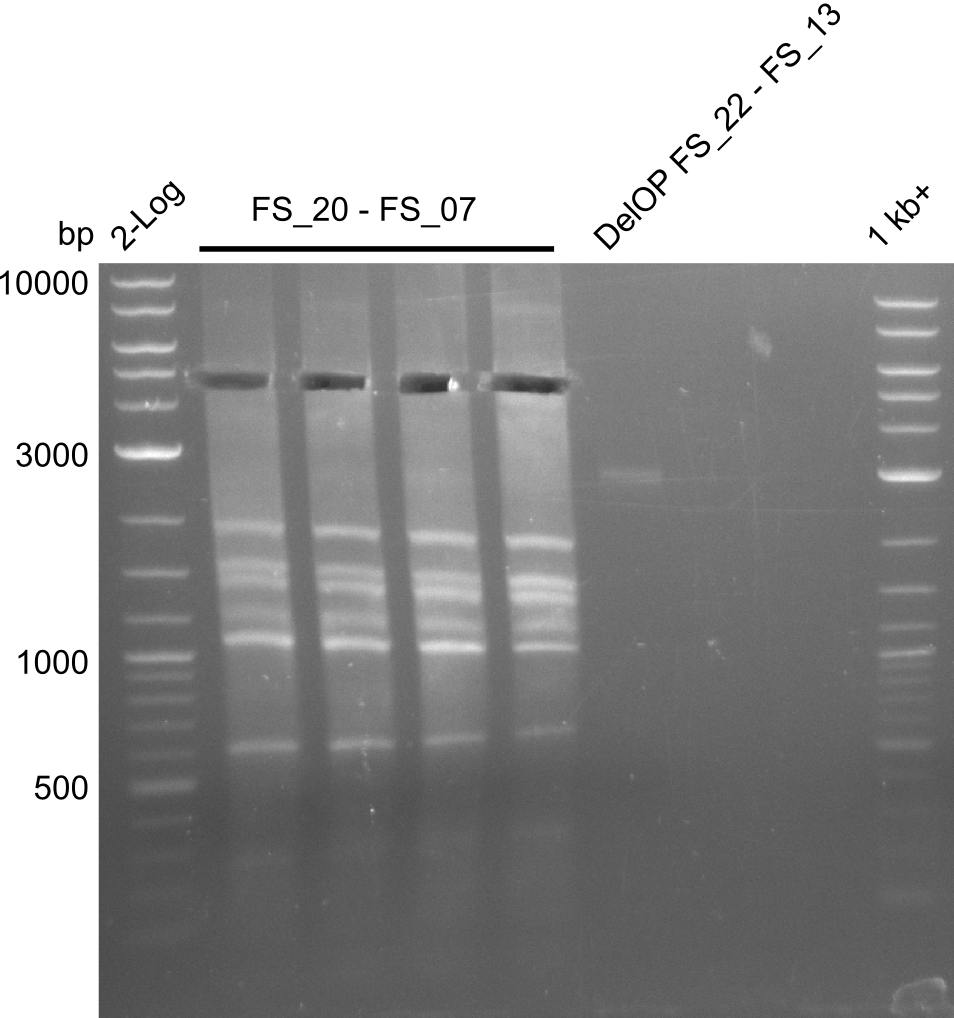
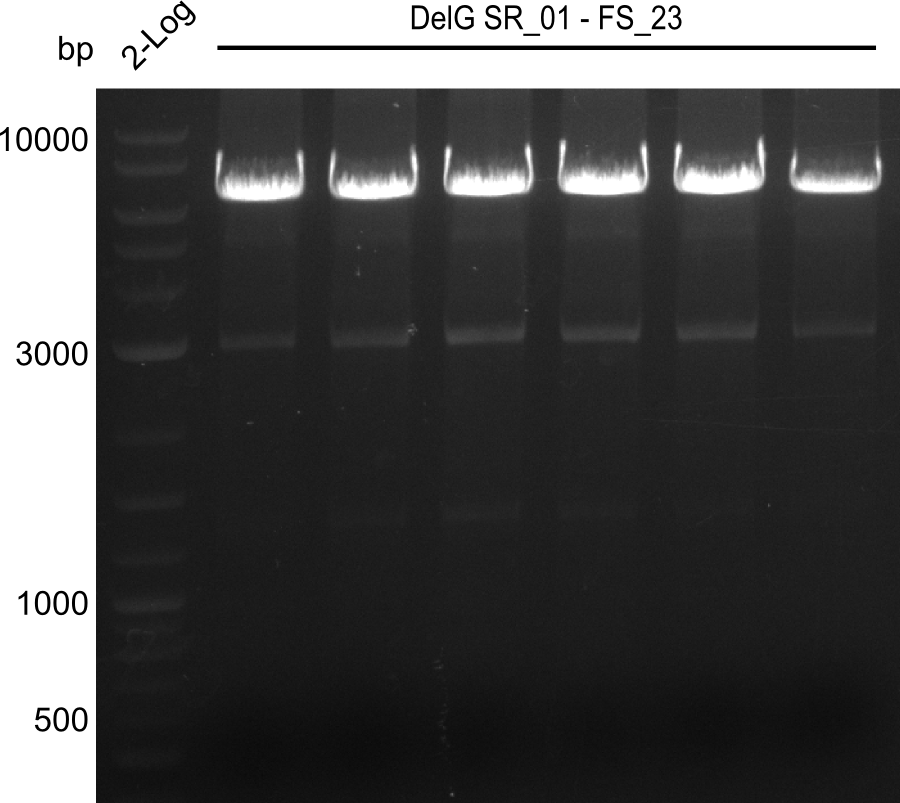
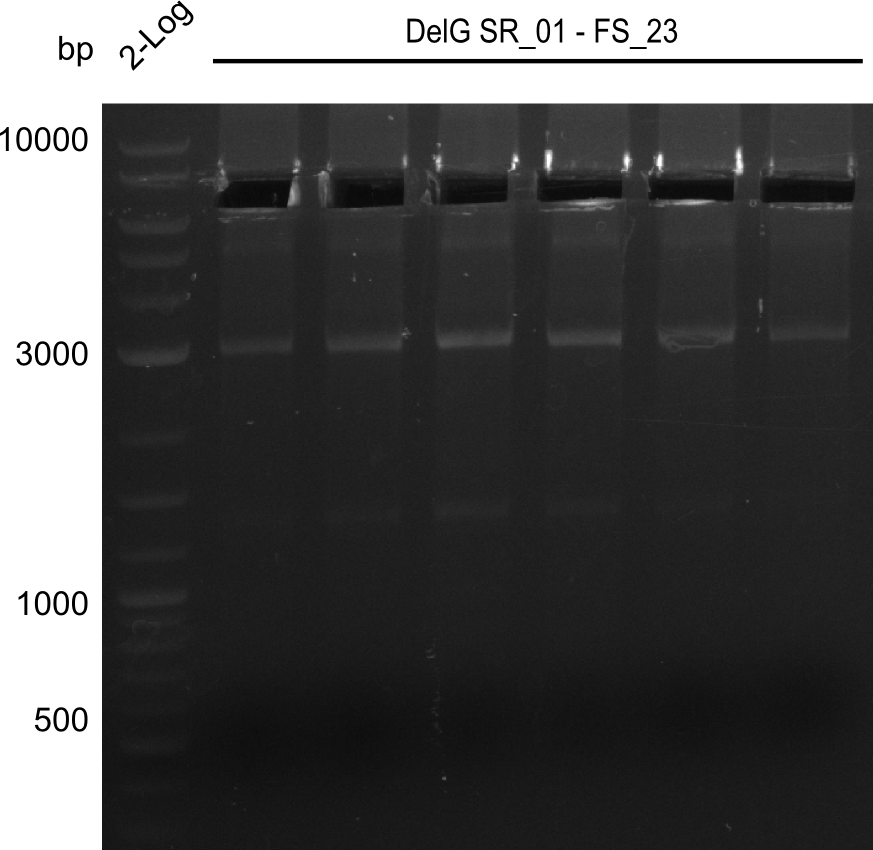
Week 18
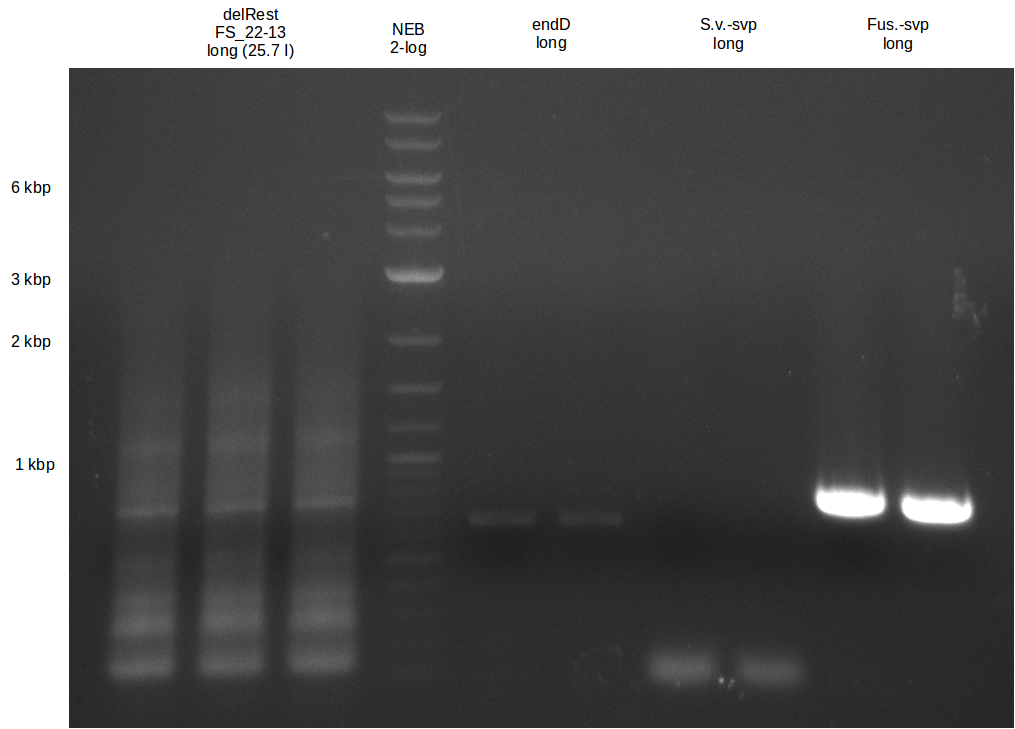
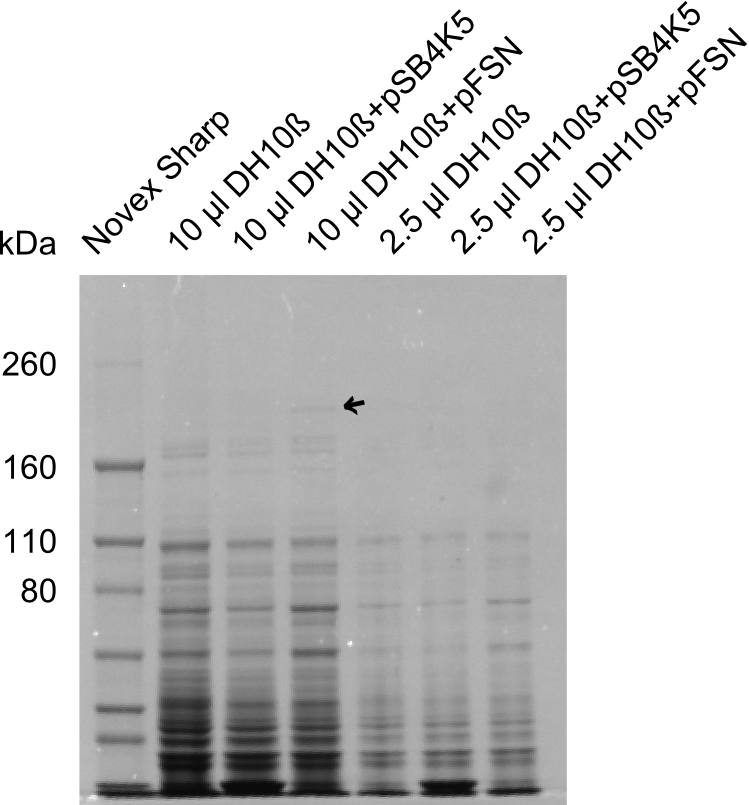
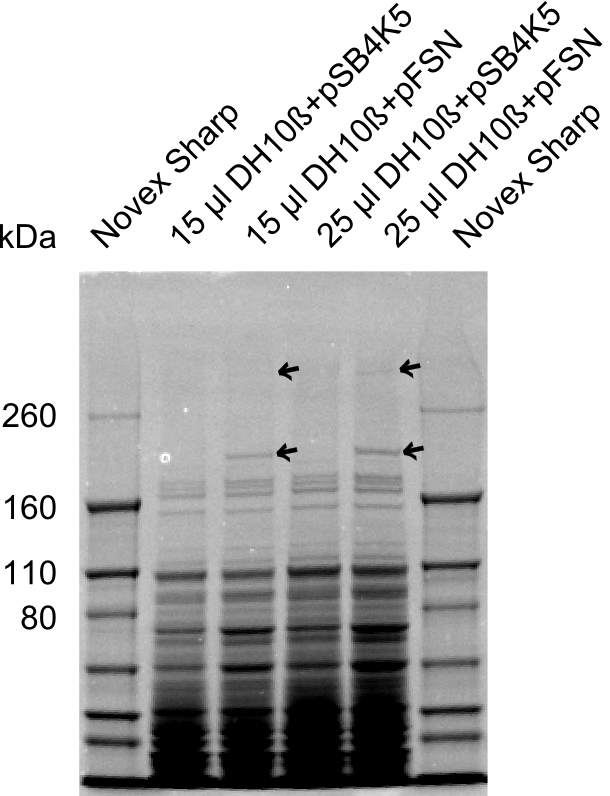
We send one of our DelRest constructs that showed the right restriction pattern in the test digest for sequencing. As it would be quite costly to sequence the whole 32 kb plasmid, we focused on the ligation sites between the different assembly fragments, which are most prone to insertion of errors. Although all insert fragments sequenced, including the ligation sites between DelA and DelF, DelO-P and DelL, were 100 % correct, we detected a mutation within the mRFP cds (note: we wanted to use mRFP in order to confirm expression from our DelRest plasmid). FACS analysis of E. coli bearing the DelRest construnct confirmed that mRFP expression was impaired, likely due to the corresponding mutation. However, as mRFP was only meant to be a general expression control on our construct, we did not start correcting out construct by mutagenesis. Instead, we started preparing samples for an SDS page in order to directly proof the expression of the Del genes by Coomassie staining.
Week 19
The Coomassie staining we carried out last week did not display all expected bands clearly. This occured most likely due to the low amount of protein loaded onto the corresponding SDS page. Therefore the SDS page was repeated. This expression analysis confirmed the results indicated by the previous SDS-page. We were able to show that DelE and DelG are expressed at a detectable level. As we could confirm the successful cloning and functioning of the DelRest expression plasmid named pFSN (for Florian-Sophie-Nils :)) the DelRest subproject was succssfully completed and the personal resources of the DelRest group were shifted to the DelH group as well as the wiki.
Strategy
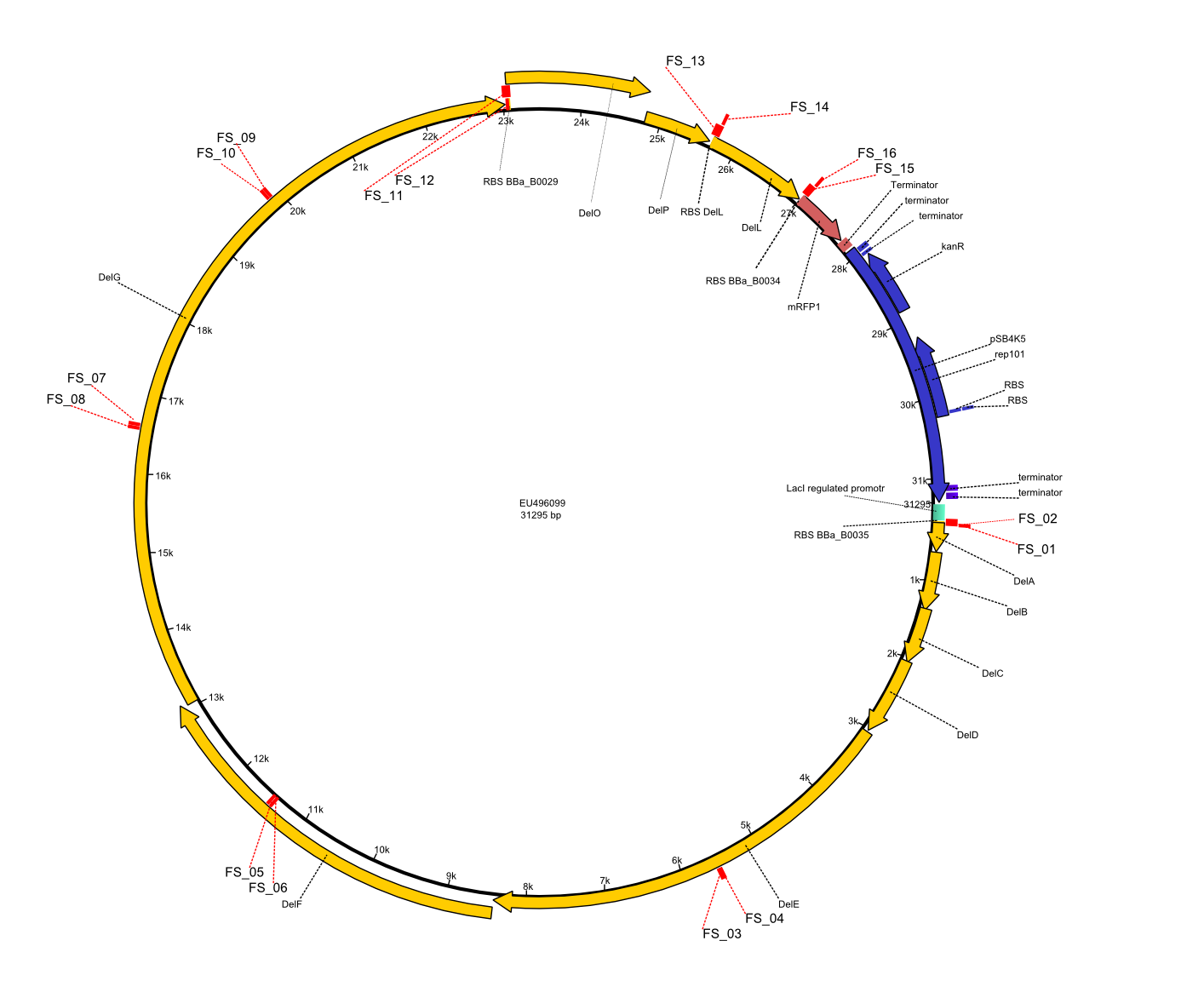
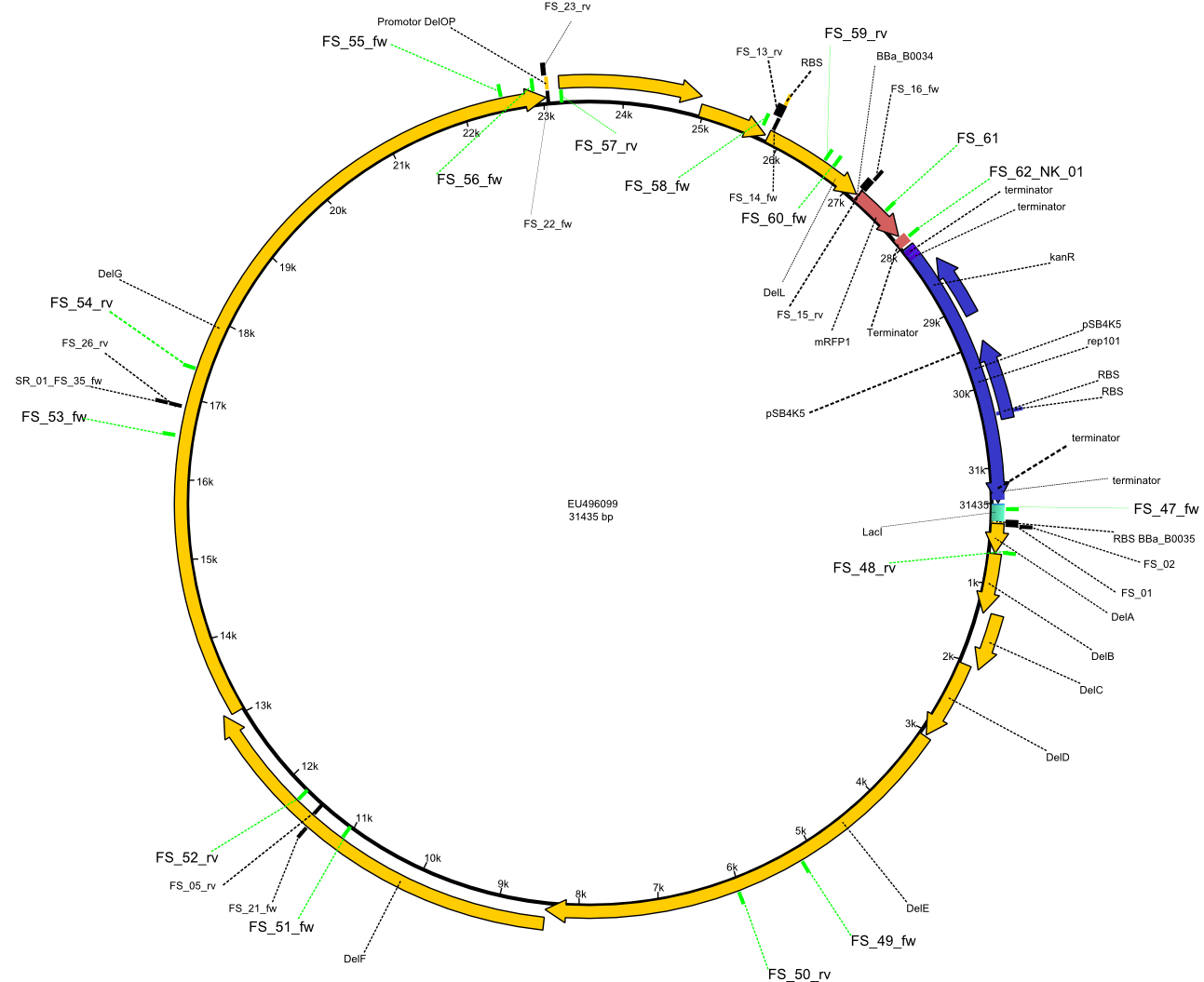
In order to transfer the delftibactin NRPS cluster from D. acidovorans in E. coli for efficient production of delftibactin, we designed the plasmid pFSN. It comprises the above listed genes as inserts. Furthermore, it includes an IPTG-inducible promoter, an origin of replication, compatible to those of the plasmids pDelH and pIK8.6 (required for delftibactin expression in E. coli) expression plasmids as well as a kanamycin resistance marker. The choice of the ori was critical, as three plasmids PFSN, pDelH and PIK8.6 would have to be cotransformed in order to obtain the final E. coli delftibactin production strain and all three plasmids needed to be stabely propagated.
Additionally, mRFP was added as last cds behind the DelRest genes onto the pFSN construct in oder to have an easy control for expression of the DelRest genes located in front of mRFP.
In the Delftibactin cluster the genes DelA, DelB, DelC, DelD, DelE, DelF and DelG are encoded as a single polycistronic operon. However, it is most likely not possible to amplify the entire coding sequence of DelA to DelG in one single PCR amplicon, as its corresponding size of 22.8 kb in combination with the complex GC-rich sequence is most likely not suitable for commonly available polymerases and PCR protocols. Consequently, we will use various primer combinations to amplify the genes in different sub-fragments. These will be assemled again using Gibson cloning
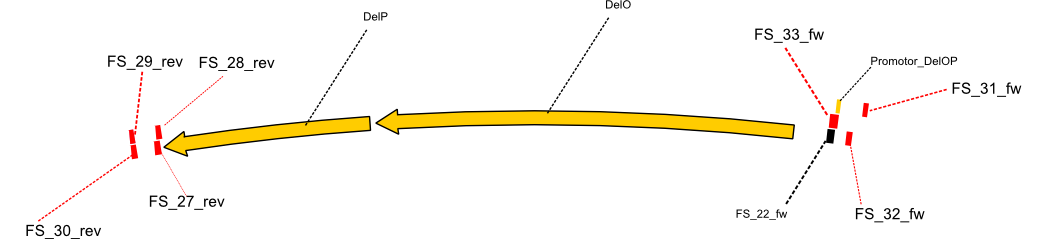
DelO and DelP are encoded in reverse-complementary direction on the Delftibactin cluster. However, in our construct these two genes will be assembled in the same orientation as the other ones and thereby added to the DelA-G operon. As DelO and DelP are also located next to each other and they will therefor be amplified together as a single 2.7 kb amplicon
DelL is the somehow lonesome rider, not surrounded by any of our desired del genes and will thus also be amplified seperately (fragment size 1.4 kb).
As complications are most likely to occure for the very long PCR amplicons, we will start with the huge DelA-G region and optimize PCR conditions for the corresponding amplicons.
We decided to use the DSM-39 substrain of Delftia acidovorans as our genomic template since our reference paper from Johnston et al. (2013) showed the gold precipitation expreiments using this substrain. The only full-length genomic sequence available for D. acidovorans is, however, the SPH-1 substrain sequence. Nevertheless, as the abovementioned paper also based their analysis on the SPH-1 strain sequence, we also based our primer designs on the available SPH-1 substrain genomic sequence, hoping the Del cluster sequence would be highly conserved among different D. acidovorans strains.
Plasmid backbone part from the partsregistry
| Backbone | Part | Distribution | Plate | Well | Usage | Resistance |
|---|---|---|---|---|---|---|
| pSB4K5 | J04450 | Spring 2012 | 1 | 5G | Backbone for DelA-G,OP,L | Kanamycin |
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_01: pSB4K5_DelA_rv | 20-13-06-28 | Amplification of pSB4K5 from the iGEM Distribution. Gibson Primer with overhang to DelA introducing the RBS BBa_B0035 | TCGCGGCGATCCGGTACTGCGCCTCTGTTGAACATCTGATATTCT CCTCTTTAATCGACAGATTGTGTGAAATTGTTATCCGCTCAC |
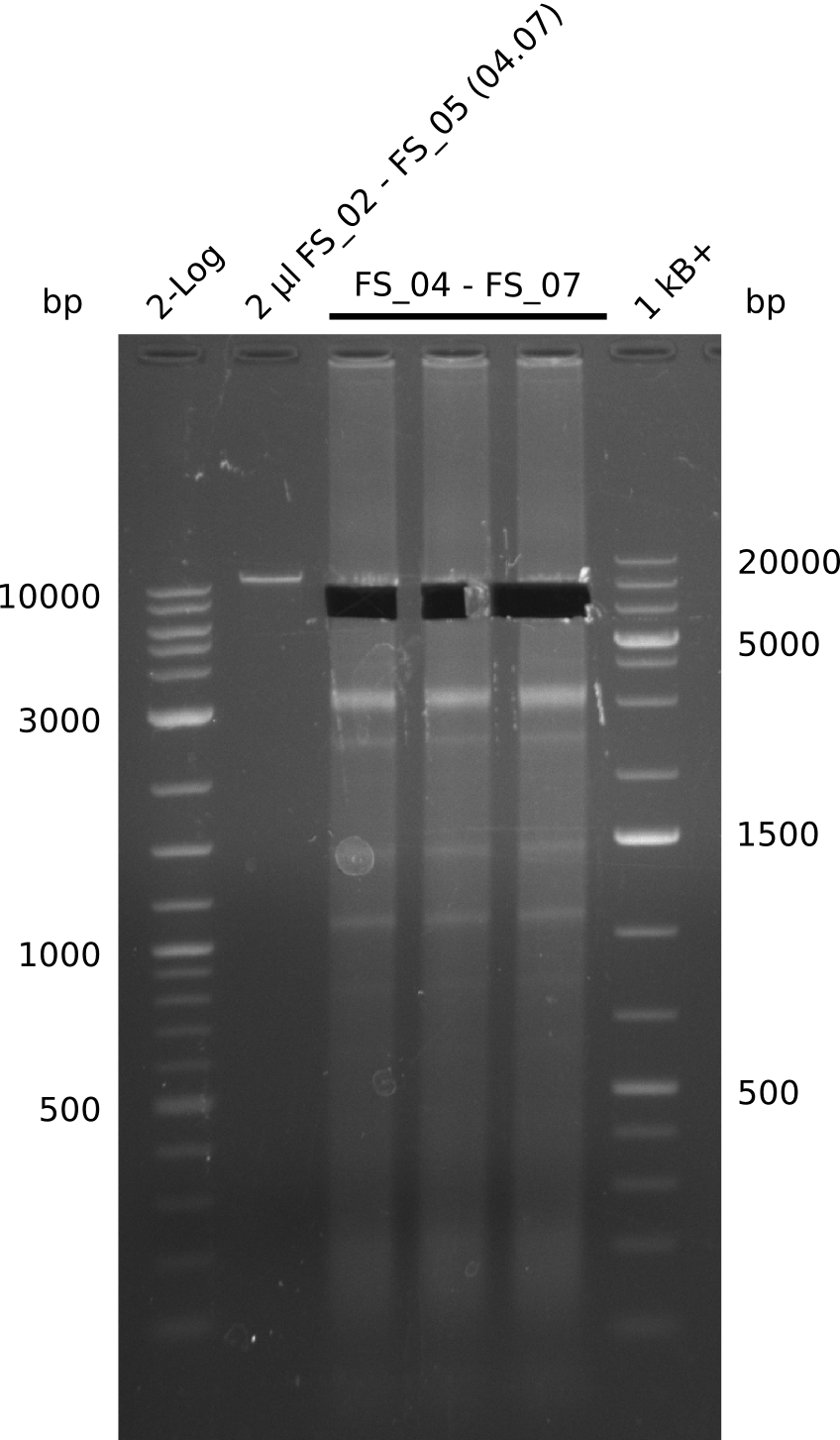
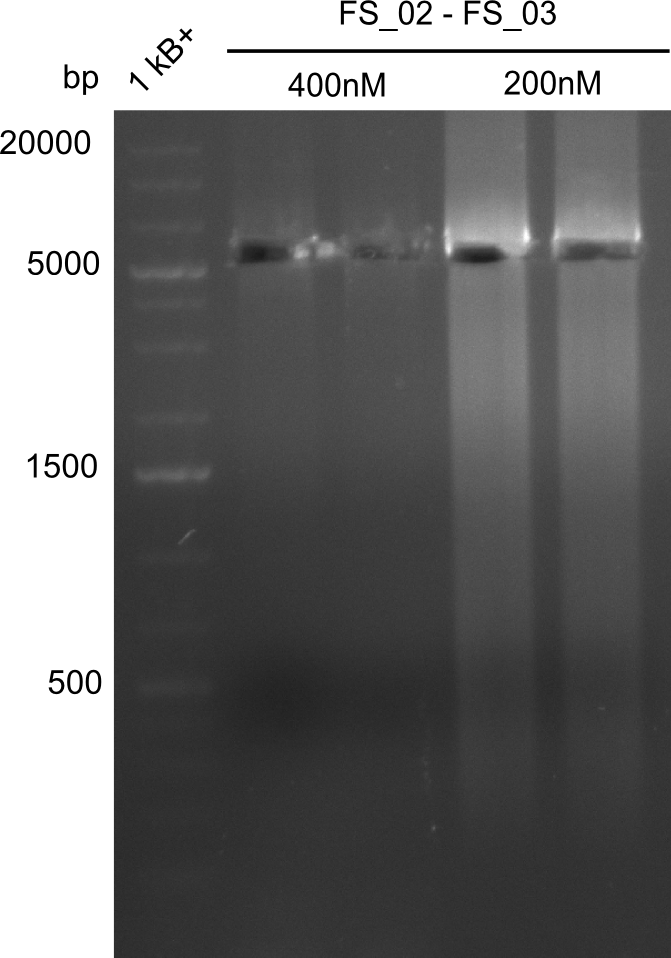
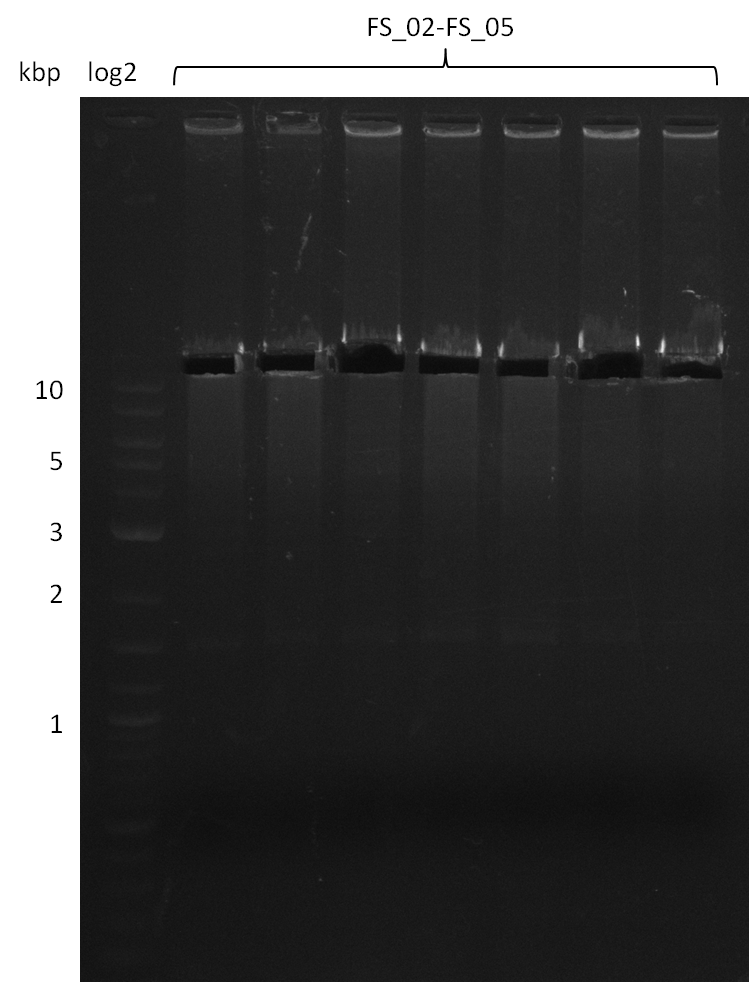
| FS_02: DelAG_1_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | TTCAACAGAGGCGCAGTACCGGATC |
| FS_03: DelAG_1_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GTCGGAGACGATGTGGTGCATCAC |
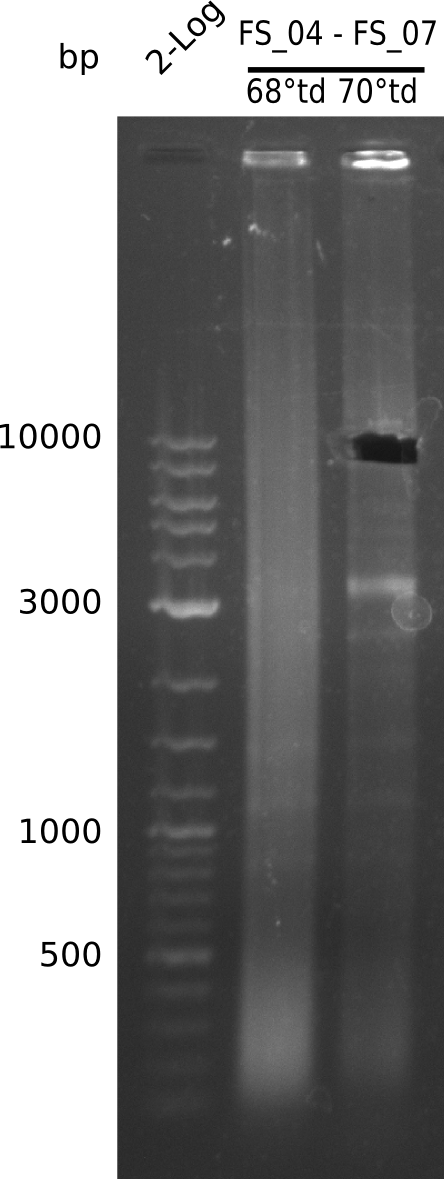
| FS_04: DelAG_2_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | CTGCAGGCCAATGAGCACATCCTG |
| FS_05: DelAG_2_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | CACAGGTGGTAGATGGCGTC |
| FS_06: DelAG_3_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | ATTGCGAGGACTTGCTCGATG |
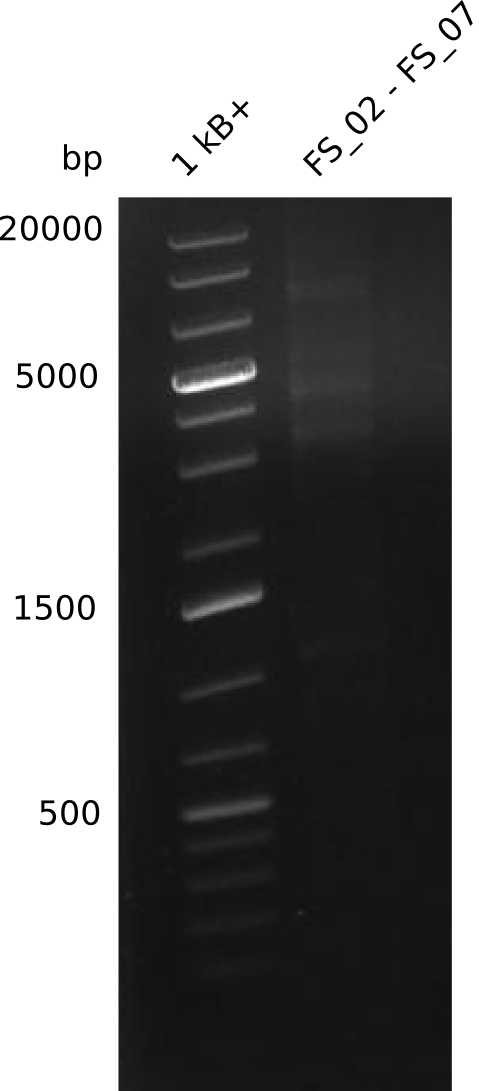
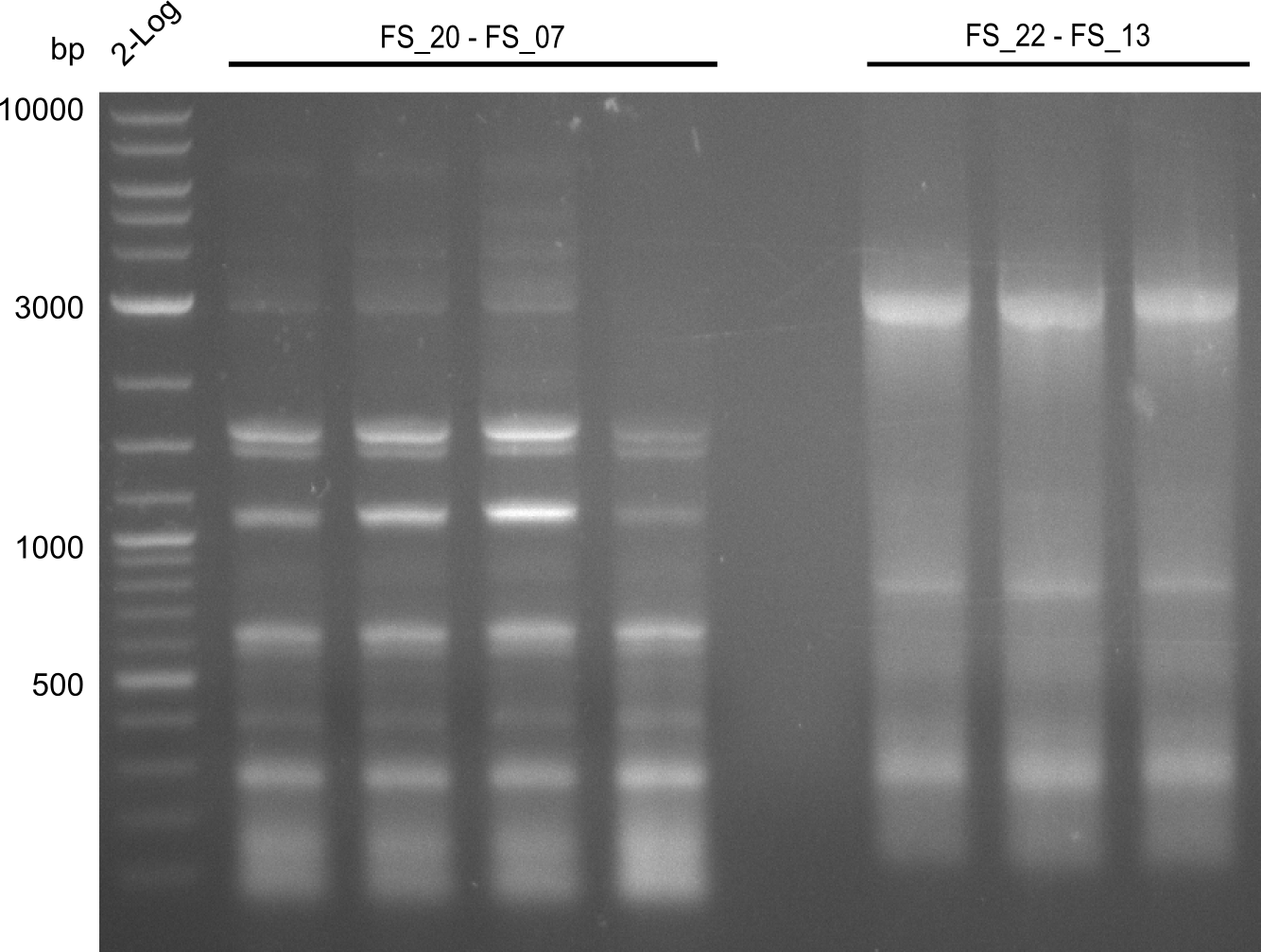
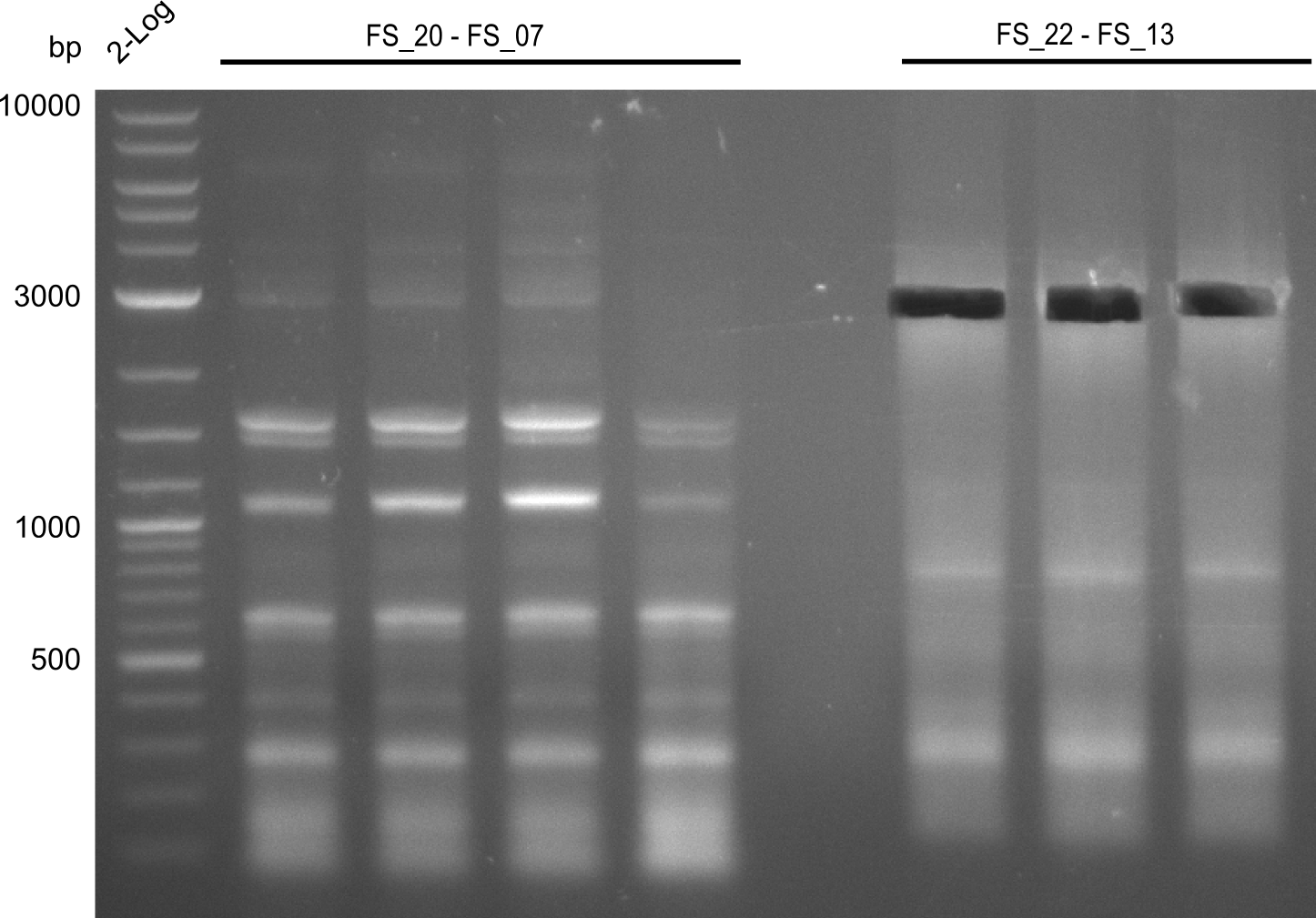
| FS_07: DelAG_3_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | TTTGCTGCAGCGCCAGCACATCGAG |
| FS_08: DelAG_4_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GTACGGCCTATCACATCAGCG |
| FS_09: DelAG_4_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GAAGCTCAGCAGGTTGGGCGAGACG |
| FS_10: DelAG_5_fw | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. | GAATTTTGTTCCACCACCTGCTG |
| FS_11: DelAG_5_rv | 2013-06-28 | Amplification of DelA-G from Delftia acidovorans genome. Gibson Primer with overhang to DelOP | CTTGAGCAGGCGCAGTACCTCGGAGGGCGGTCGGCTGGCGTTTTCCATGATTCAGG TTTCCTGTGTGAAGCTCATCTCAGATATCTCCCAGAGTTTCGAGAAAG |
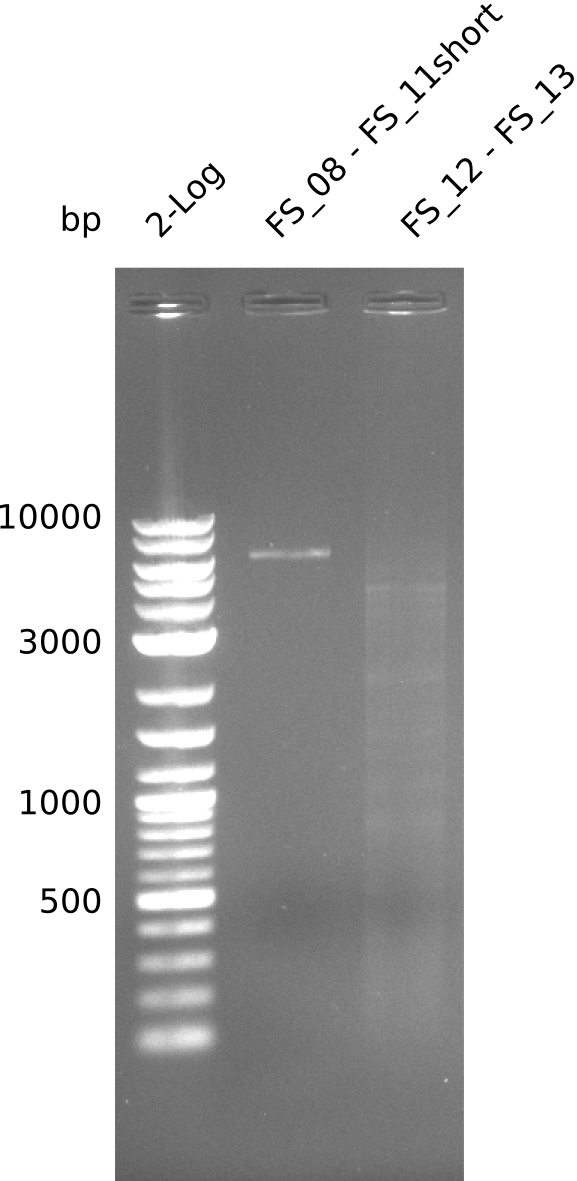
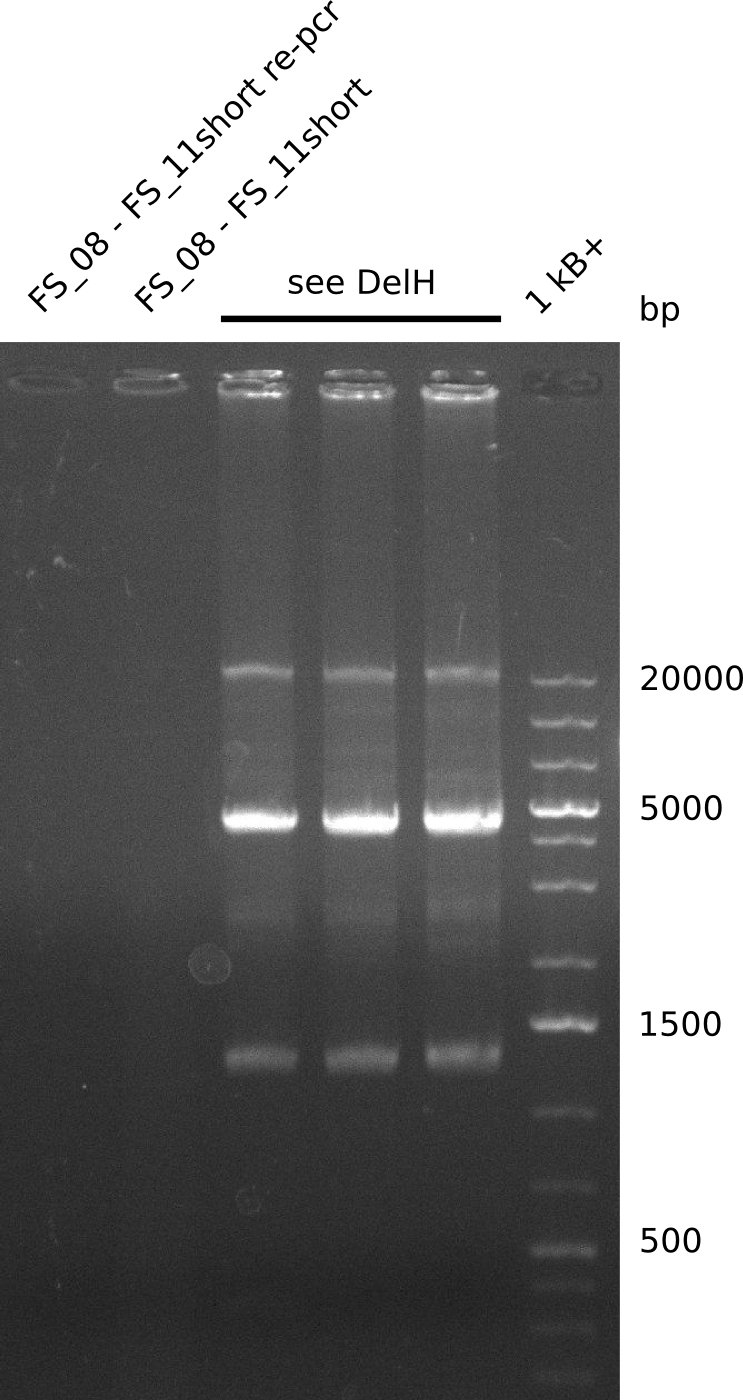
| FS_11: DelAG_5_short_rv | 2013-08-02 | Amplification of DelA-G from Delftia acidovorans genome. | TCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_12: DelOP_fw | 2013-06-28 | Amplification of DelO-P from Delftia acidovorans genome. | GAATCATGGAAAACGCCAGCCGAC |
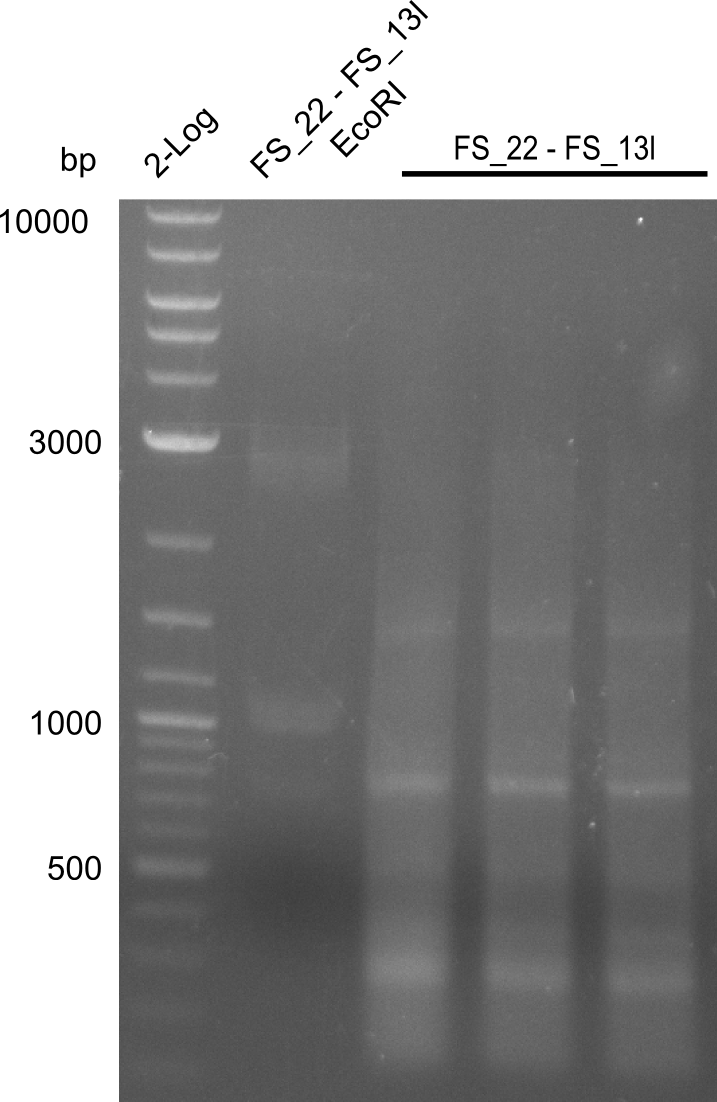
| FS_13: DelOP_rv | 2013-06-28 | Amplification of DelO-P from Delftia acidovorans genome. Gibson Primer with overhang to DelL | CAATGTTGGAGGGGCCGAAGCCGATGCCGATCAGCGGGTGGGTTTGCATGGAAGGT CCTTTCATTGGGTCGATGCGTCCAGTGTCACACCGTGGTGTCTGCAGGCG |
| FS_13: DelOP_short_rv | 2013-06-28 | Amplification of DelO-P from Delftia acidovorans genome. Gibson Primer with overhang to DelL | TCACACCGTGGTGTCTGCAGGCG |
| FS_14: DelL_fw | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome. | CAAACCCACCCGCTGATCGGCATC |
| FS_15: DelL_mRFP_pSB4K5_rv | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome. Gibson Primer with overhang to BBa_J04450 | GAAACGCATGAACTCTTTGATAACGTCTTCGGAGGAAGCCATCTAGTATTTCTCCTC TTTCTCTAGTATCAGTCCTGCAGCGCCAGCTGTTCTGTG |
| FS_15: DelL_mRFP_pSB4K5__short_rv | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome. Gibson Primer with overhang to BBa_J04450 | TCAGTCCTGCAGCGCCAGCTGTTCTGTG |
| FS_16: mRFP_pSB4K5_fw (Del) | 2013-06-28 | Amplification of pSB4K5 from iGEM Distribution. Gibson Primer | GCTTCCTCCGAAGACGTTATC |
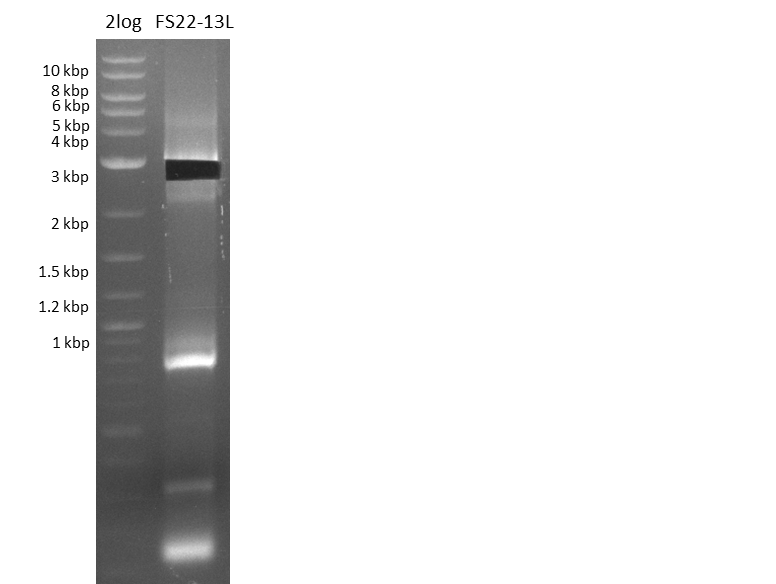
Amplification of Del Genes from DSM-39 genome
Goals
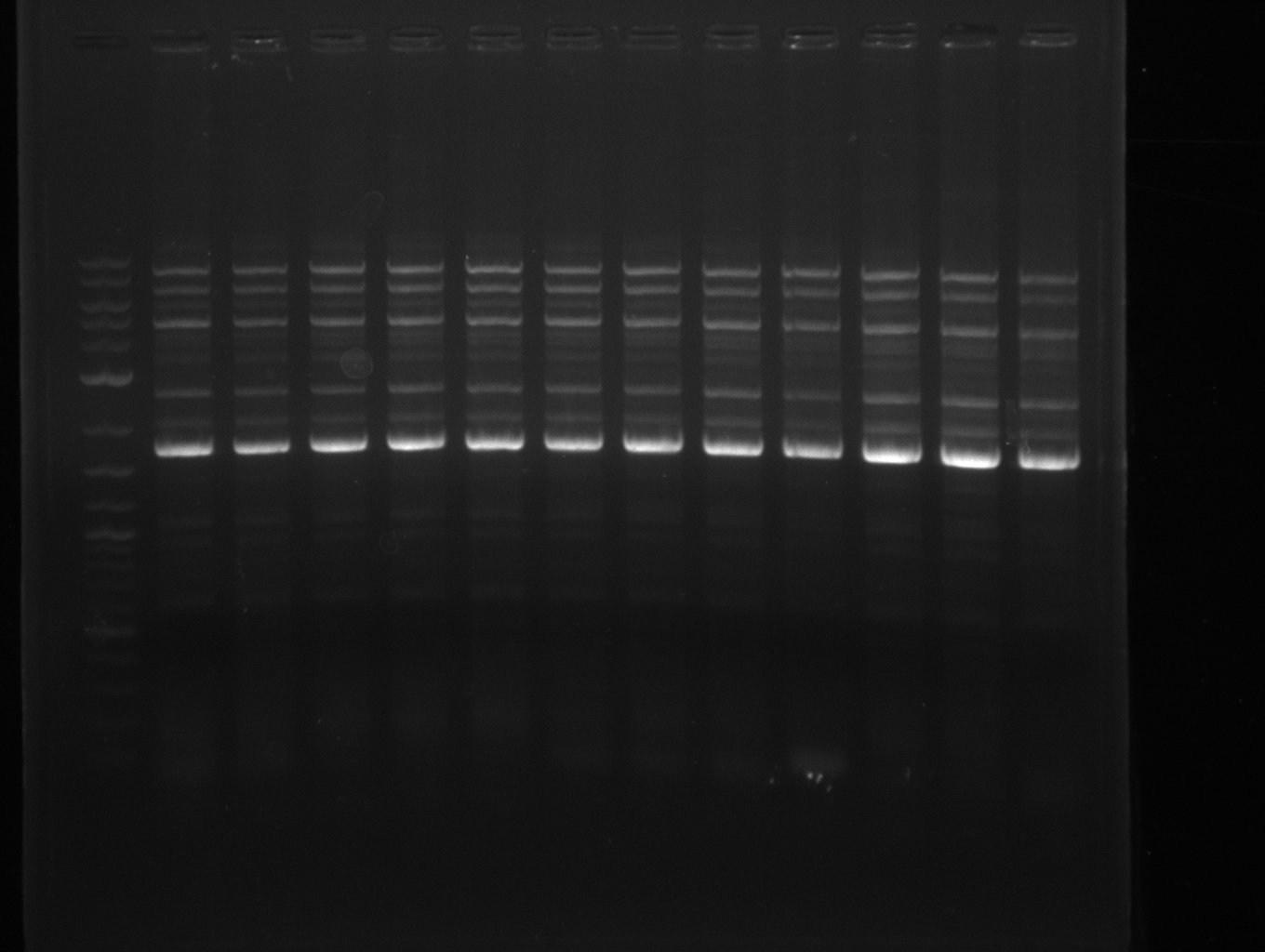
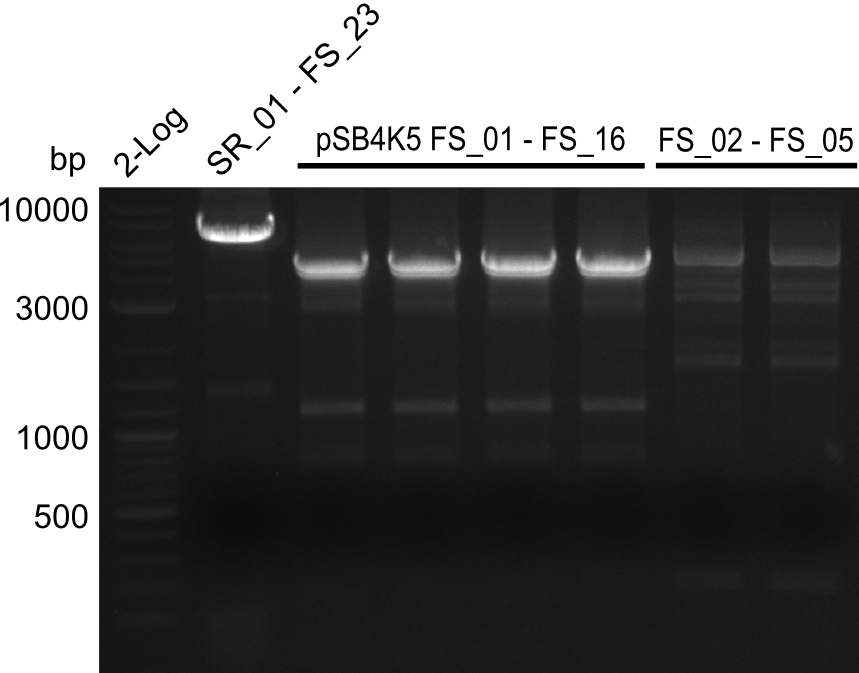
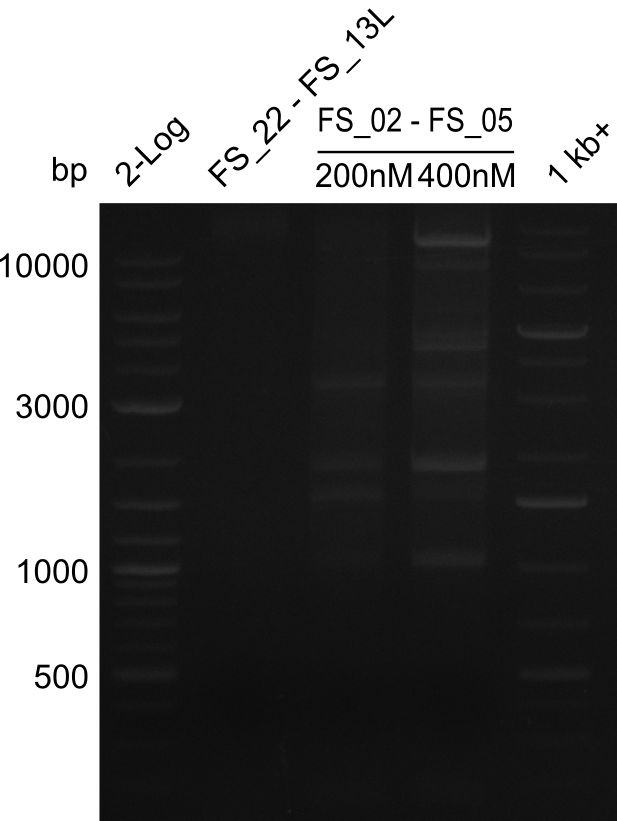
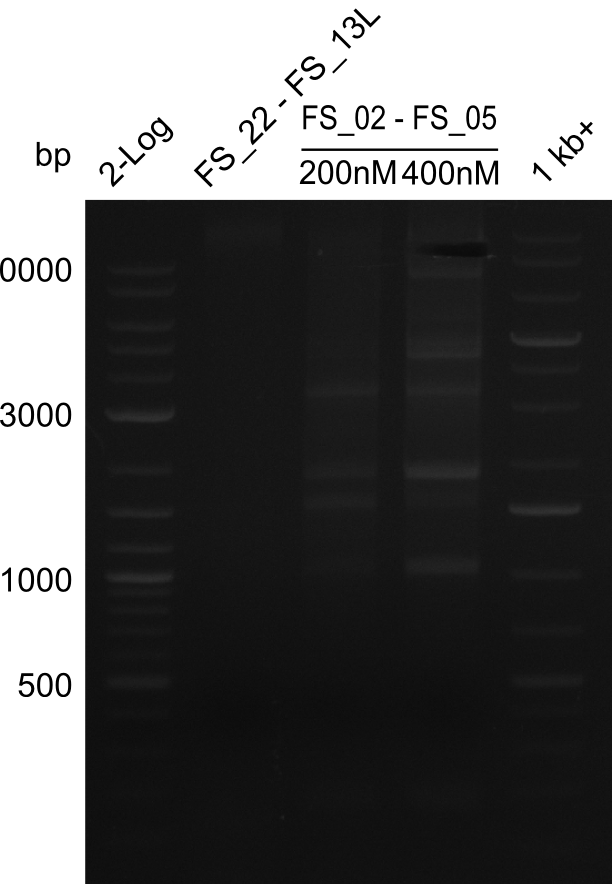
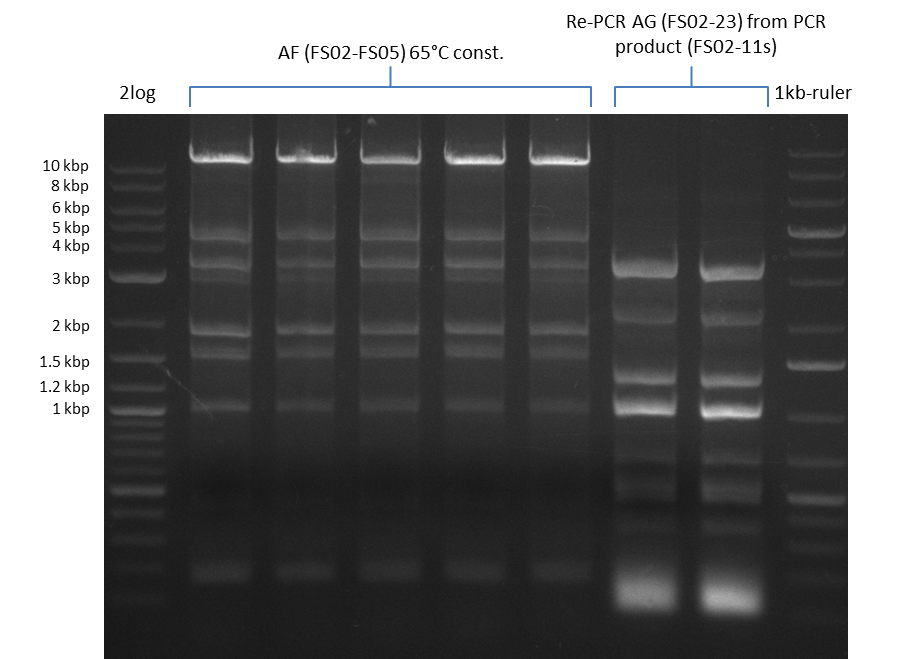
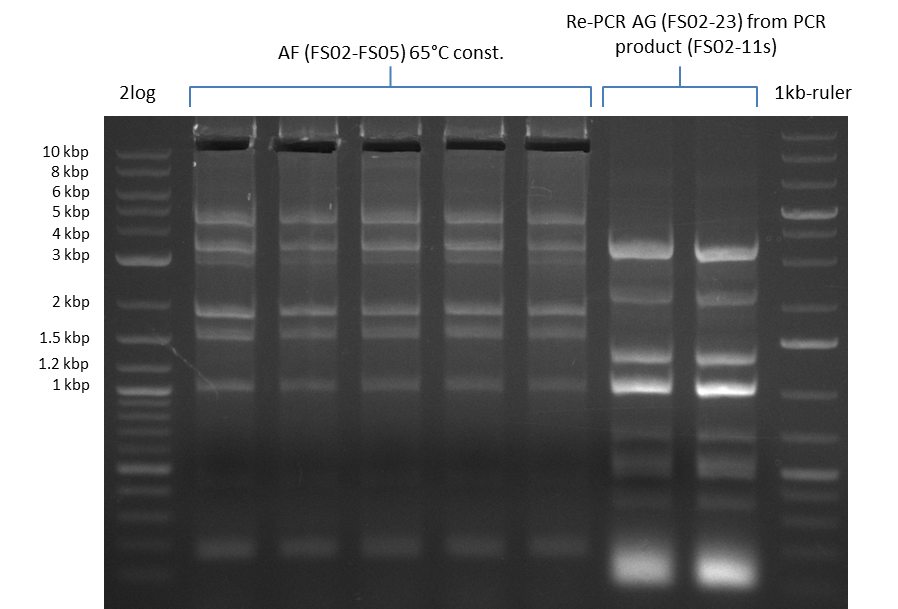
This week primers FS_01 to FS_16 arrived. As the aim of this week is to amplify the backbone pSB4K5 with Gibson overlaps matching our initial and very last insert fragment, we will firstly use the pSB4K5 backbone including the mRFP cassette as template for this purpose. Furthermore amplification of the desired genes from the Del-cluster of D.acidovorans DSM-39 will be carried out in parallel.
Results
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
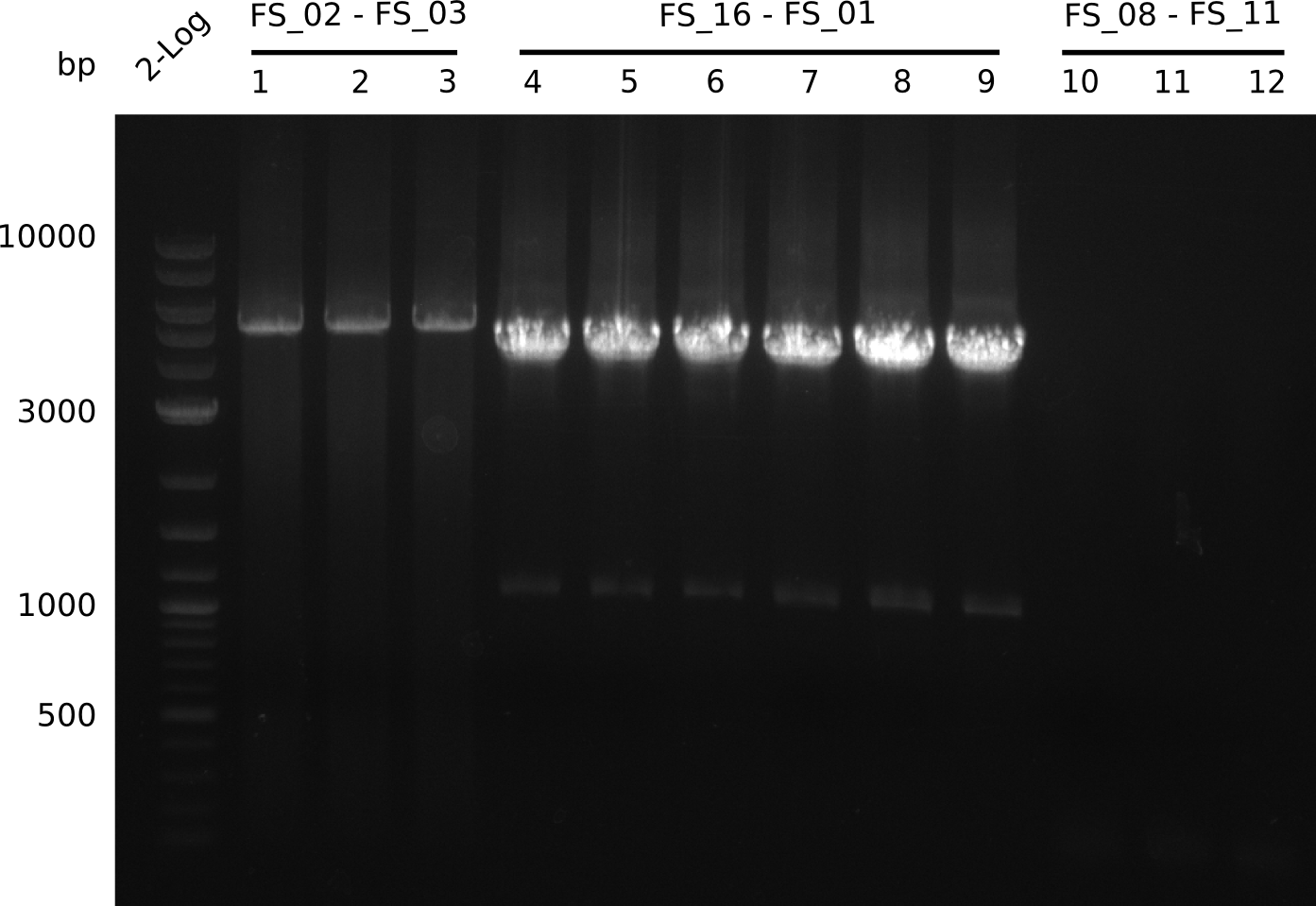
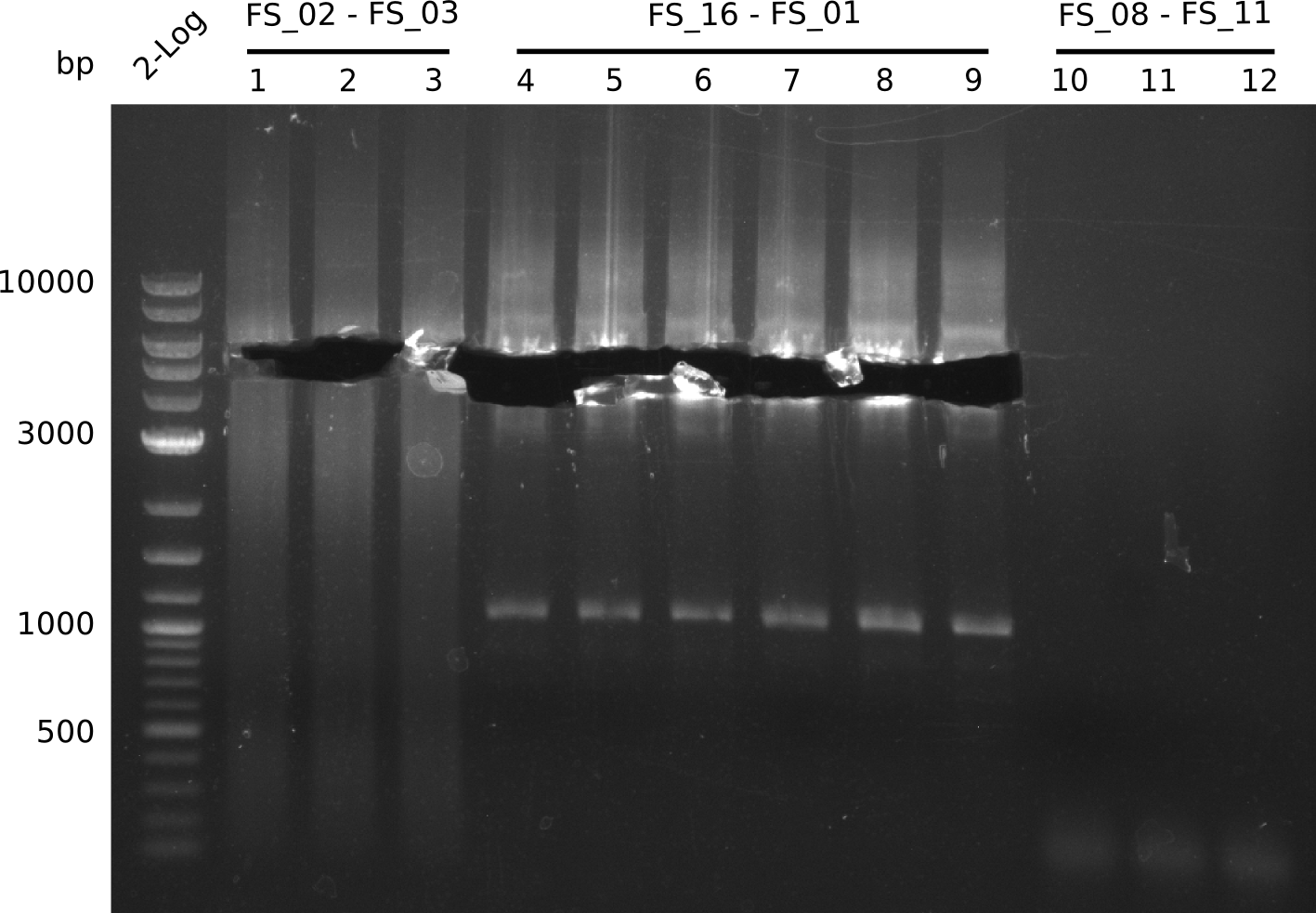
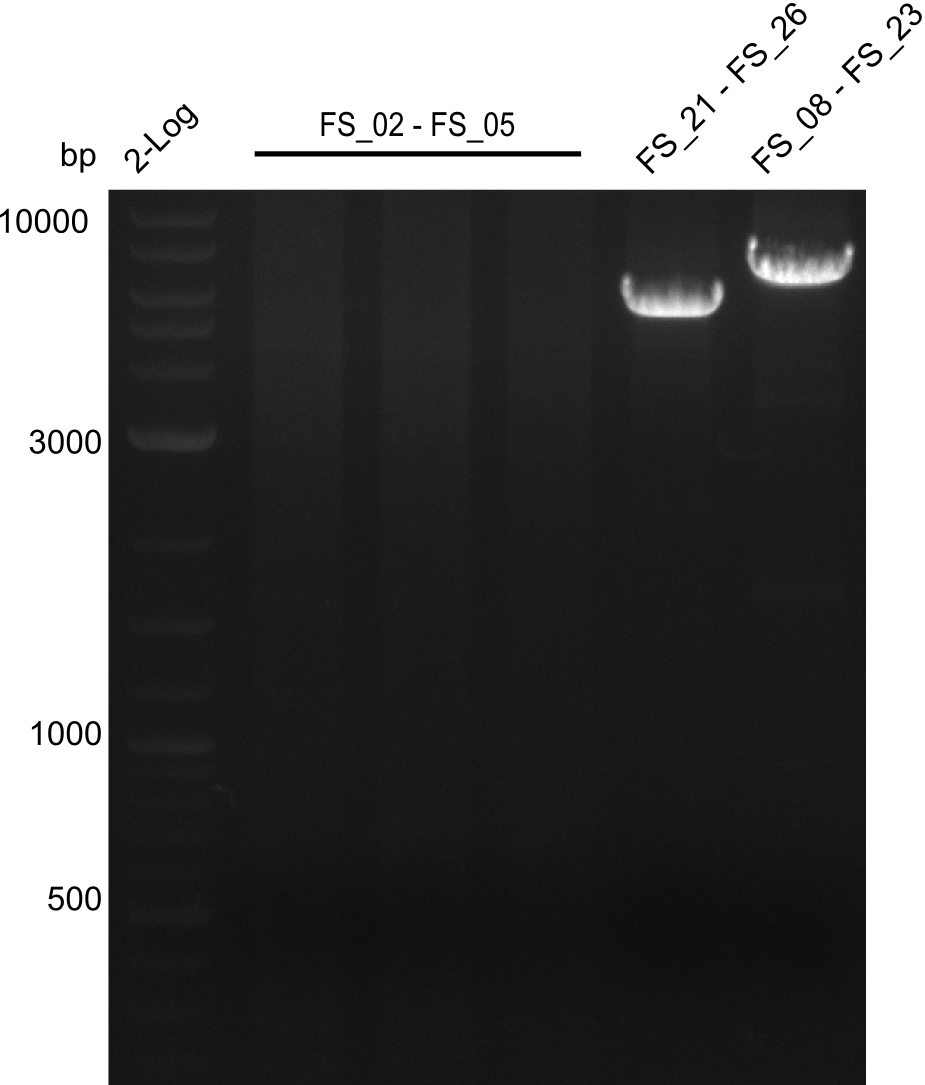
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | FS_02 and FS_03 | ||
| DelA-F | FS_02 and FS_05 | |||
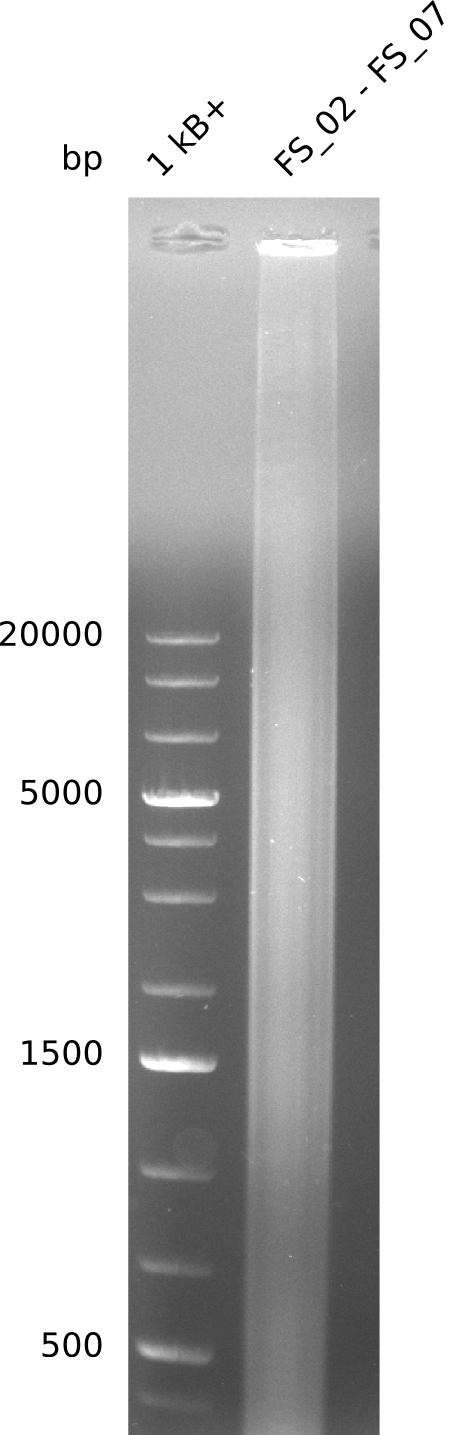
| DelA-G | FS_02 and FS_07 | |||
| DelE-G | FS_04 and FS_07 | |||
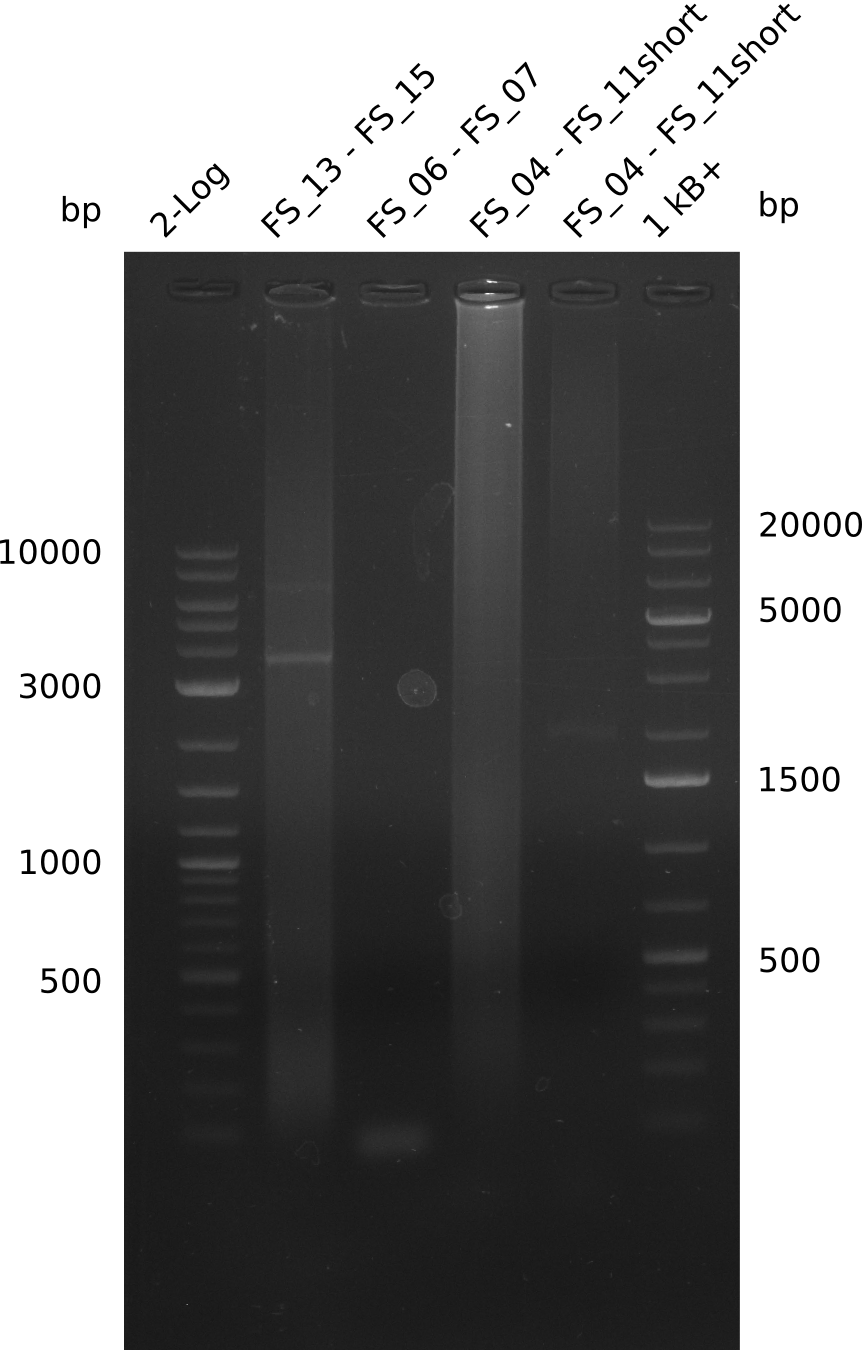
| DelF-G | FS_06 and FS_07 | |||
| FS_06 and FS_09 | ||||
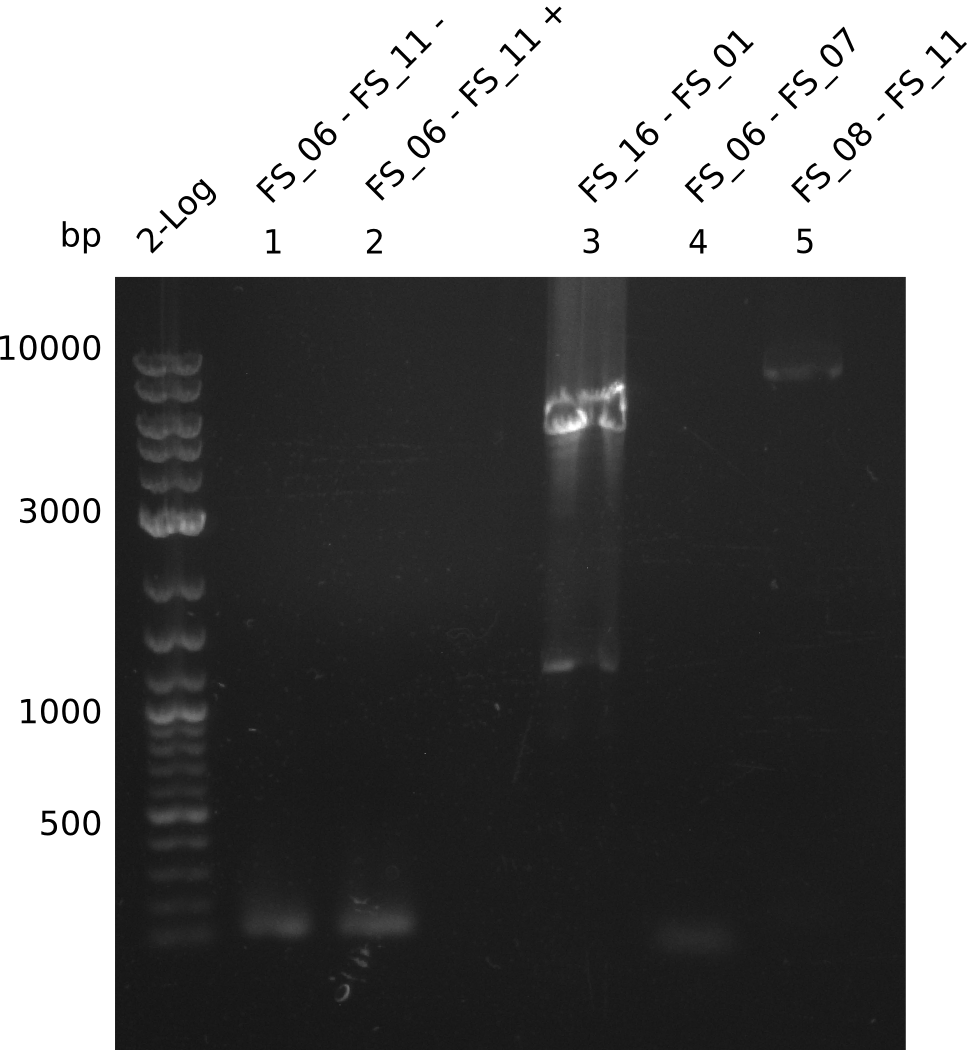
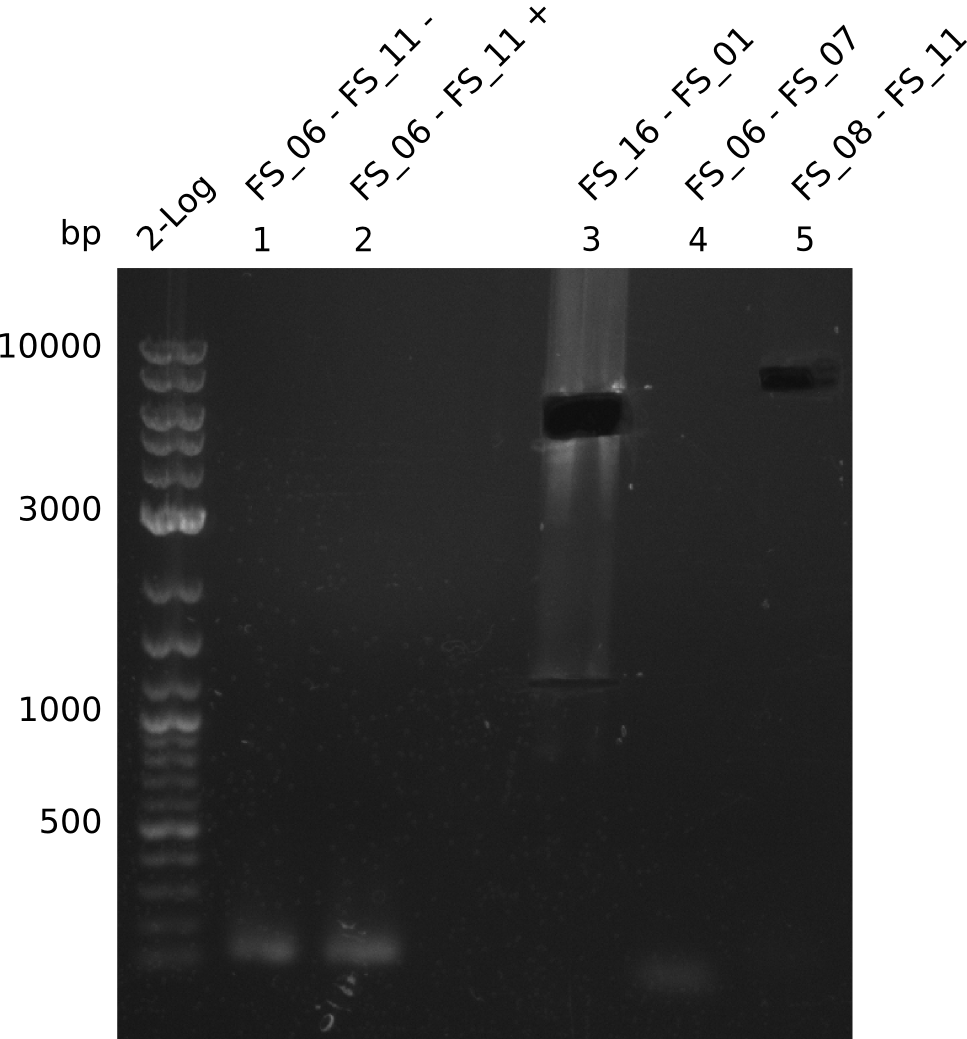
| FS_06 and FS_11 | ||||
| DelG | FS_08 and FS_11 | |||
| DelO DelP DelL | DelO-P | FS_12 and FS_13 | ||
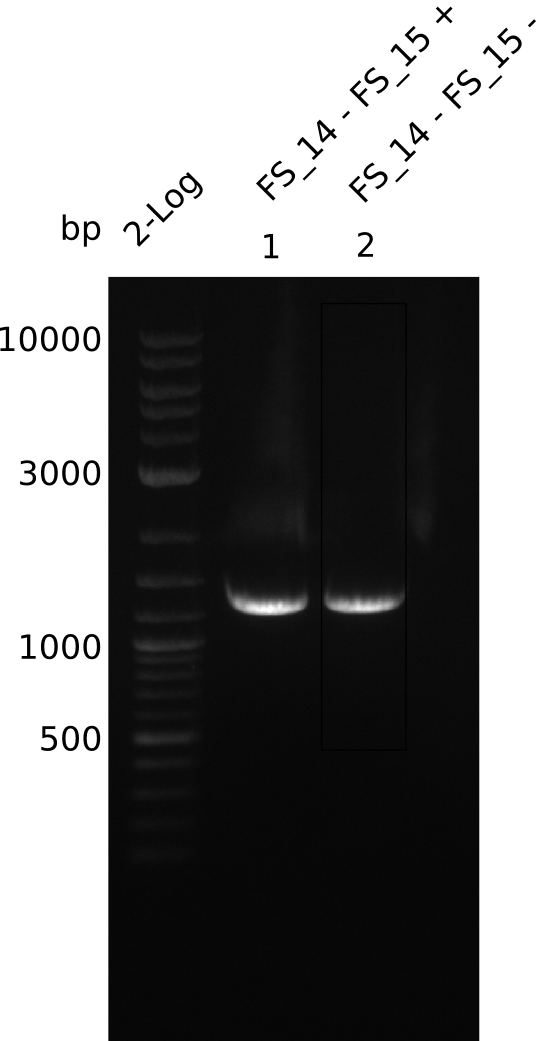
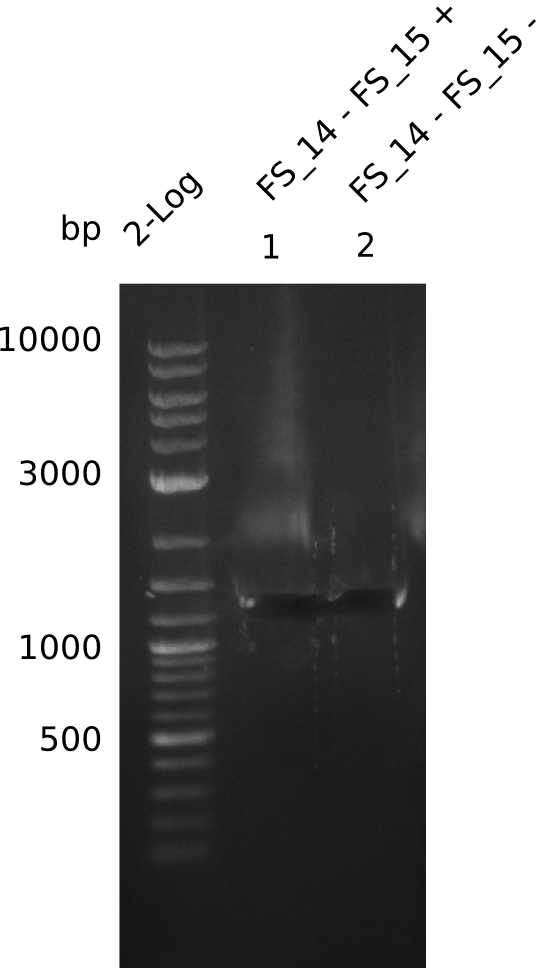
| DelL | FS_14 and FS_15 | |||
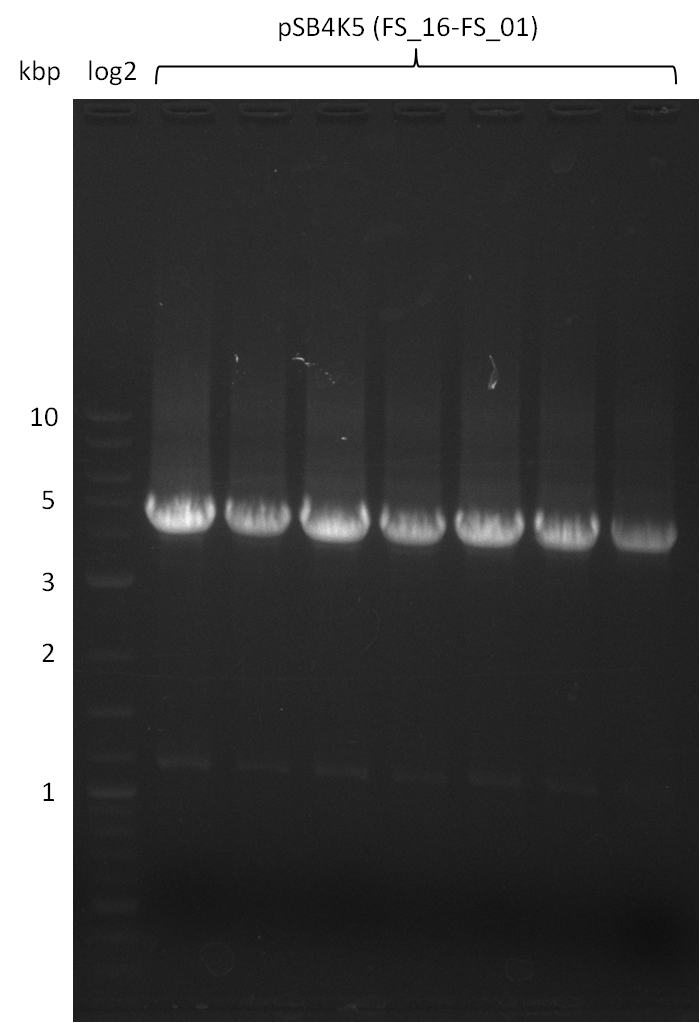
| pSB4K5 | pSB4K5 | FS_01 and FS_16 | ||
06-07-2013
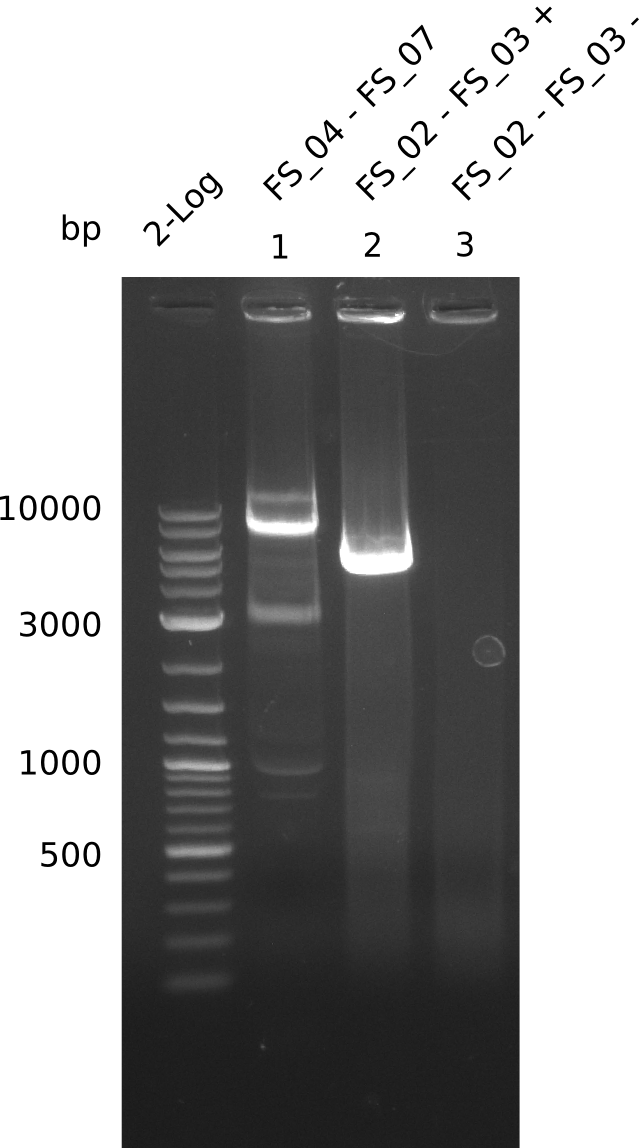
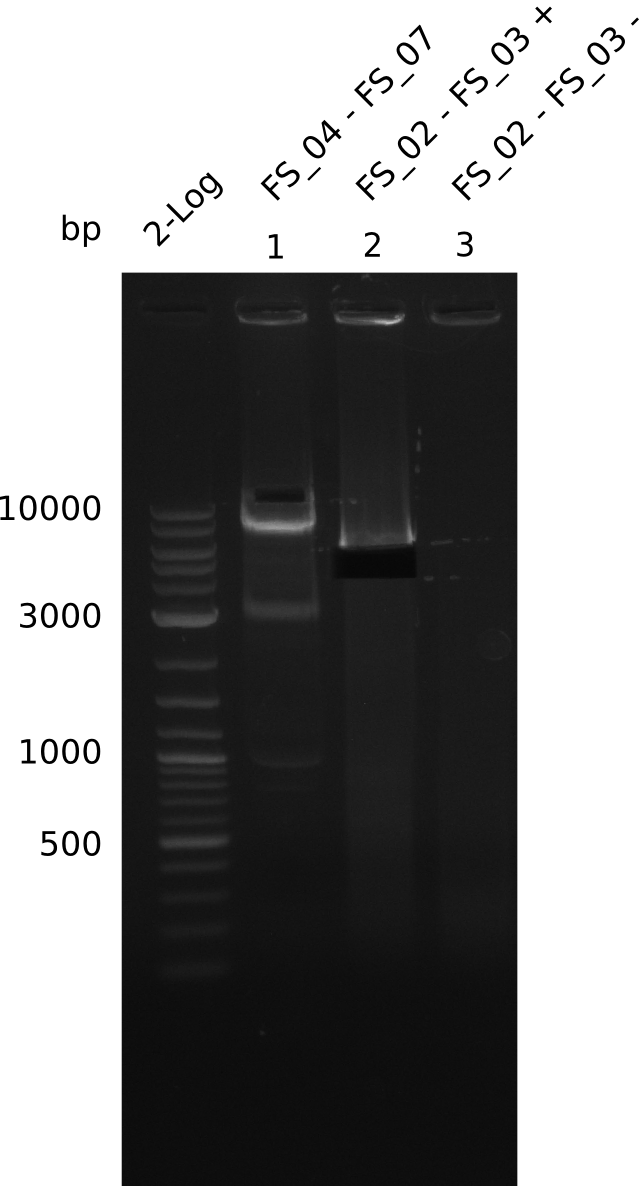
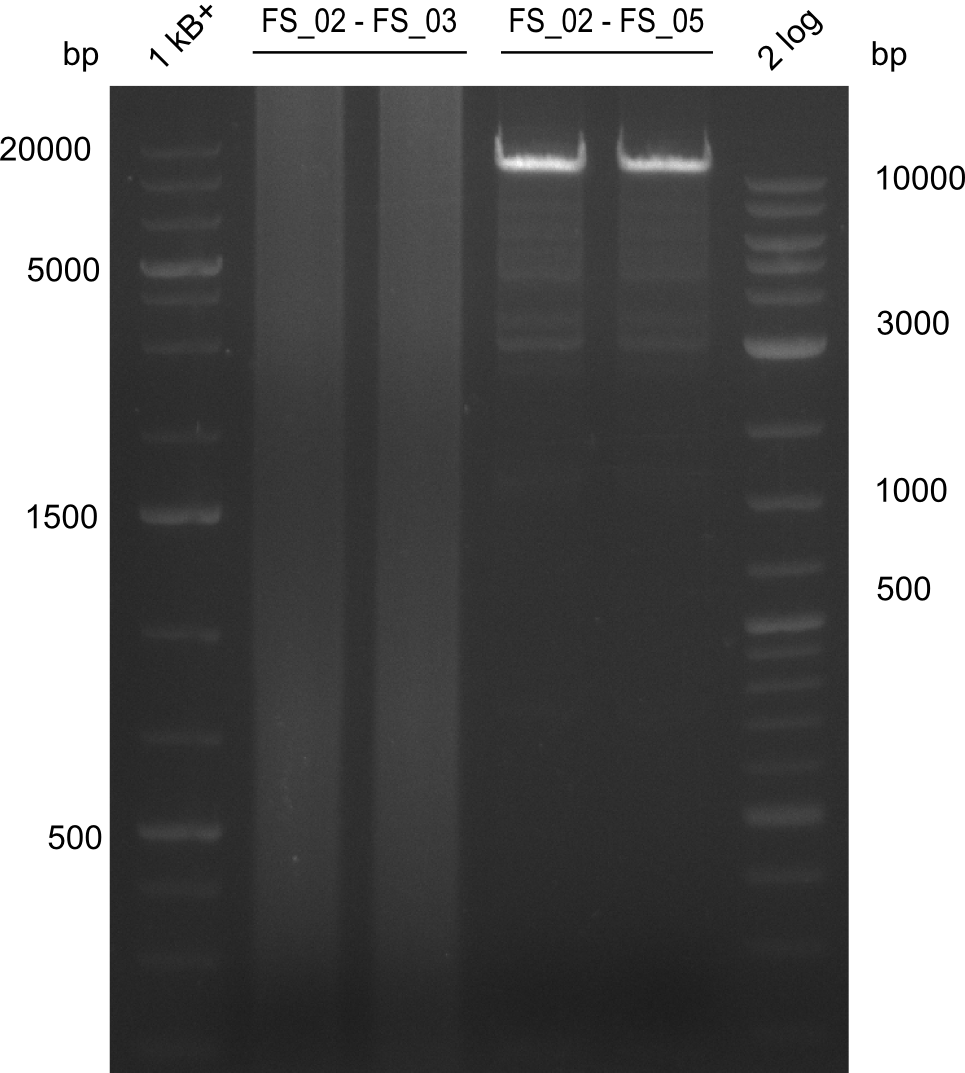
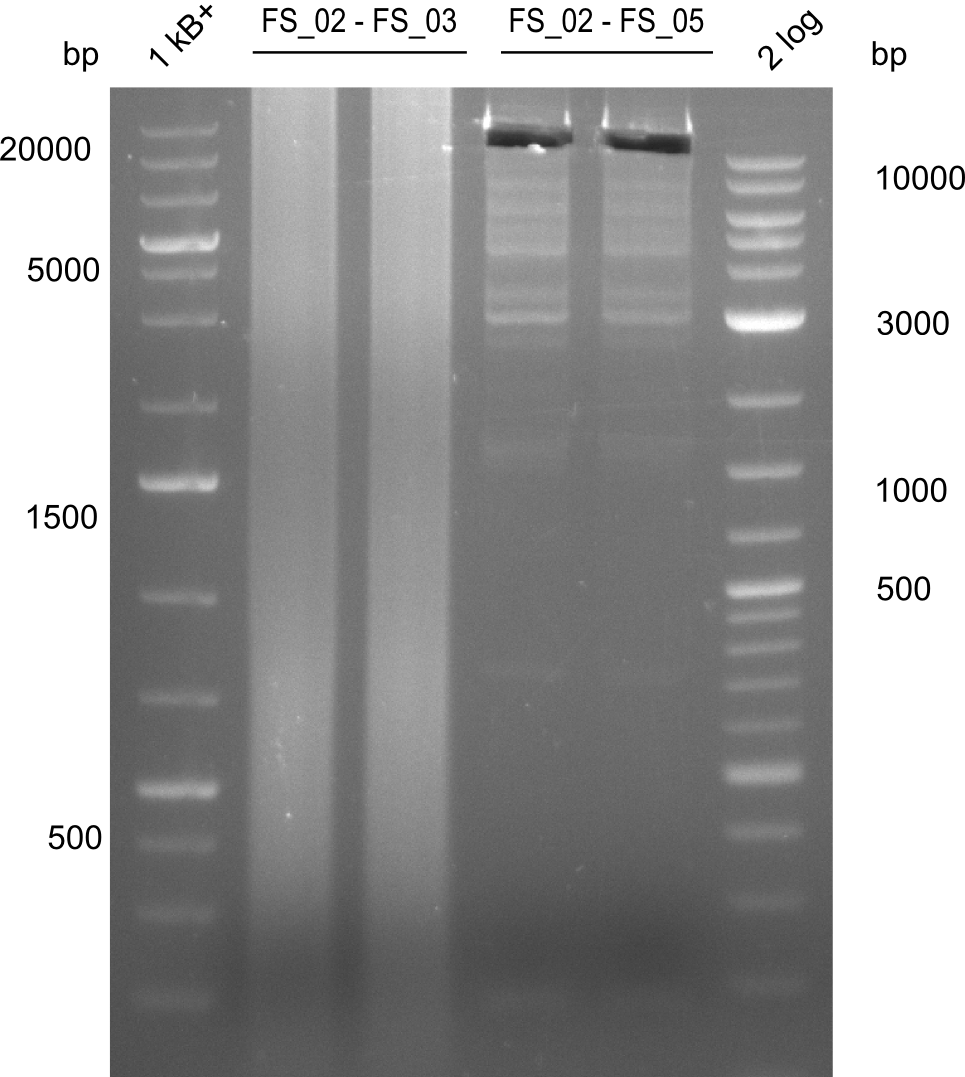
Amplification from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 1 |
| FS_03: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 6/7 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification worked with 5% DMSO
- Repeat Amplification with the same protocol to increase concentration when DNA is extracted from gel slices
03-07-2013
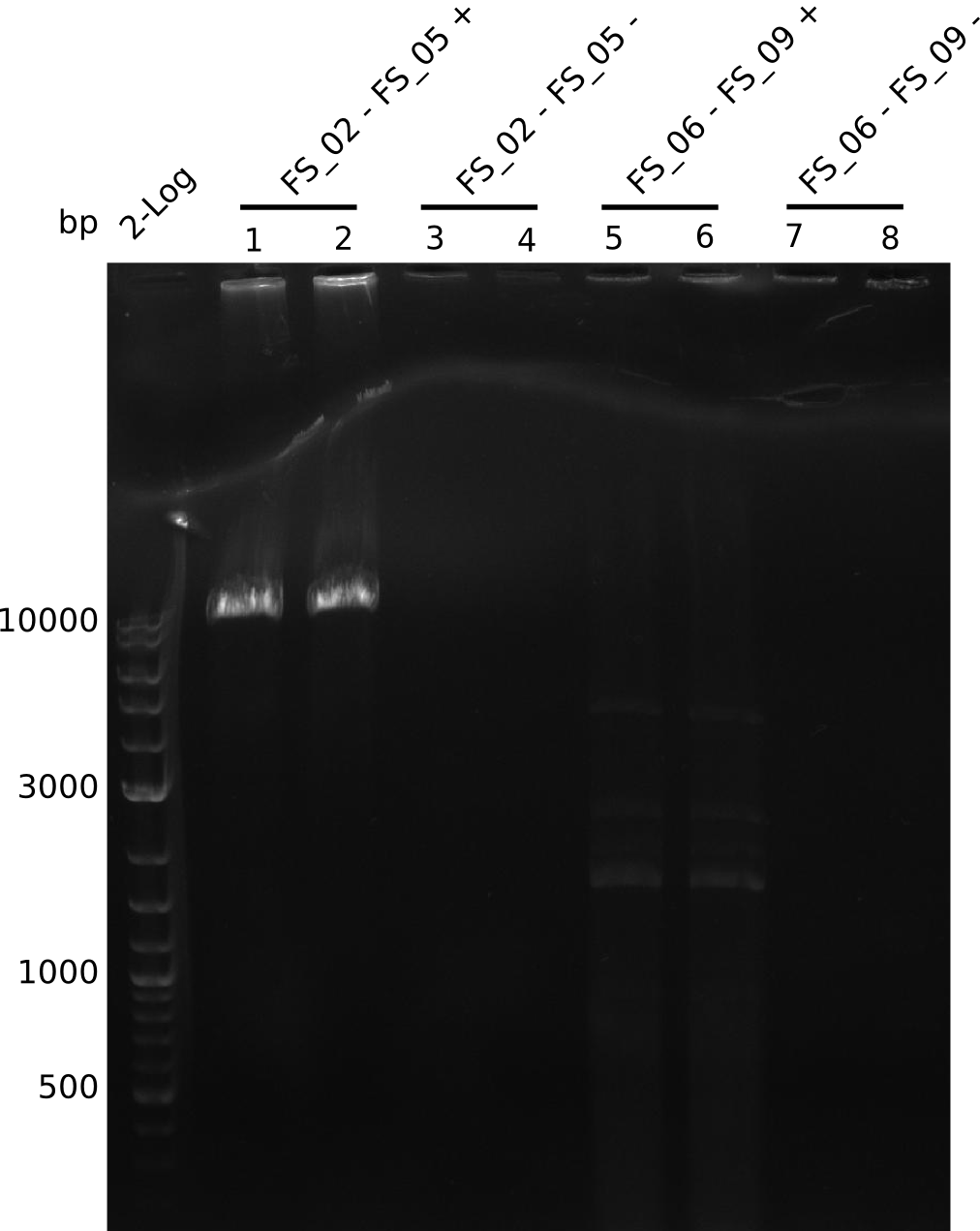
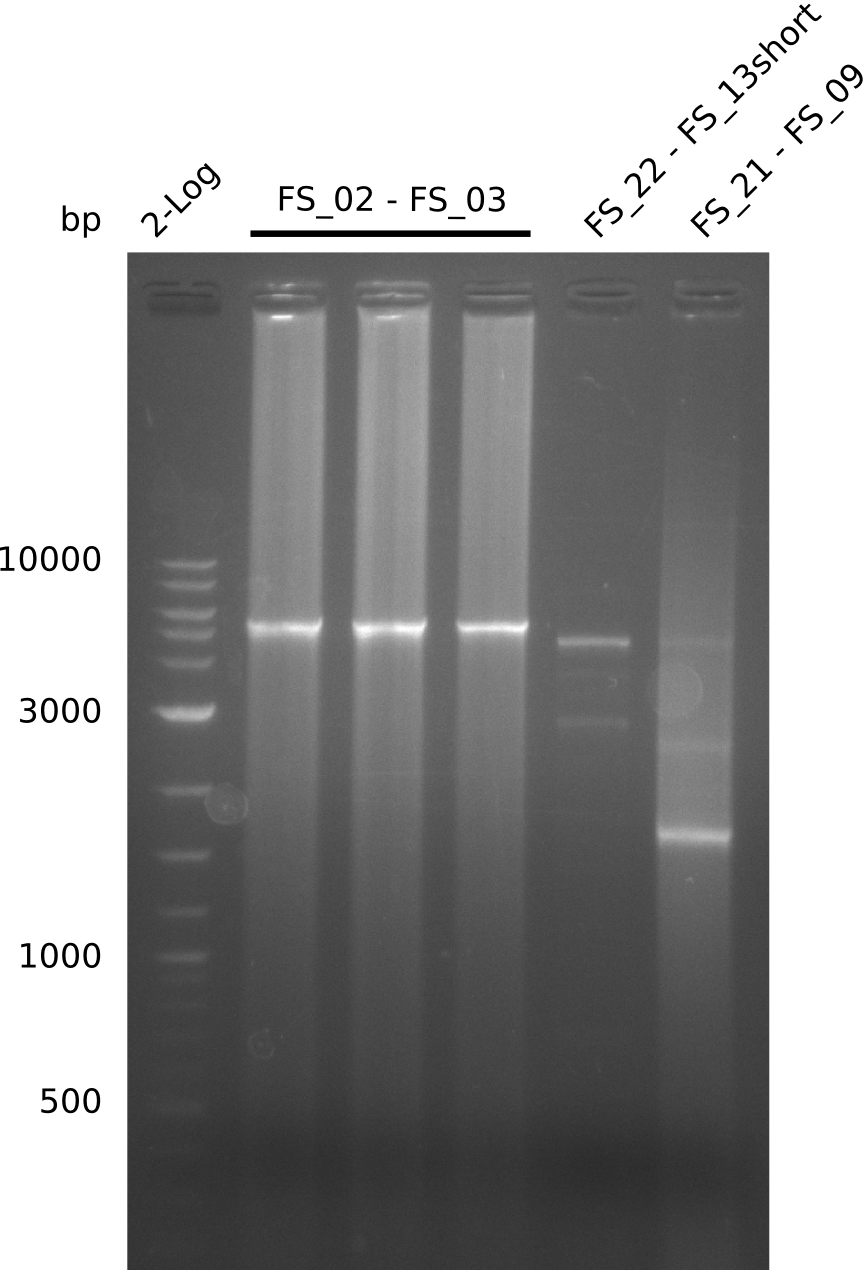
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| µL 1st PCR | what | µL 2nd PCR |
|---|---|---|
| 1 | D. acidovorans DSM-39 | 1 |
| 2.5 | FS_02 (1/10) | 2.5 |
| 2.5 | FS_05 (1/10) | 2.5 |
| 25 | Phusion Master Mix | 25 |
| - | DMSO | 2.5 |
| 19 | dd H2O | 19 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAE worked
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- sample has to be purified before further use since contamination with propanol was present
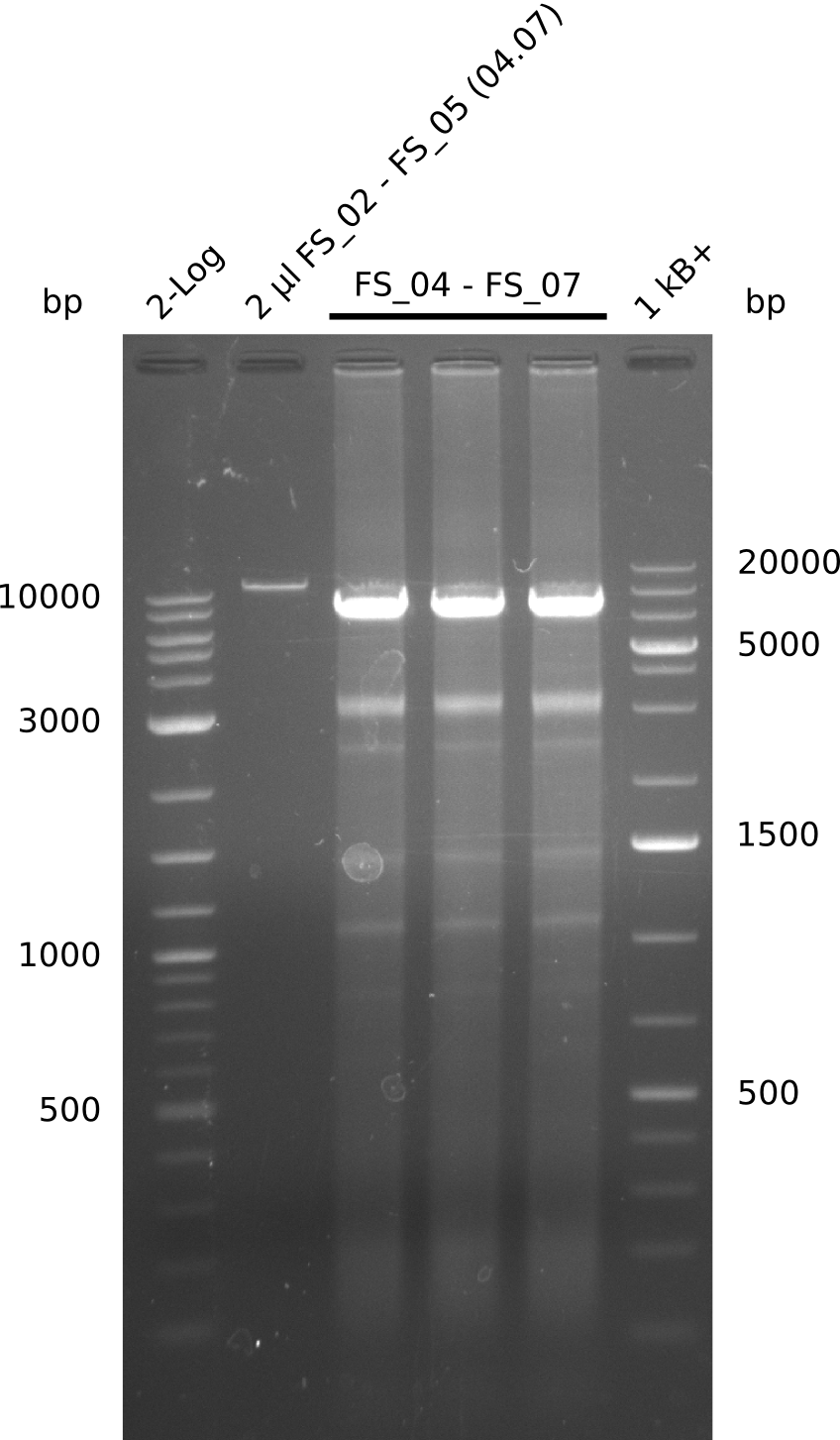
04-07-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| what | µL 2nd PCR |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02 (1/10) | 2.5 |
| FS_05 (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAE worked
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit resulting in a final concentratio of 10ng/µL
- PCR will be repeated and gel slices purified with QIAX II Gel Extraction Kit, which is specifically designed to deliver higher yields when purifying fragments with sizes over 10 kbp
05-08-2013
Amplification from FS_02 to FS_07; 16.4 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 6 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 5min 40s | |
| 24 | 98 | 1 |
| 60 | 5 | |
| 72 | 5min 40s | |
| 1 | 72 | 17 min |
| 1 | 4 | inf |
Results:
- amplification of DelAG did not work
- repeat PCR with higher annealing temperature to increase specifity
06-07-2013
Amplification from FS_04 to FS_07; 11.1 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 6/7 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelEG worked but another unexpected product was amplified as well
- band was cut out avoiding contamination with other product and DNA purified using QIAquick Gel Extraction Kit
03-07-2013
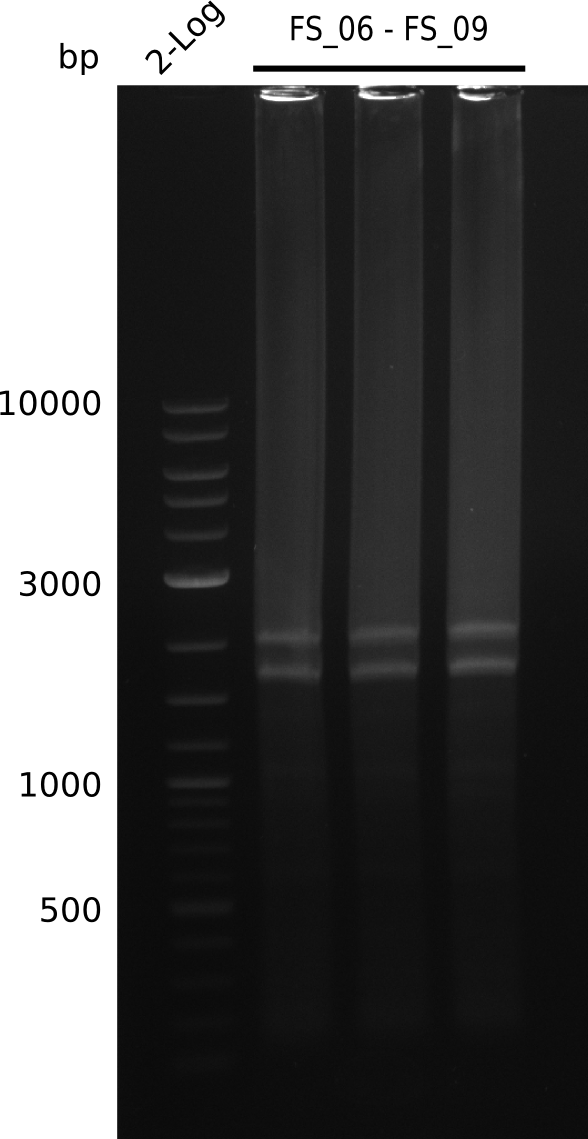
Amplification from FS_06 to FS_09; 8.5 kb
- Reaction
| µl 1st PCR | what | µl 2nd PCR |
|---|---|---|
| 1 | D. acidovorans DSM-39 | 1 |
| 2.5 | FS_06 (1/10) | 2.5 |
| 2.5 | FS_09 (1/10) | 2.5 |
| 25 | Phusion Master Mix | 25 |
| - | DMSO | 2.5 |
| 19 | dd H2O | 19 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelFG did not work
- PCR will be repeated with a lower annealing temperature of 68°C (touchdown)
Amplification from FS_06 to FS_09; 8.5 kb
- Reaction
| what | µl 2nd PCR |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06 (1/10) | 2.5 |
| FS_09 (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelFG did not work, many different side product occured, PCR will be repeated with other primers
Amplification from FS_06 to FS_11; 11.6 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 1 |
| FS_11: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
05-07-2013
Amplification from FS_06 to FS_07; 5.2 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 14 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 3:20 min | |
| 16 | 98 | 1 |
| 60 | 5 | |
| 72 | 3:20 min | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
05-07-2013
Amplification from FS_08 to FS_11; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 1 |
| FS_11: (1/10) | 1 |
| Phusion Master Mix | 10 |
| dd H2O | 6 |
| DMSO | 1 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 14 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 3:20 min | |
| 16 | 98 | 1 |
| 60 | 5 | |
| 72 | 3:20 min | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
Results:
- A weak band was visible, but it was on the wrong height.
- Accidently the band was cut anyway.
- Either the primers did not bind or the DNA still had to many secondary structures --> the consequence is to change the annealing temperature.
07-07-2013
Amplification from FS_08 to FS_11; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2.5 |
| FS_11: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| dd H2O | 19 |
| DMSO | 2.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- There was no band visible on the gel.
- Either the primers did not bind or the DNA still had to many secondary structures --> the consequence is to change the annealing temperature.
04-07-2013
Amplification from FS_12 to FS_13; 2.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_12: (1/10) | 1 |
| FS_13: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 40 | |
| 1 | 72 | 420 |
| 1 | 4 | inf |
Results:
- Amplification of DelOP did not work
- Amplification will be repeated to exclude that pipetting errors were the reason for the failure
04-07-2013
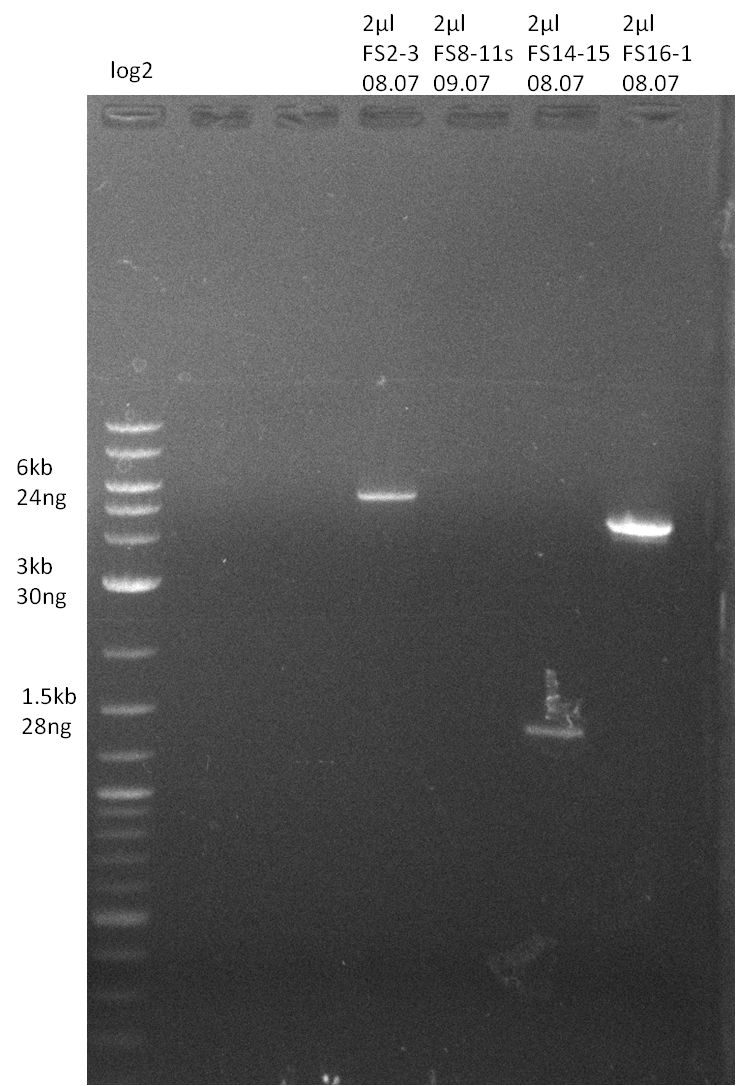
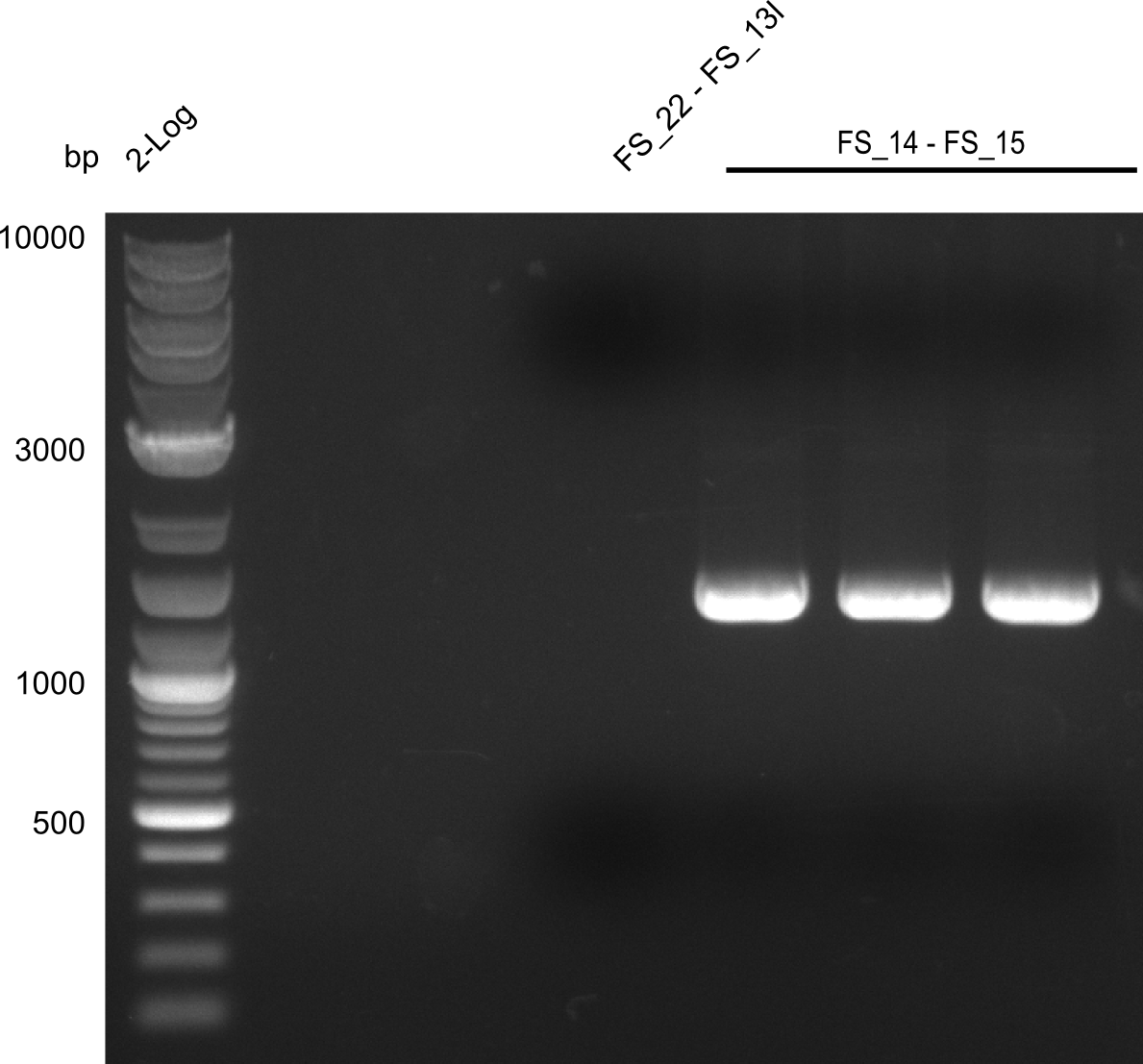
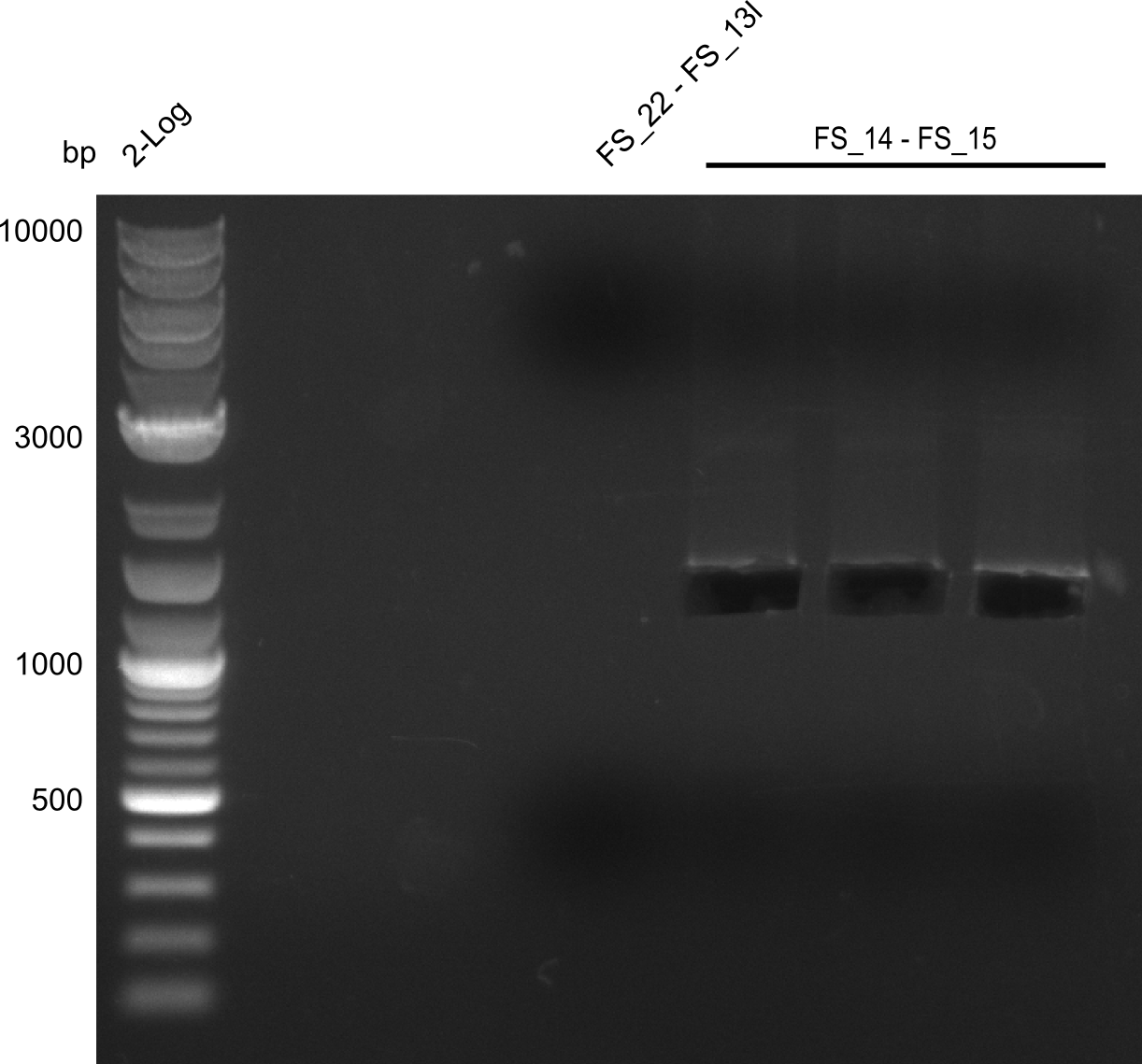
Amplification from FS_14 to FS_15; 1.4 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_14: (1/10) | 1 |
| FS_15: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 40 | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Results:
- Amplification worked very well, we had a bright band on the right height.
- Band was cut out and DNA purified using QIAquick Gel Extration Kit.
06-07-2013
Amplification from FS_14 to FS_15; 1.4 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_14: (1/10) | 1 |
| FS_15: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 6/7 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 42 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 42 | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
05-07-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_16: (1/10) | 1 |
| FS_01: (1/10) | 1 |
| Phusion Master Mix | 10 |
| dd H2O | 6 |
| DMSO | 1 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 14 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 3:20 min | |
| 16 | 98 | 1 |
| 60 | 5 | |
| 72 | 3:20 min | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
Results:
- Amplification worked well and band was at expected height. Nevertheless the gel band did not run properly on the gel.
Strategy
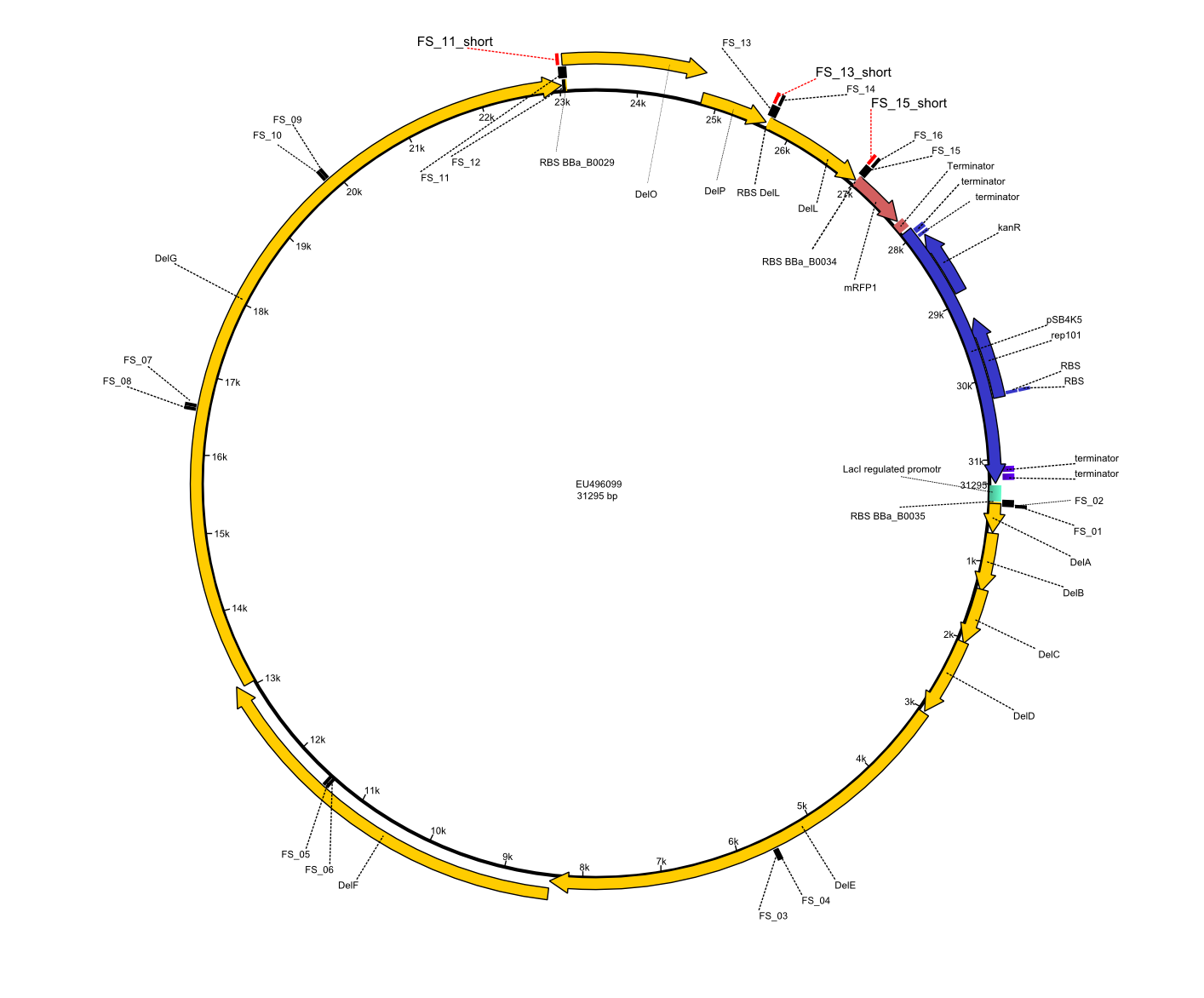
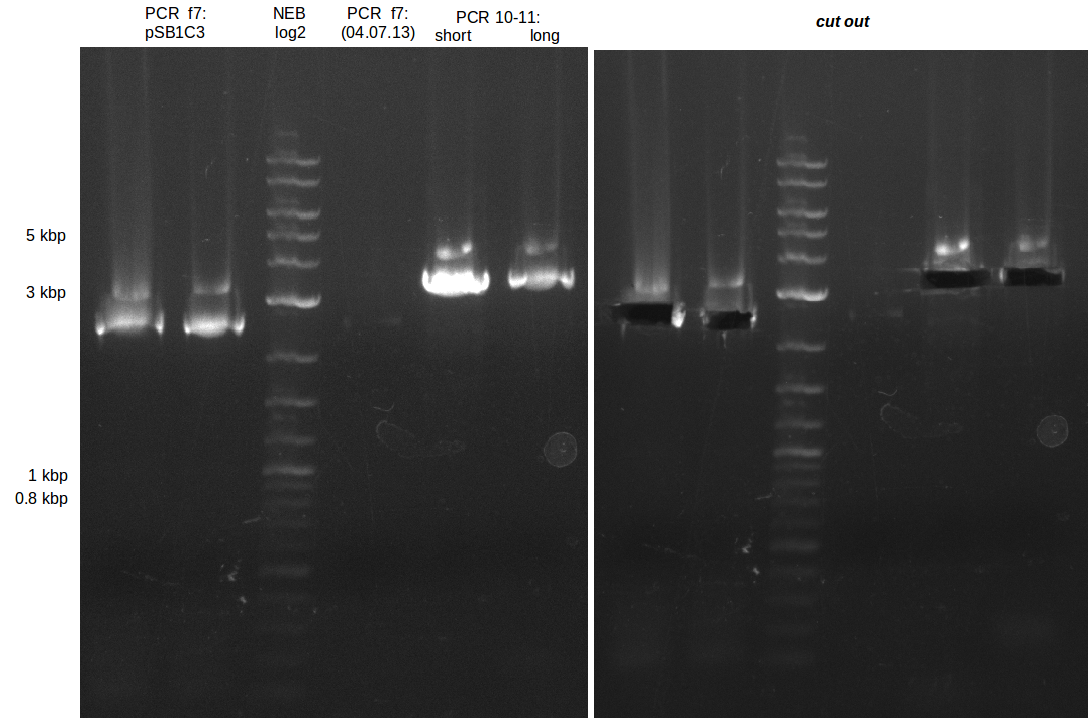
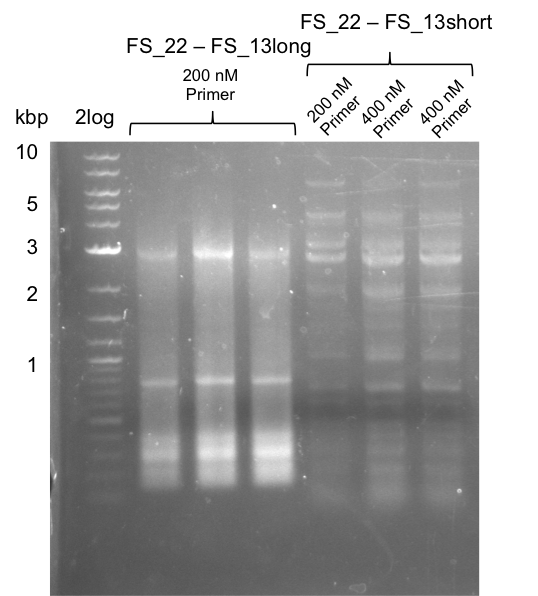
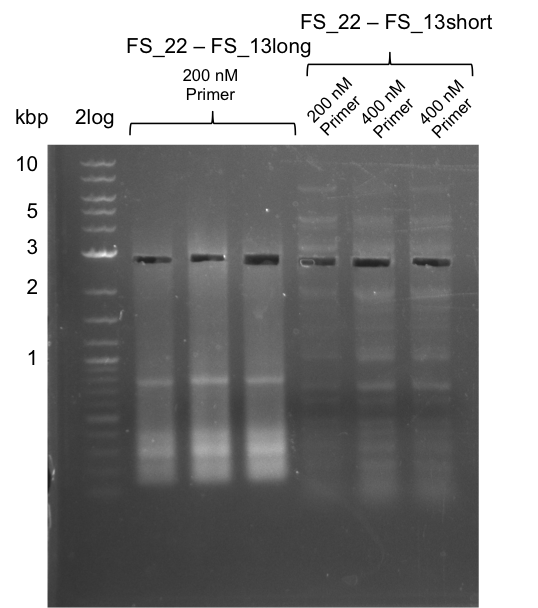
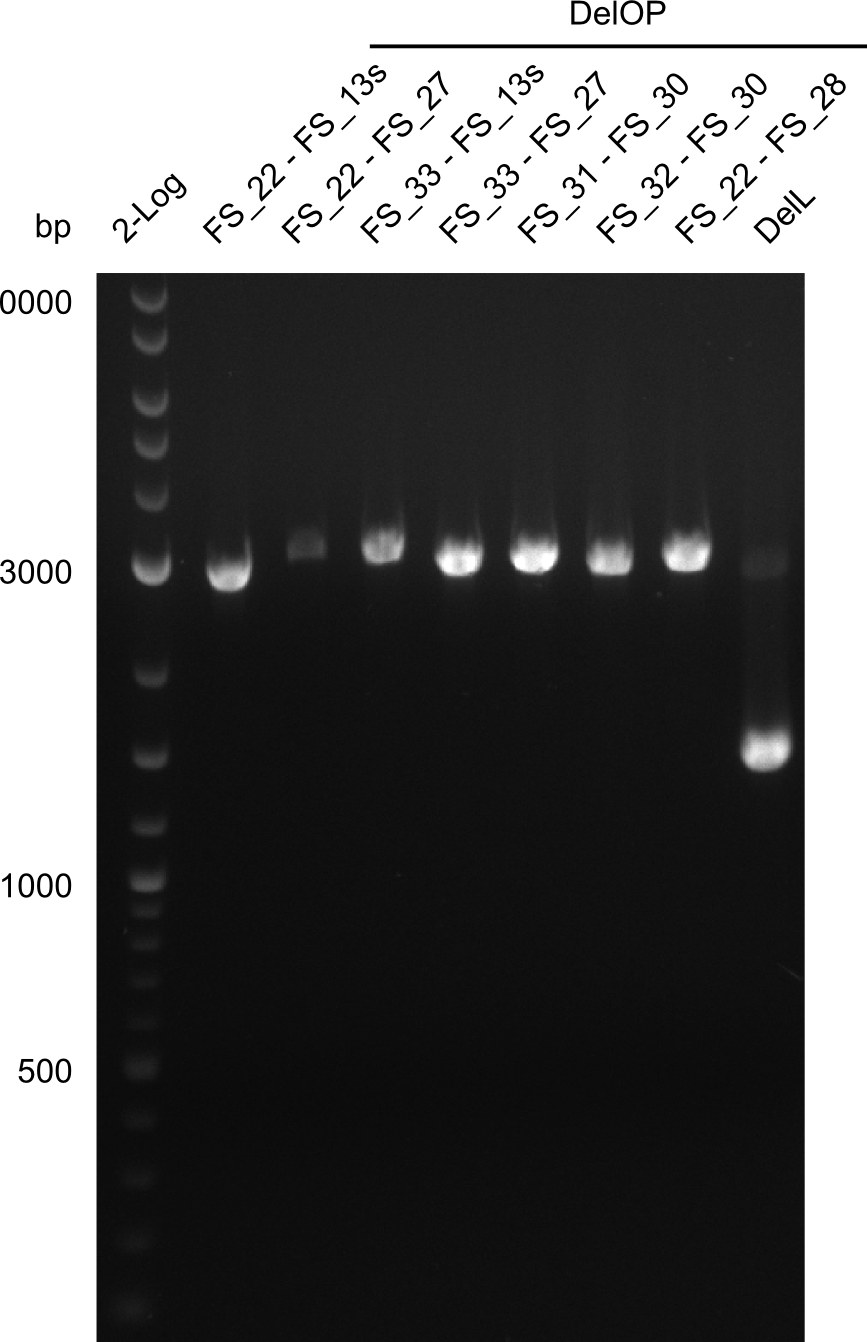
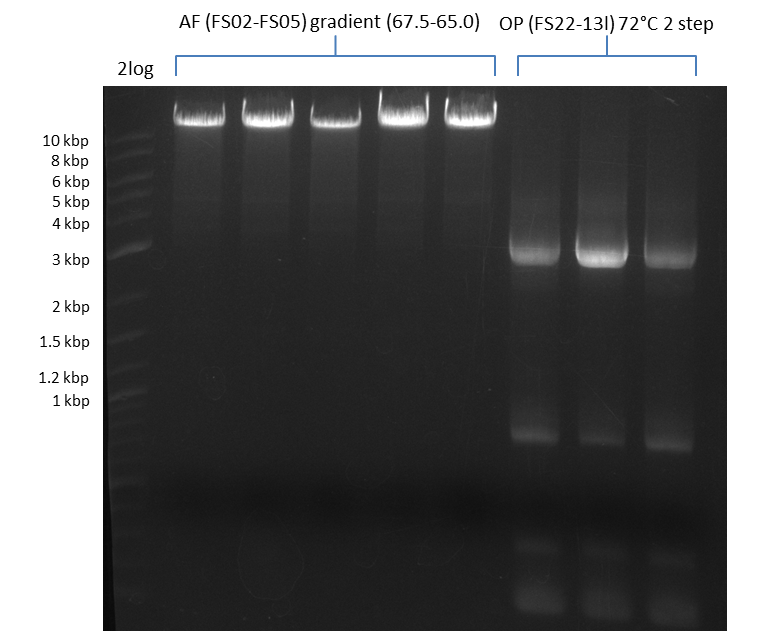
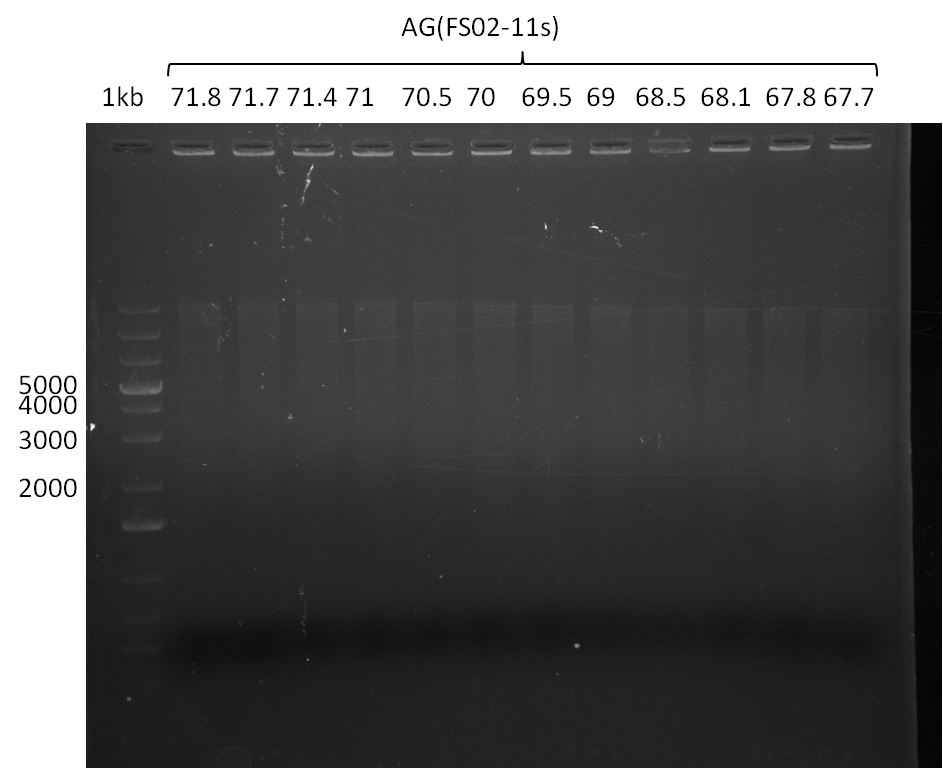
We had various problems with the amplification of the fragments located in theregion DelF-G and DelO-P. For these particular PCRs long primers containing the Gibson overlaps had to be used. As these long primers often form complex secondary structures decreasing annealing efficiency we designed short version of these primers, having the same sequence region complementary to the genomic target, but lacking any longer overhangs. Consequently these primesrs will serve only for amplification of the desired genes from the genome of D. Acidovorans to obtain specific templates. This specific template afterwards will be used to introduce the designated overlap using the long primers again. In total 3 short primer versions FS_11s, FS_13s and FS_15s were ordered.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_11: DelAG_5_short_rv | 2013-05-07 | Amplification of DelAG from Delftia acidovorans. Gibson Primer | TCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_13: DelOP_short_rv | 2013-05-07 | Amplification of DelOP from Delftia acidovorans. Gibson Primer with overhang to DelL | TCACACCGTGGTGTCTGCAGGCG |
| FS_15: DelL_mRFP_pSB4K5__short_rv | 2013-05-07 | Amplification of DelL. Gibson Primer with overhang to BBa_J04450 | TCAGTCCTGCAGCGCCAGCTGTTCTGTG |
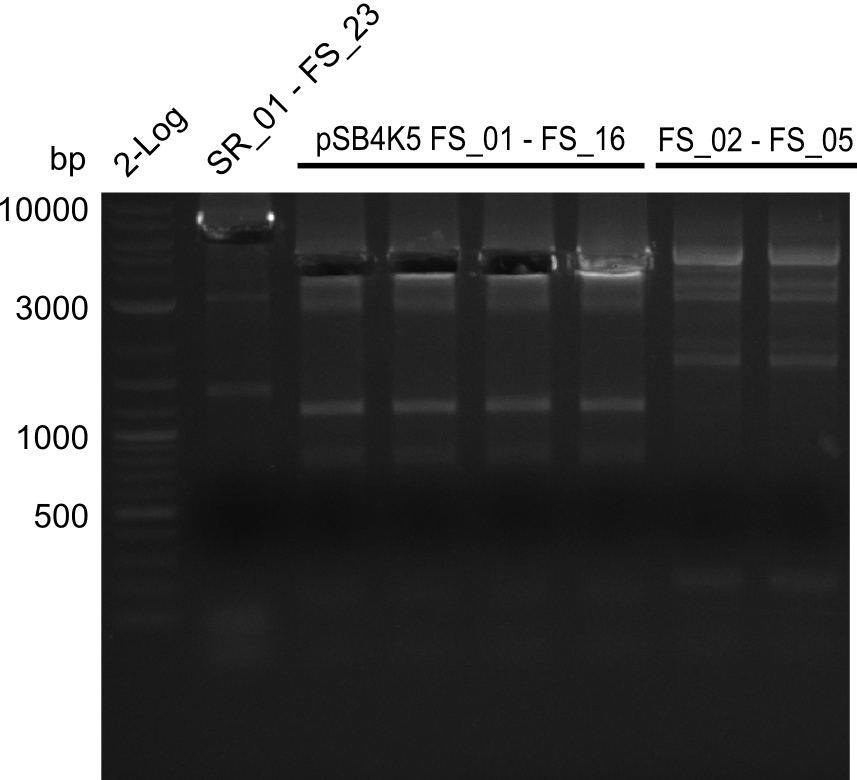
Amplification of Del Genes from DSM-39 genome
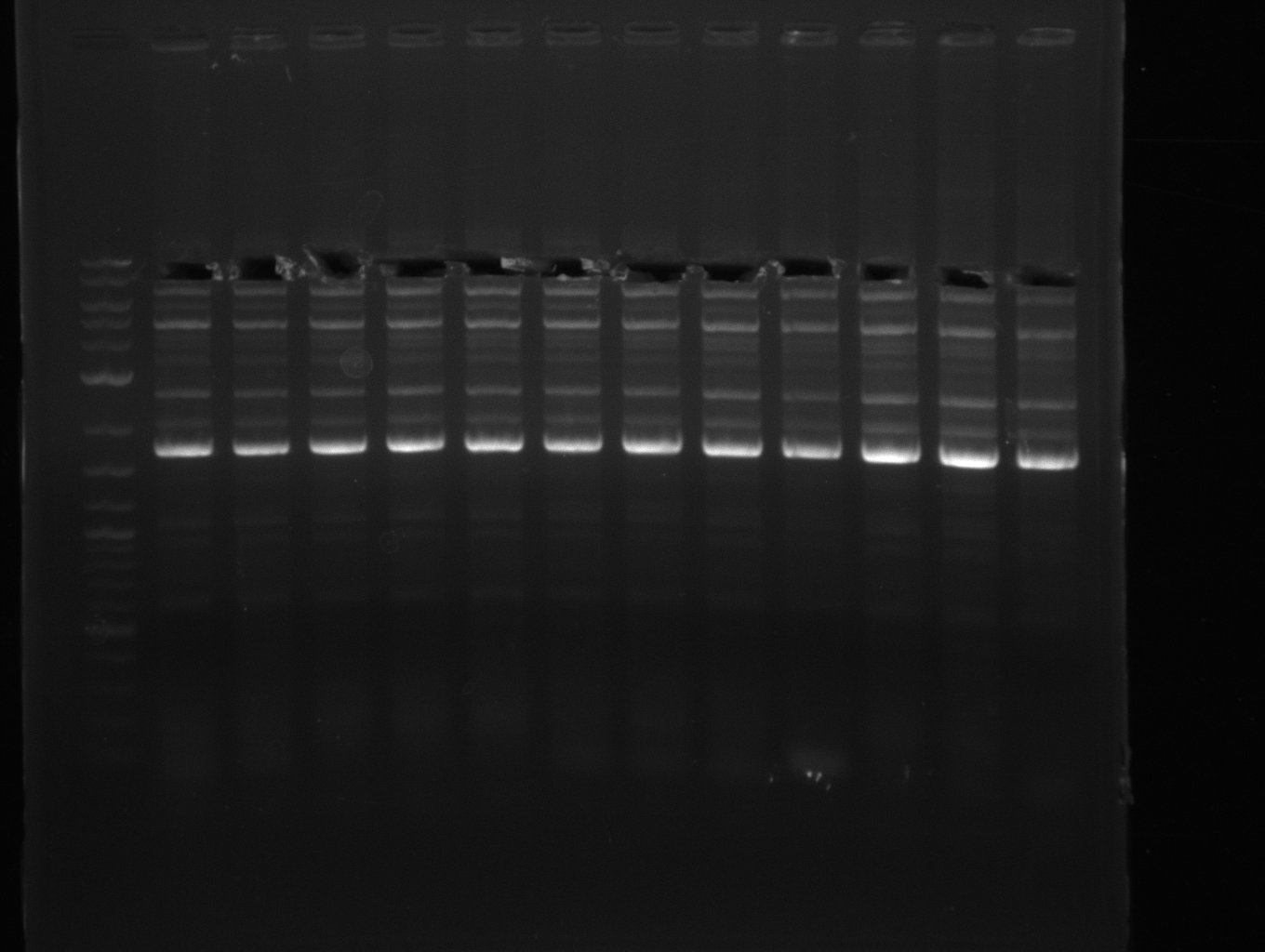
Goals
The PCRs which worked in the previous week will be repeated to obtain higher product yields, as high concentrations of the long amplicons are needed for a successfull Gibson Assembly.
Furthermore we will try to establish PCRs for the obtained short Primers and thereby provide specific templates for the second amplification step using the corresponding primers introducing the required Gibson overlaps.
Amplifications of the sequence region DelF-G will be tried with different protocols to optimize PCR conditions, as we did not obtain any amplicons at all last week. Therefore it is likely that these primers are only able to bind in a very small frame of annealing temperature or even that the used primer combination do not posses a common annealing temperature and therefore amplification with the desired combinations is not possible.
As the amplification of DelO-P also turned out to be difficult, we decided to try a backup strategy which not only includes the desired genes DelO and DelP but also DelL and, since they are located between DelP and DelL, DelM and DelN. These two genes would increase our amplicon size by only 2.2 kb, but we hoped that on the other hand the corresponding primers would work better.
Results
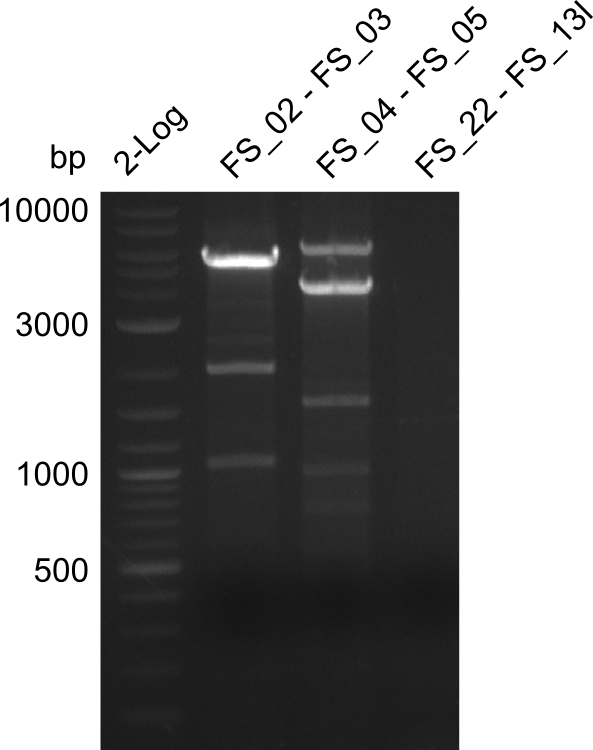
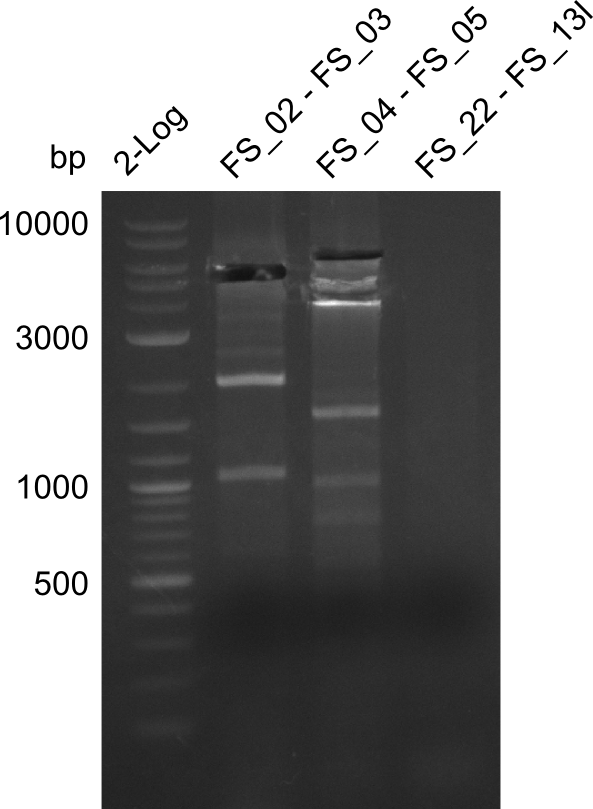
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | FS_02 and FS_03 | ||
| DelA-G | FS_02 and FS_07 | |||
| DelE-G | FS_04 and FS_07 | |||
| FS_04 and FS_11s | ||||
| DelF-G | FS_06 and FS_07 | |||
| FS_06 and FS_09 | ||||
| FS_06 and FS_11 | ||||
| FS_06 and FS_11s | ||||
| DelG | FS_08 and FS_11s | |||
| FS_10 and FS_11 | ||||
| FS_10 and FS_11s | ||||
| DelO DelP DelL | DelO-P | FS_12 and FS_13 | ||
| FS_12 and FS_13s | ||||
| DelL | FS_14 and FS_15 | |||
| DelL-P | FS_13 and FS_15s | |||
| pSB4K5 | pSB4K5 | FS_01 and FS_16 | ||
| Re-PCRs | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelF DelG | DelF-G | Primer FS_06 and FS_09 | ||
| DelO DelP DelL | DelL-P | Primer FS_13_short and FS_15_short | ||
08-07-2013
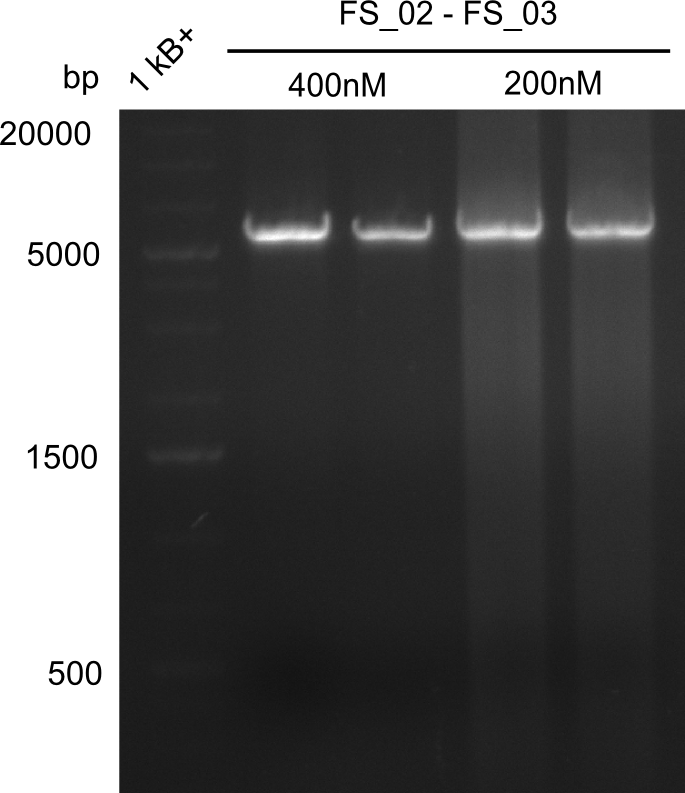
Amplification from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2.5 |
| FS_03: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 19 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAE worked
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
12-07-2013
Amplification from FS_02 to FS_07; 16.4 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2 |
| FS_07: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 5:30 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 5:30 | |
| 1 | 72 | 15min |
| 1 | 12 | inf |
Results:
- only a smear occured, no specific product was amplified
- PCR will be repeated with a lower, constant annealing temperature
09-07-2013
Amplification from FS_04 to FS_07; 11.1 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions I
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Conditions II
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 68 | 5 | |
| 72 | 3 min | |
| 25 | 98 | 1 |
| 72 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelEG did not work
11-07-2013
Amplification from FS_04 to FS_11s; 17.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 1 |
| FS_11_short: (1/10) | 1 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 5:30 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 5:30 | |
| 1 | 72 | 15min |
| 1 | 12 | inf |
Results:
- Amplification of DelEG did not work
- Experiment will be repeated with NEB Phusion II Polymerase as Phusion II is not provided as mastermix and therefore GC-buffer can be used
Amplification from FS_04 to FS_11; 17.5 kb; Phusion II
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 1 |
| FS_11_short: (1/10) | 1 |
| Phusion II | 0.2 |
| DNTP | 0.4 |
| Buffer | 4 |
| DMSO | 0.6 |
| dd H2O | 11.8 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 30 |
| 12 | 98 | 5 |
| 68 ↓ 0.5 | 30 | |
| 72 | 8:10 | |
| 18 | 98 | 5 |
| 66 | 30 | |
| 72 | 8:10 | |
| 1 | 72 | 15min |
| 1 | 17 | inf |
Results:
- Amplification of DelEG did not work
- it seems not to be possible to amplify the desired 17 kbp fragent with the chosen primers and the given template, primercombination will be changed in further amplification attempts
14-07-2013
Amplification from FS_04 to FS_07; 11.1 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 2 |
| FS_07: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
Cycler incubation room right
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Conditions II
Cycler incubation room left
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelEG worked with a touchdown PCR starting from 70°C annealing temperature
- band was cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be repeated to increase the amount of DNA and gather the concentrations necessary for Gibson Assembly
09-07-2013
Amplification from FS_08 to FS_11_short; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08 (1/10) | 1 |
| FS_11_short (1/10) | 1 |
| Phusion Master Mix | 10 |
| dd H2O | 6 |
| DMSO | 1 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 24 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 10min |
| 1 | 4 | inf |
Results:
- A weak band was visible at the right height.
- Band was cut out and DNA purified using QIAquick Gel Extration Kit.
- Concentration after gel extration was too low
- Maybe increasing the temperature further will result in higher yield.
13-07-2013
Amplification from FS_10 to FS_11s; 3.3 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_10: (1/10) | 2 |
| FS_11_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 4 |
| DMSO | 1 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 70 | 5 | |
| 72 | 1:10 min | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Results:
- No band was visible on the gel.
- The PCR conditions of the 09-07-2013 should be further optimized.
Amplification from FS_10 to FS_11(s); 3.3 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_10: (1/10) | 2 |
| FS_11 (short or long): (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 4 |
| DMSO | 1 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:10 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:10 min | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Results:
- Bright bands were visible in both, the PCR with the short and the long primer.
- The PCR with the short primers worked better than the one with the long primer.
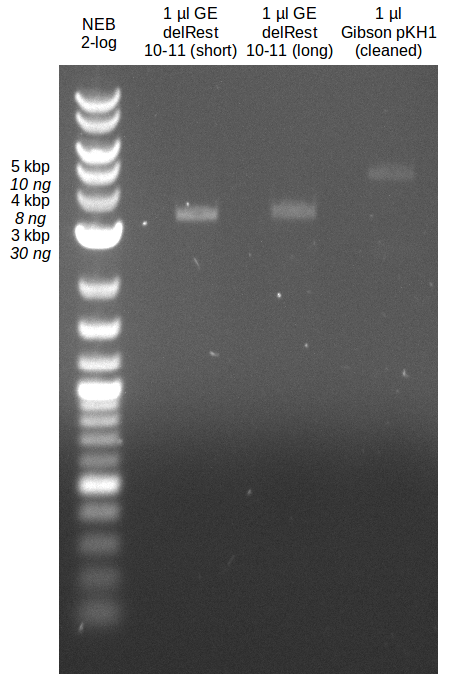
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
- Concentration after gel extraction with primer FS_11short=6ng/µl in 18µl
- Concentration after gel extraction with primer FS_11long=4ng/µl in 18µl
11-07-2013
Amplification from FS_13s to FS_15s; 6.4 kb
- Reaction of DelLP
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_13_short: (1/10) | 1 |
| FS_15_short: (1/10) | 1 |
| Phusion flash Master Mix | 10 |
| dd H2O | 6 |
| DMSO | 1 |
- Conditions of Del LP
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- There was a very weak band visible on the right height.
- The band was cut out anyway, as we wanted to use it as template for a Re-PCR
- The DNA was purified using QIAquick Gel Extration Kit.
12-07-2013
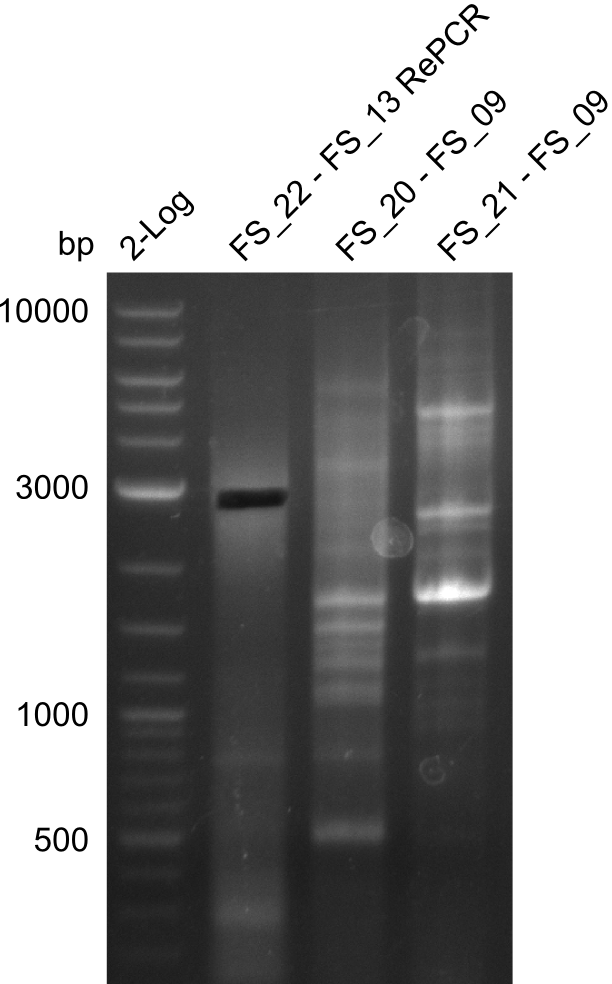
Re-PCR of DelLP (FS_13s to FS_15s; 6.4 kb; 11-07-2013)
- Reaction of DelLP
| what | µl |
|---|---|
| Gel extracted fragment LP (11-07-2013) | 2 |
| FS_13_short: (1/10) | 1 |
| FS_15_long: (1/10) | 1 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
| DMSO | 1 |
- Conditions of Del LP
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- The Re-PCR did not work, no bands were visible on the gel.
09-07-2013
Amplification I from FS_12 to FS_13; 2.7 kb
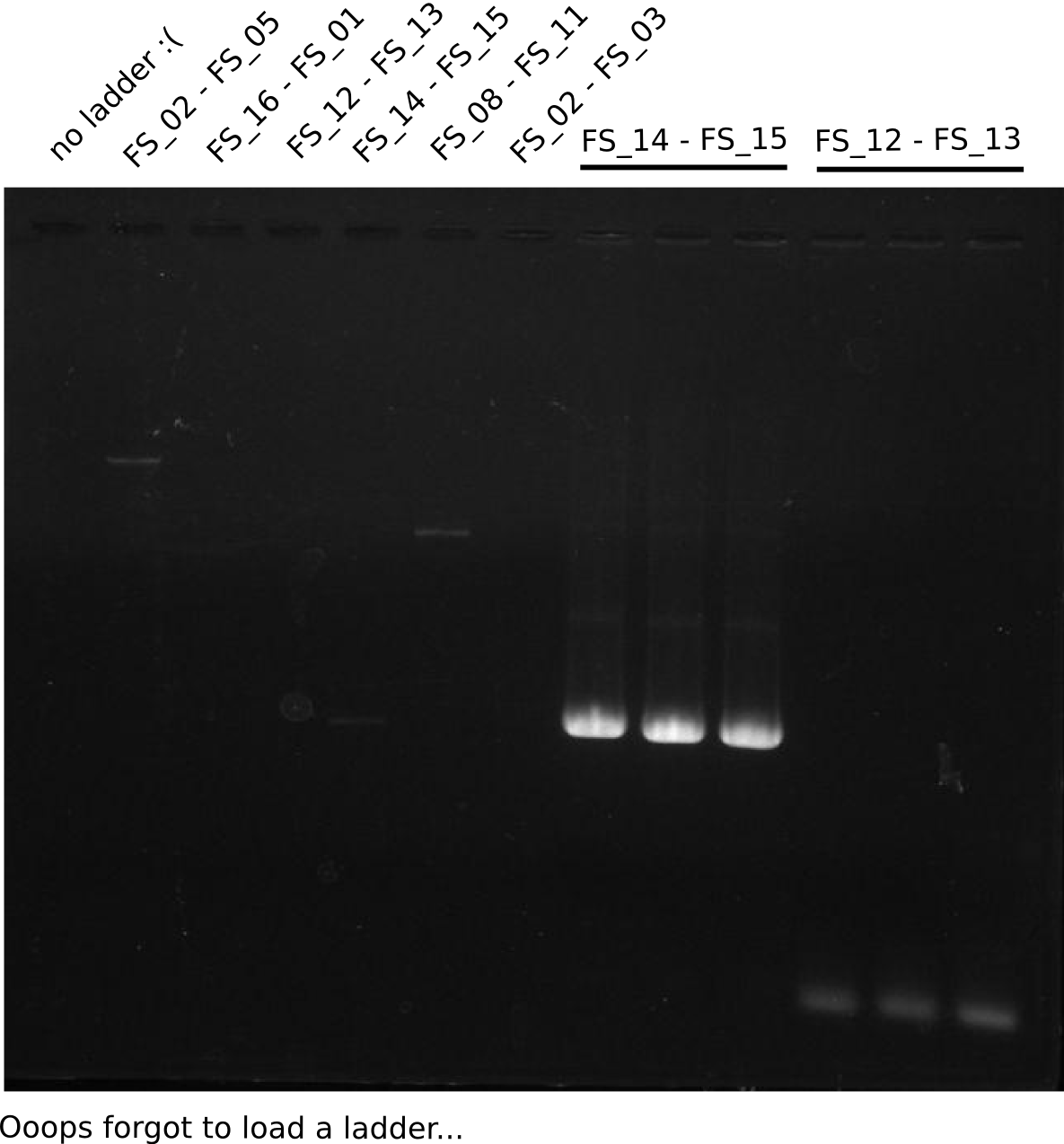
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_12: (1/10) | 1 |
| FS_13: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 40 | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP did not work
- PCR was repeated, Annealing was carried out as touchdown starting from 68°C
Amplification II from FS_12 to FS_13; 2.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_12: (1/10) | 1 |
| FS_13short: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 6 |
| 8 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 50 | |
| 24 | 98 | 1 |
| 72 | 5 | |
| 72 | 50 | |
| 1 | 72 | 11min |
| 1 | 4 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated with the newly ordered short version of primer FS_13, testing different DMSO settings
10-07-2013
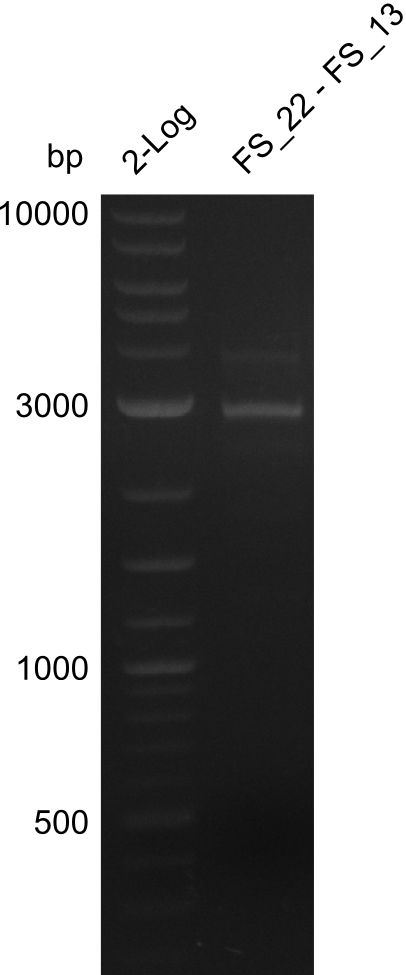
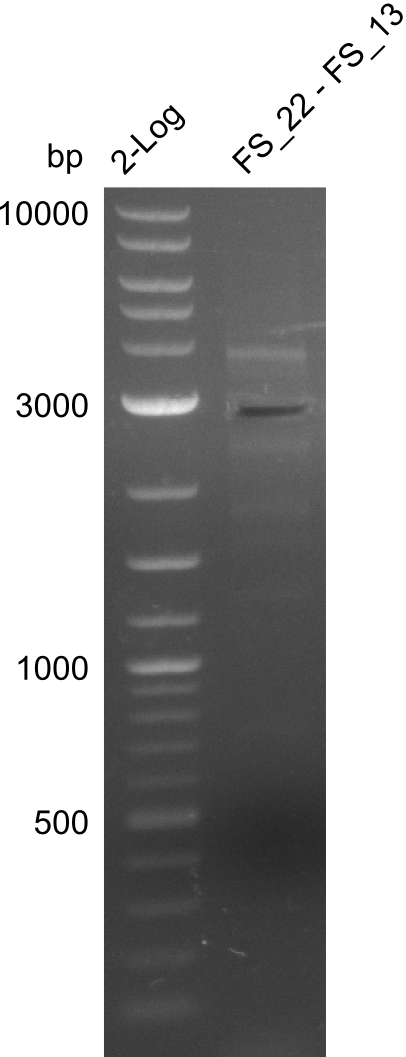
Amplification from FS_12 to FS_13(s); 2.7 kb
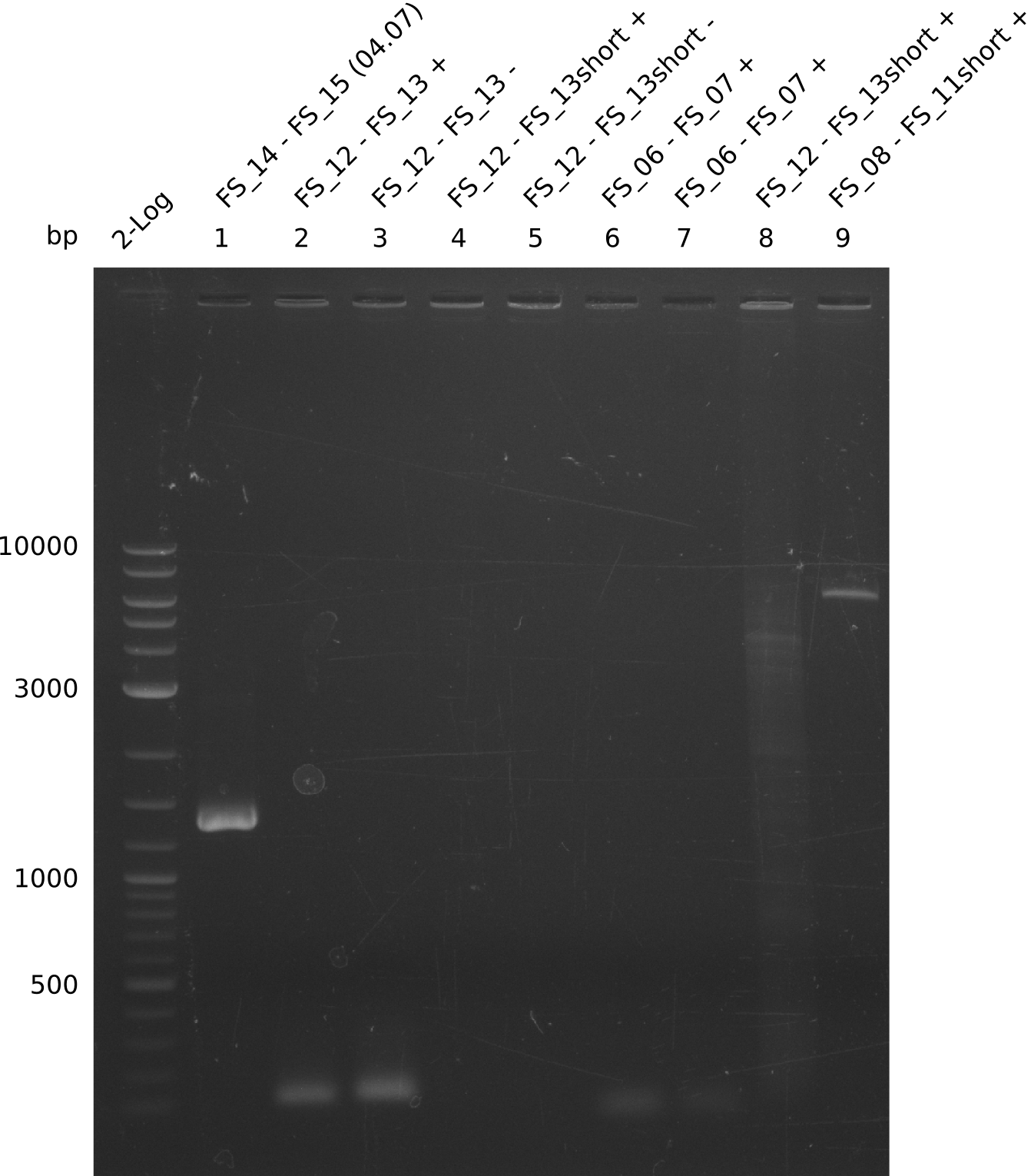
- Reaction of DelO-P (2.6 kb)
4 reactions with different conditions: with/without DMSO, short/long Primer FS13
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_12: (1/10) | 1 |
| FS_13(long or short): (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 6/7 |
- Conditions of Del O-P
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 40 | |
| 1 | 72 | 420 |
| 1 | 12 | inf |
Results:
- None of the Amplification neither with primer FS_13 nor with FS_13short worked out, nevertheless did addition of DMSO change the amplification result
- PCR will be carried out as 2-step in another more precise cycler
- PCR will be set up with as touchdown PCR, annealing starting at 68°C
- Furthermore a modified version of the forward primer will be order. This primer (FS_22) includes a recently in the Del Cluster predicted promotor as well as a likewise predicted ribosome binding site
08-07-2013
Amplification from FS_14 to FS_15; 1.4 kb

- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_14: (1/10) | 2.5 |
| FS_15: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 42 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 42 | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
08-07-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(2x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 14 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 3:20 min | |
| 16 | 98 | 1 |
| 60 | 5 | |
| 72 | 3:20 min | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
10-07-2013
Amplification I from FS_06 to FS_07; 5.2 kb

- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 6/7 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:10 min | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:10 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification did not work, not product is visible
- PCR will be repeated with lower annealing temperature to increase primer binding
Amplification II from FS_06 to FS_07; 5.2 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work
- PCR will be repeated with higher annealing temperature as reaction might not have worked due to secondary structures of primers
Amplification III from FS_06 to FS_07; 5.2 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 60 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 58 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
11-07-2013
Amplification from FS_06 to FS_07; 5.2 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 1 |
| FS_07: (1/10) | 1 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 6 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work
- other primers will be designed in order to amplify the desired sequence from D. acidovorans
12-07-2013
Amplification from FS_06 to FS_11(s); 11.6 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 2 |
| FS_11 (long or short): (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
Attention: 4 cycles were accidently carried out with an elongation time of 3:00 min
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 11 | 98 | 1 |
| 73 ↓ 0.5 | 5 | |
| 72 | 5:00 | |
| 19 | 98 | 1 |
| 68 | 5 | |
| 72 | 5:00 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, neither with the short nor with the long version of FS_11, consequently PCR will be repeated with different primers
13-07-2013
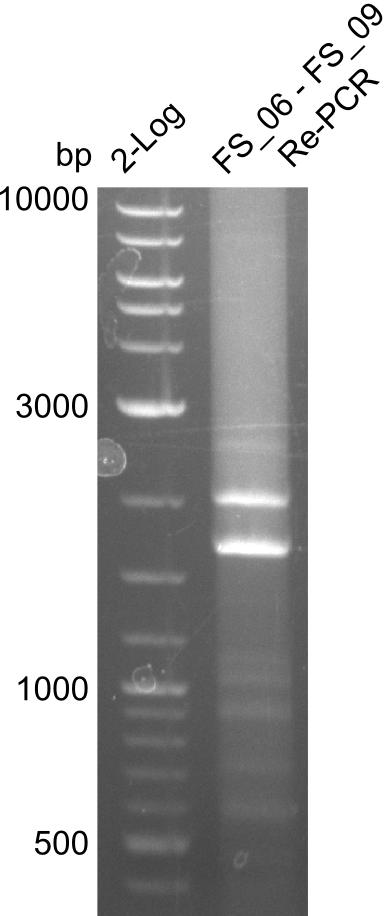
Amplification from FS_06 to FS_09; 8.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
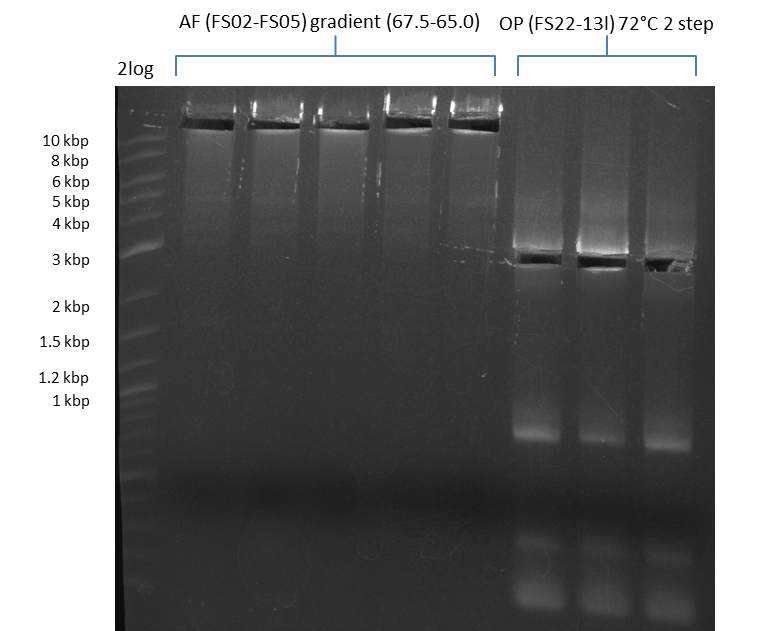
a gradient PCR combined with touchdown was carried out. 12 wells, gradient in the annealing temperature from 74°C - 66°C, resulting in a gradient of 73°C - 65°C in the constant program
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 74 ↓ 0.5 to 65 ↓ 0.5 | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 73 to 65 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
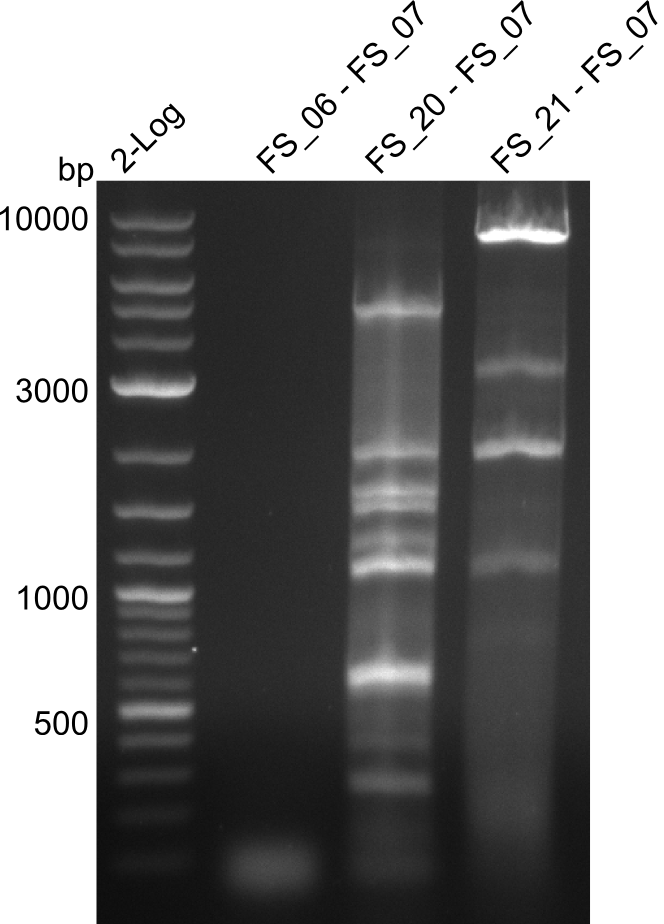
- Amplification of DelFG worked, though several other bands occured, indicating low primer specifity
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- final concenctration after QIAquick Nucleotide Removal Kit: 5ng/µl in 18µl
14-07-2013
Re-Amplification from FS_06 to FS_09; 8.5 kb; 13-07-2013
- Reaction
| what | µl |
|---|---|
| Template of gel extraction (13-07-2013) | 1 |
| FS_06: (1/10) | 5 |
| FS_09: (1/10) | 5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 11.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 71 ↓ 0.5 | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 70 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, no product was detectable
Strategy

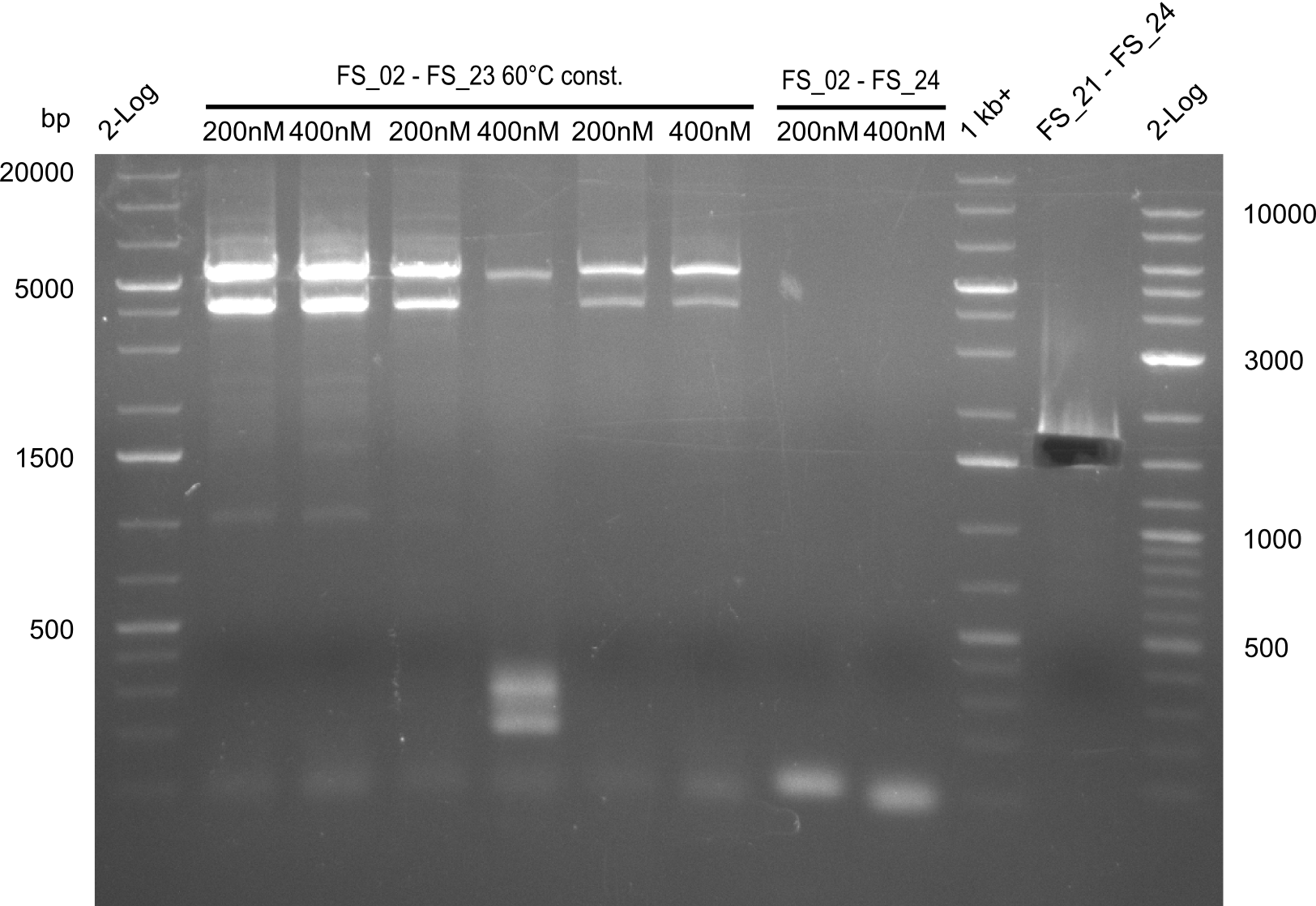
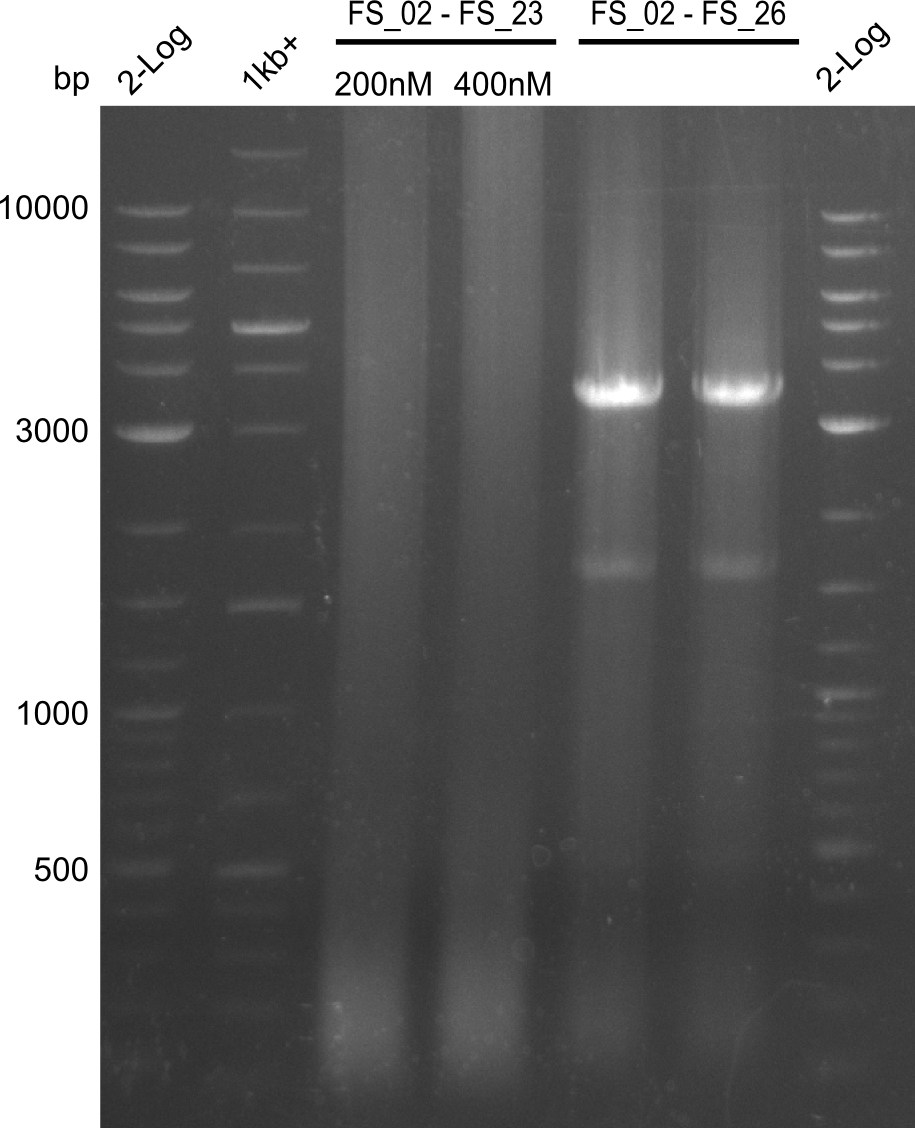
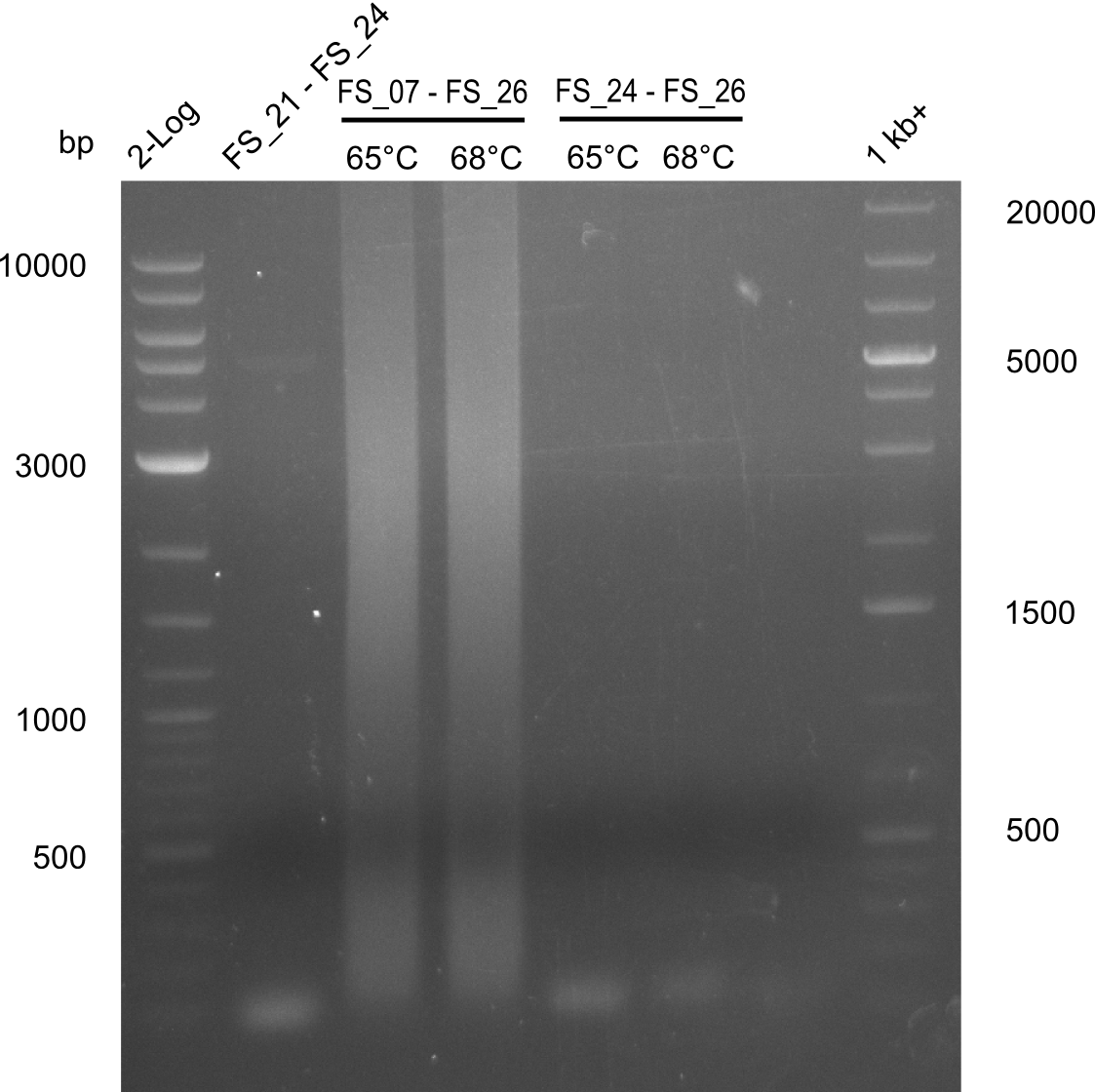
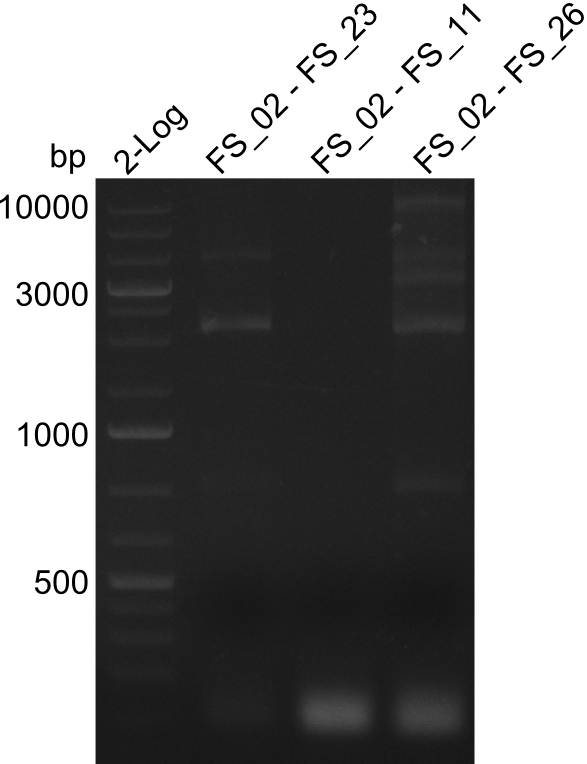
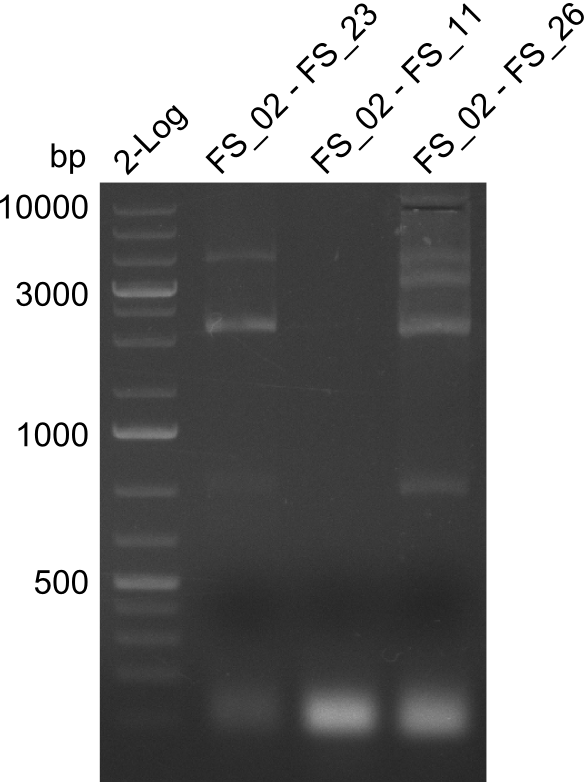
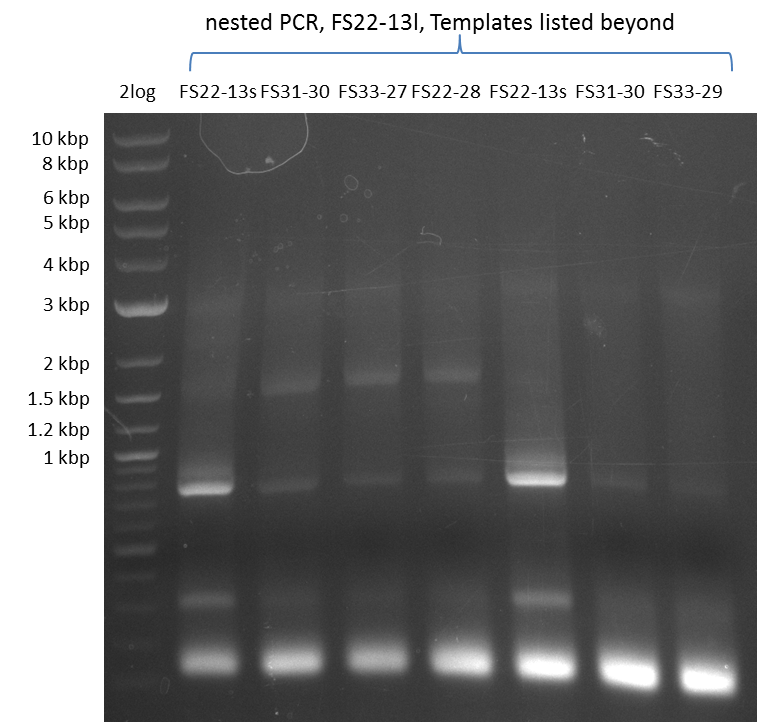
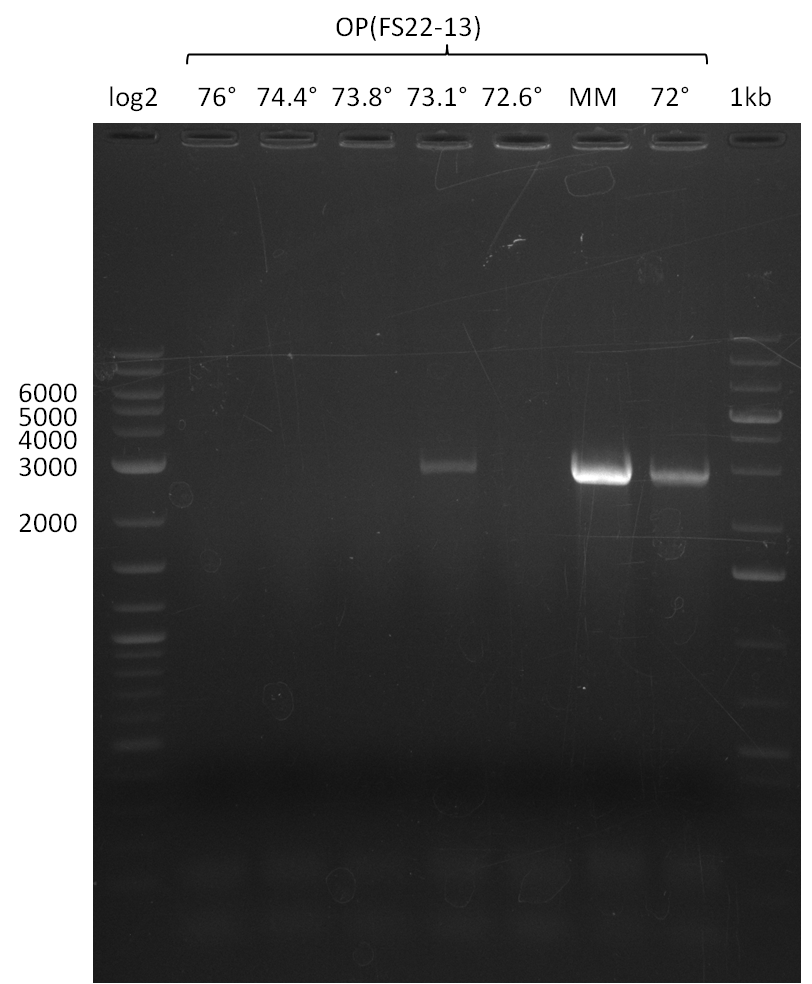
As we discovered that there is a predicted promoter located right in front of DelO, we decided to include this promotor into our construct. This should ensure that the genes DelO, DelP and DelL are still expressed in the case that there is a terminator by the end of DelG. For this we designed primer FS_22 which binds right at the predicted promoter. However, this also meant, that we had to change the reverse primer for the DelG fragment in order to enable Gibson assembly of these fragments next to each oterh. Furthermore we created new primers for amplifying DelE, DelF and DelG as we were not successful in amplifying these fragments so far. These primers now offer more potential fragment combinations which can be tested. We hope to get the missing fragments with this new strategy.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
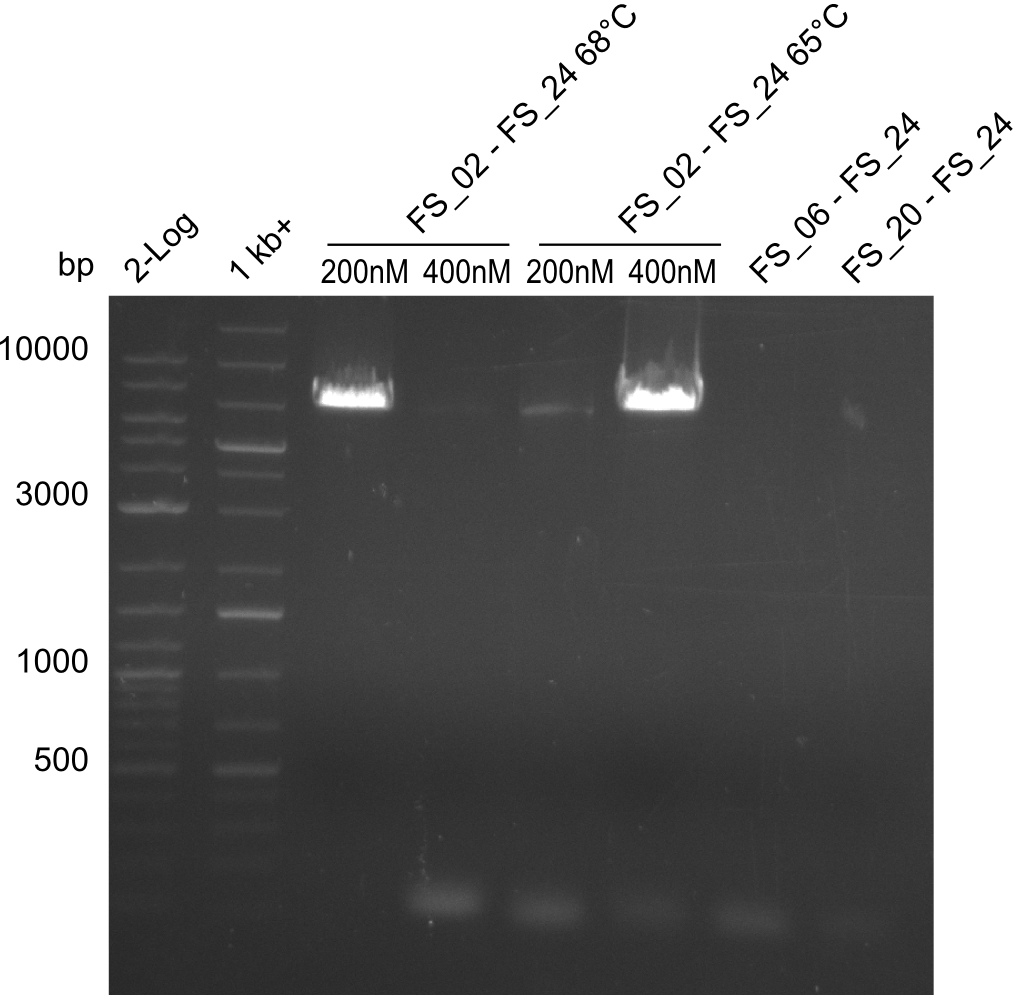
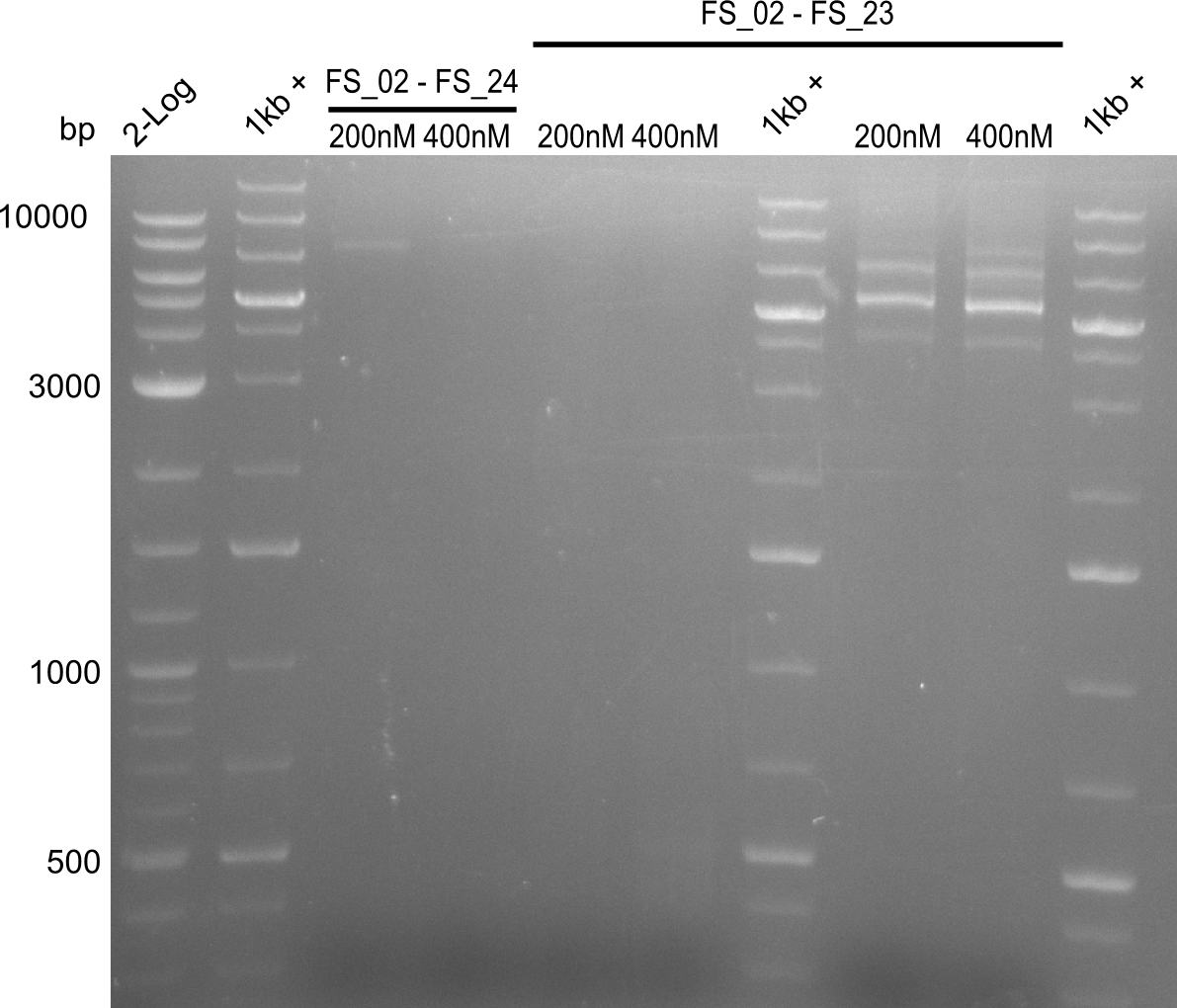
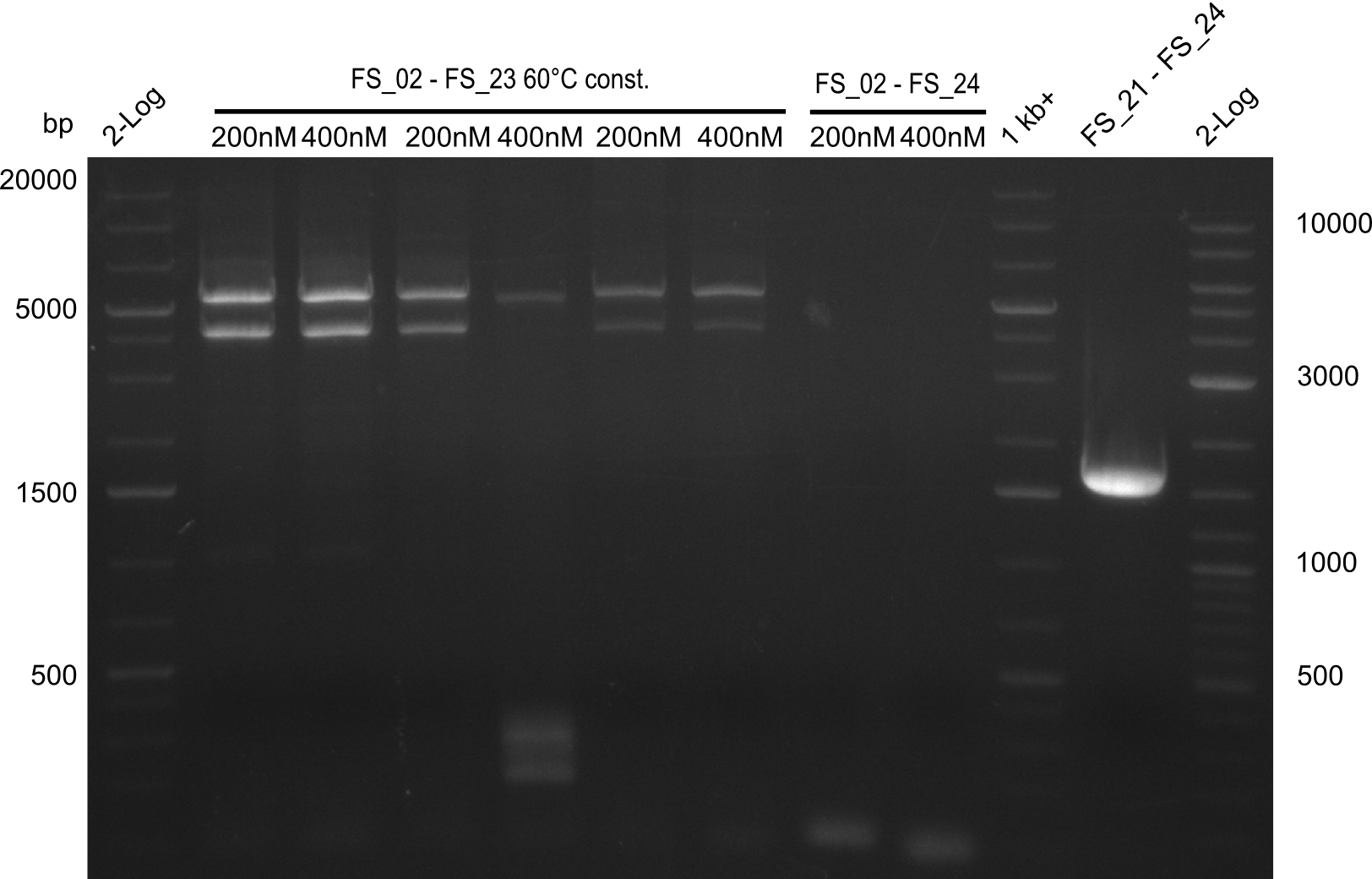
| FS_21: DelF_fw | 2013-07-13 | Amplification of DelF from Delftia acidovorans Gibson Primer | GACGCCATCTACCACCTGTG |
| FS_22: DelOP_short_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans inlcuding the recently predicted endogenous Promotor for DelOP Gibson Primer | GATGACGCAGGGCGGCGGAATTTGTTCATC |
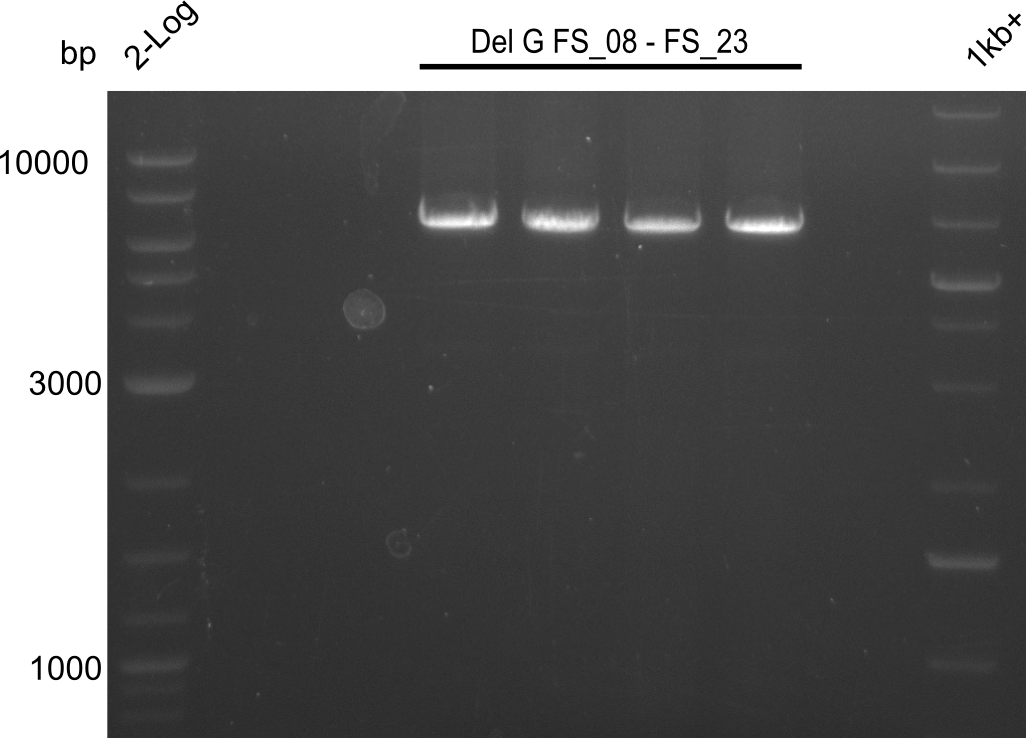
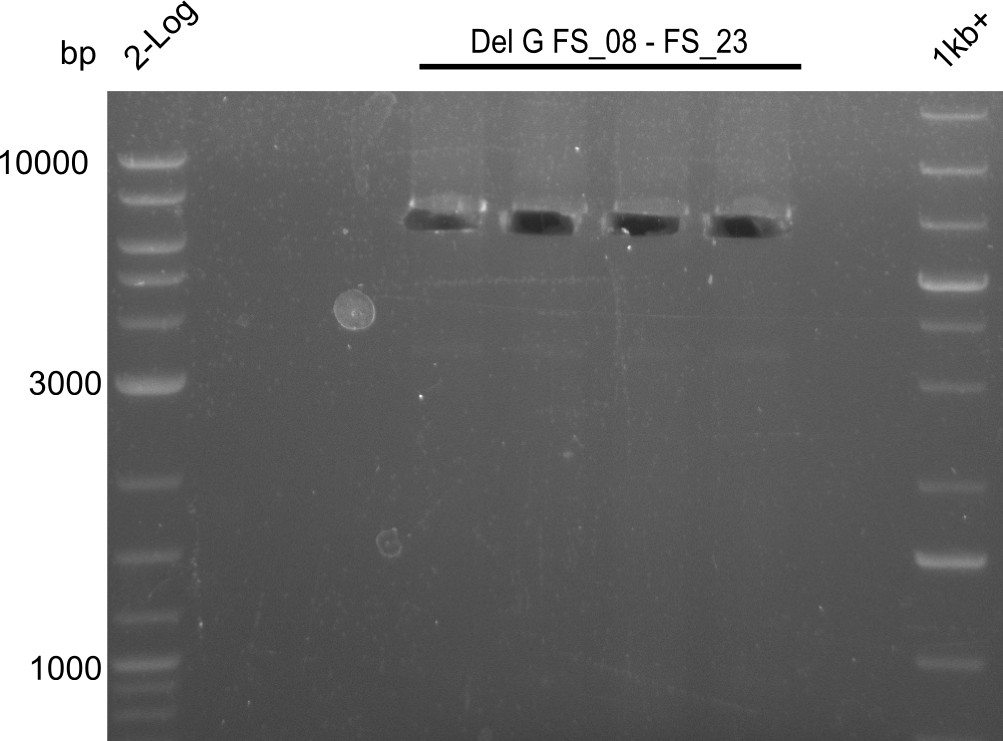
| FS_23: DelG_long_rv | 2013-07-13 | Amplification of DelG from Delftia acidovorans Gibson primer with overhang to DelOP element including the recently predicted endogenous Promotor | GATGAACAAATTCCGCCGCCCTGCGTCATCTCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_24: DelAE_rv | 2013-07-13 | Amplification of DelAE from Delftia acidovorans Gibson Primer | CAGAAGAATTCCCAGAAGGAGATGTCGAAG |
| FS_26: DelFG_rv | 2013-07-13 | Amplification of DelFG from Delftia acidovorans Gibson Primer | GAATTCATCCACGATGATCTGCATG |
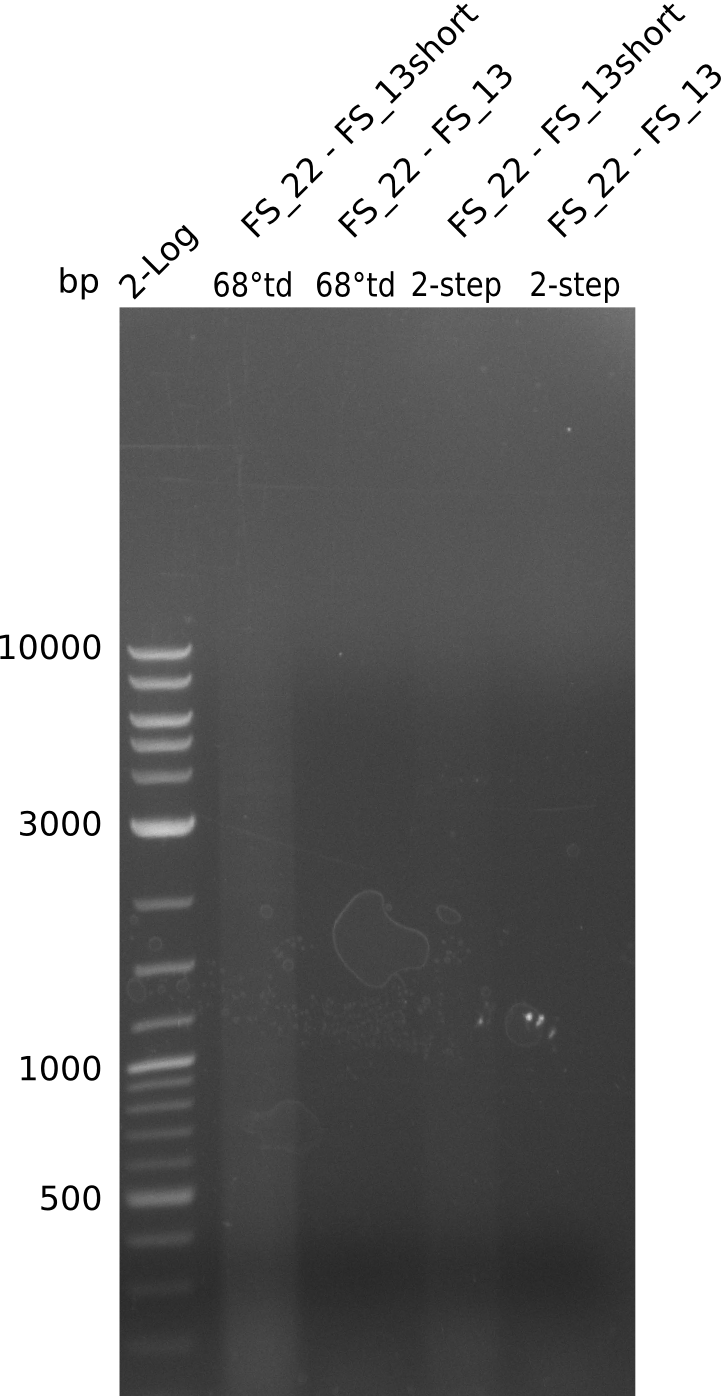
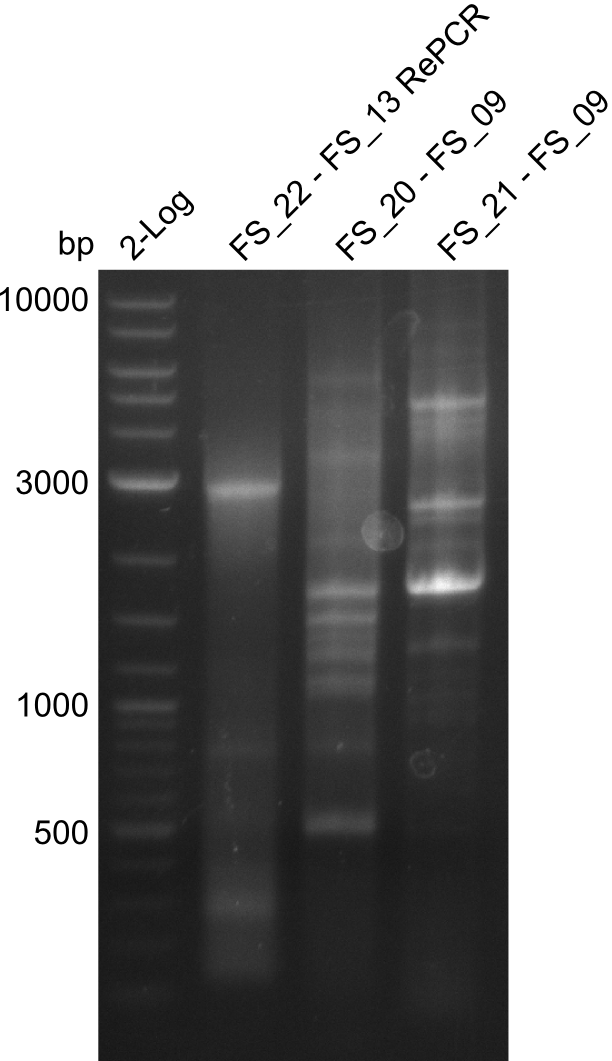
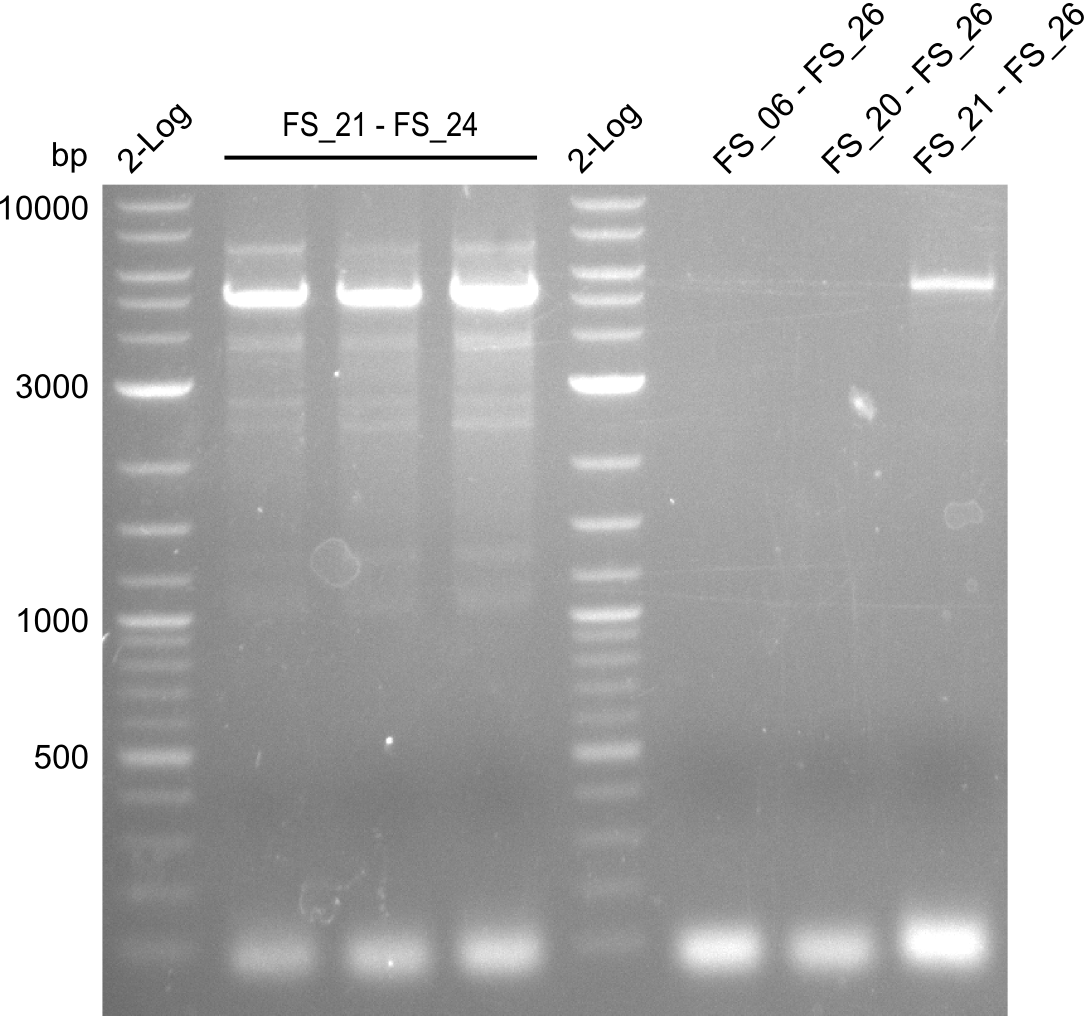
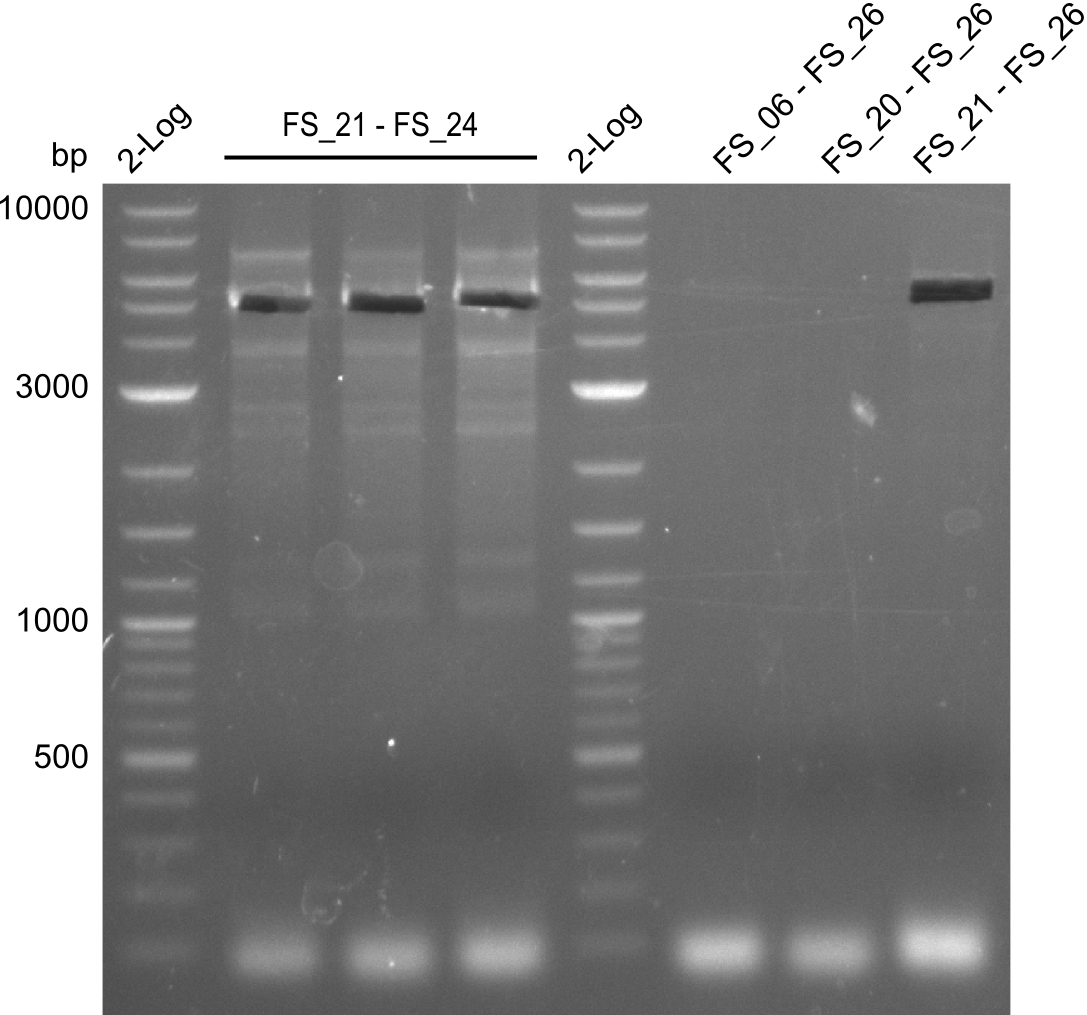
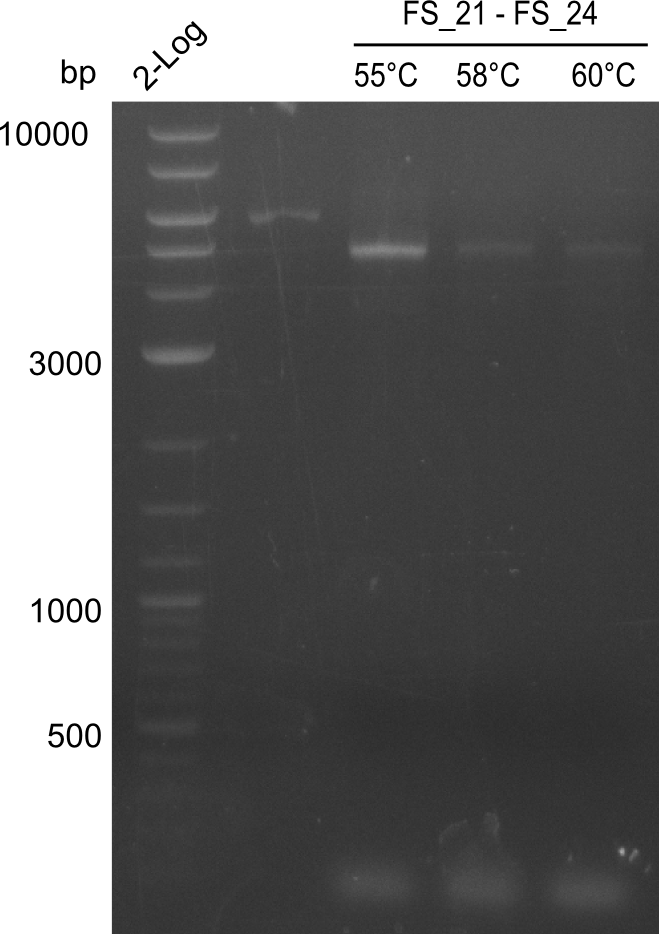
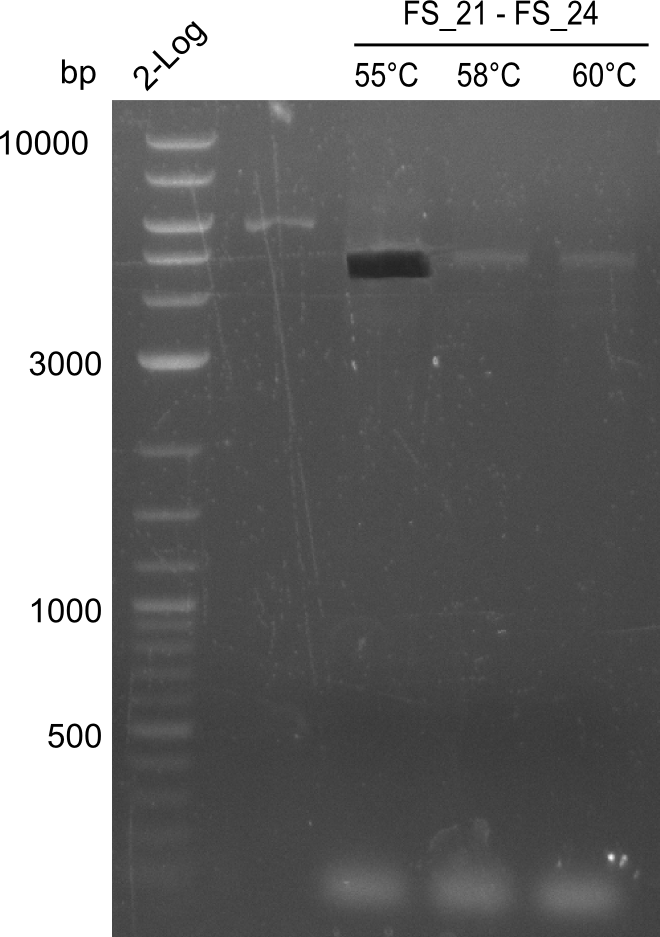
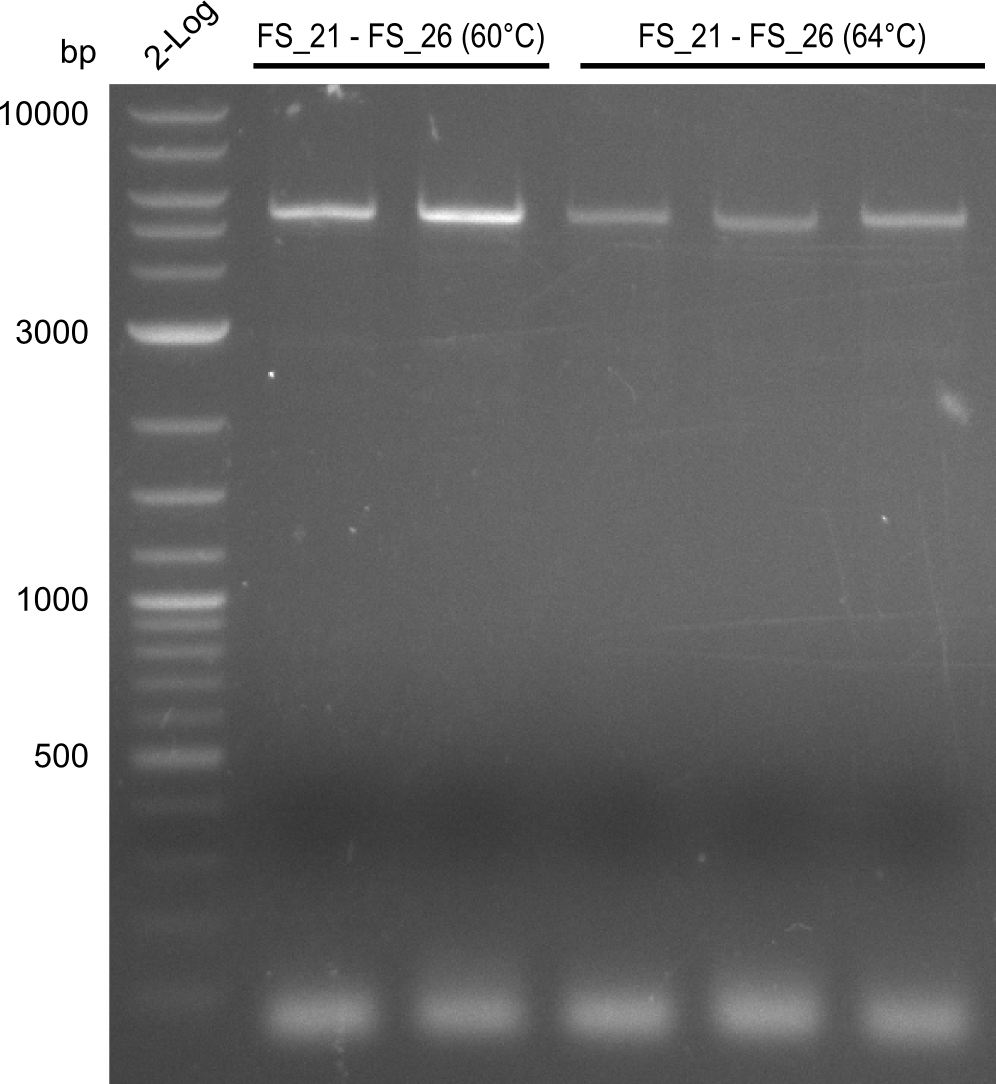
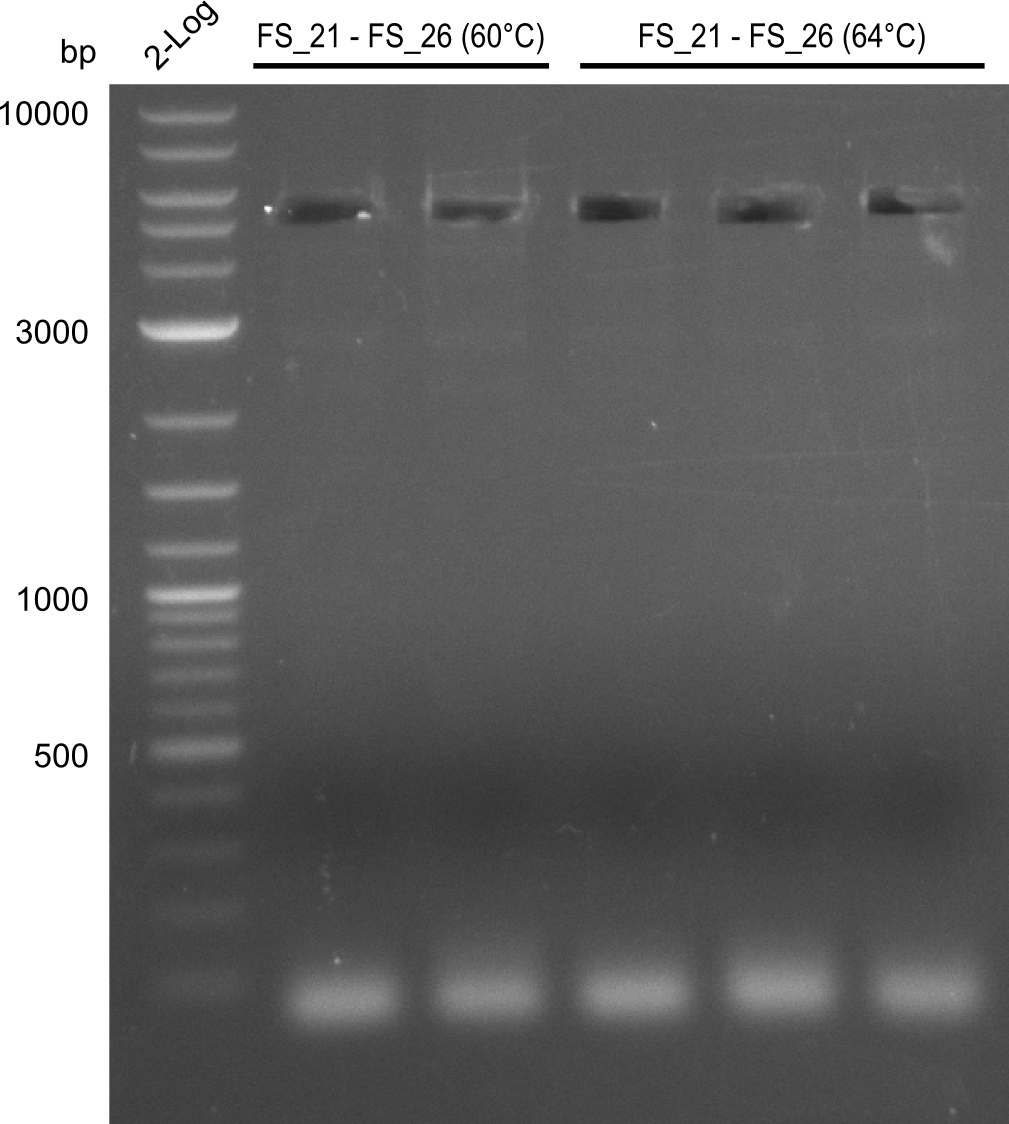
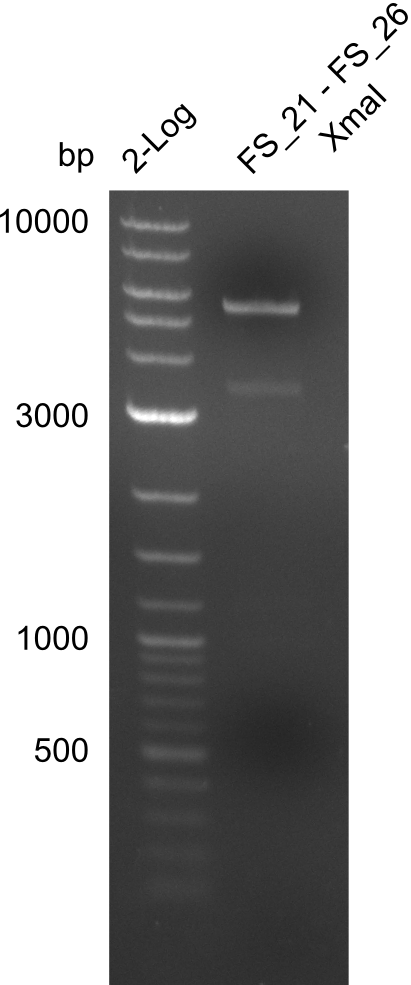
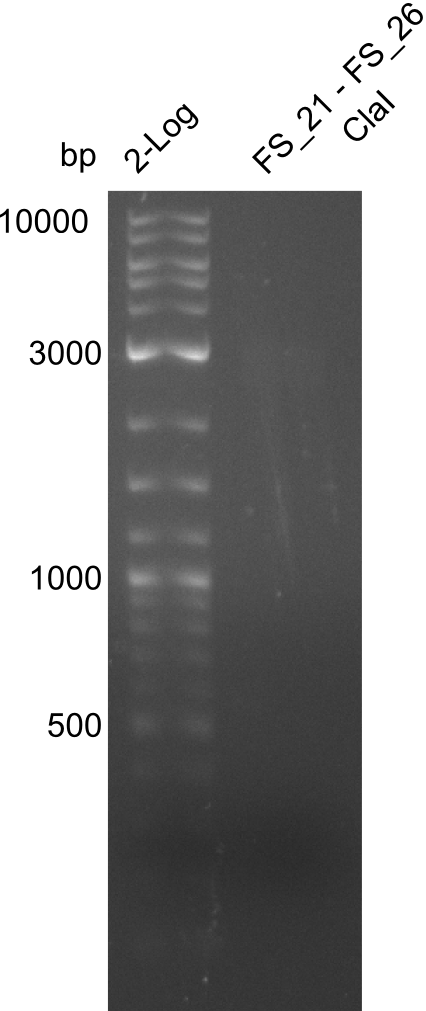
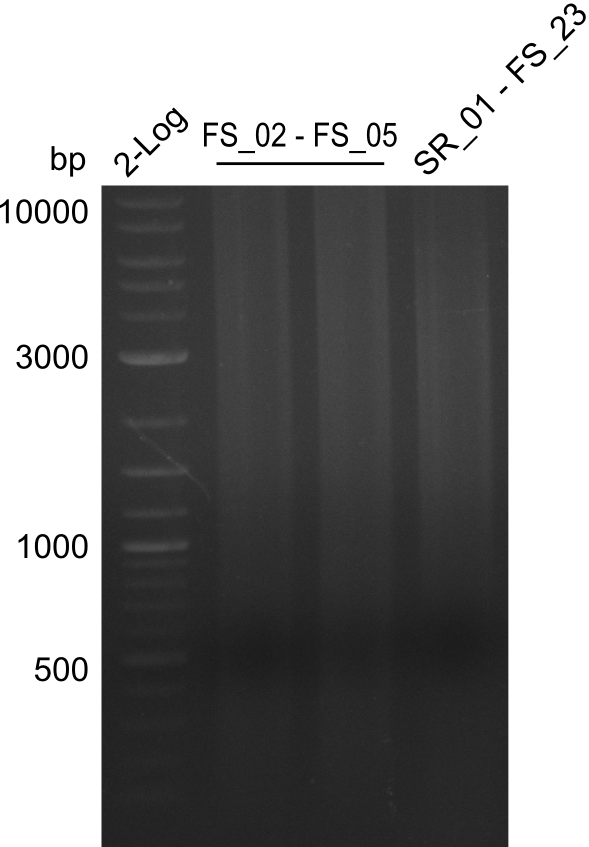
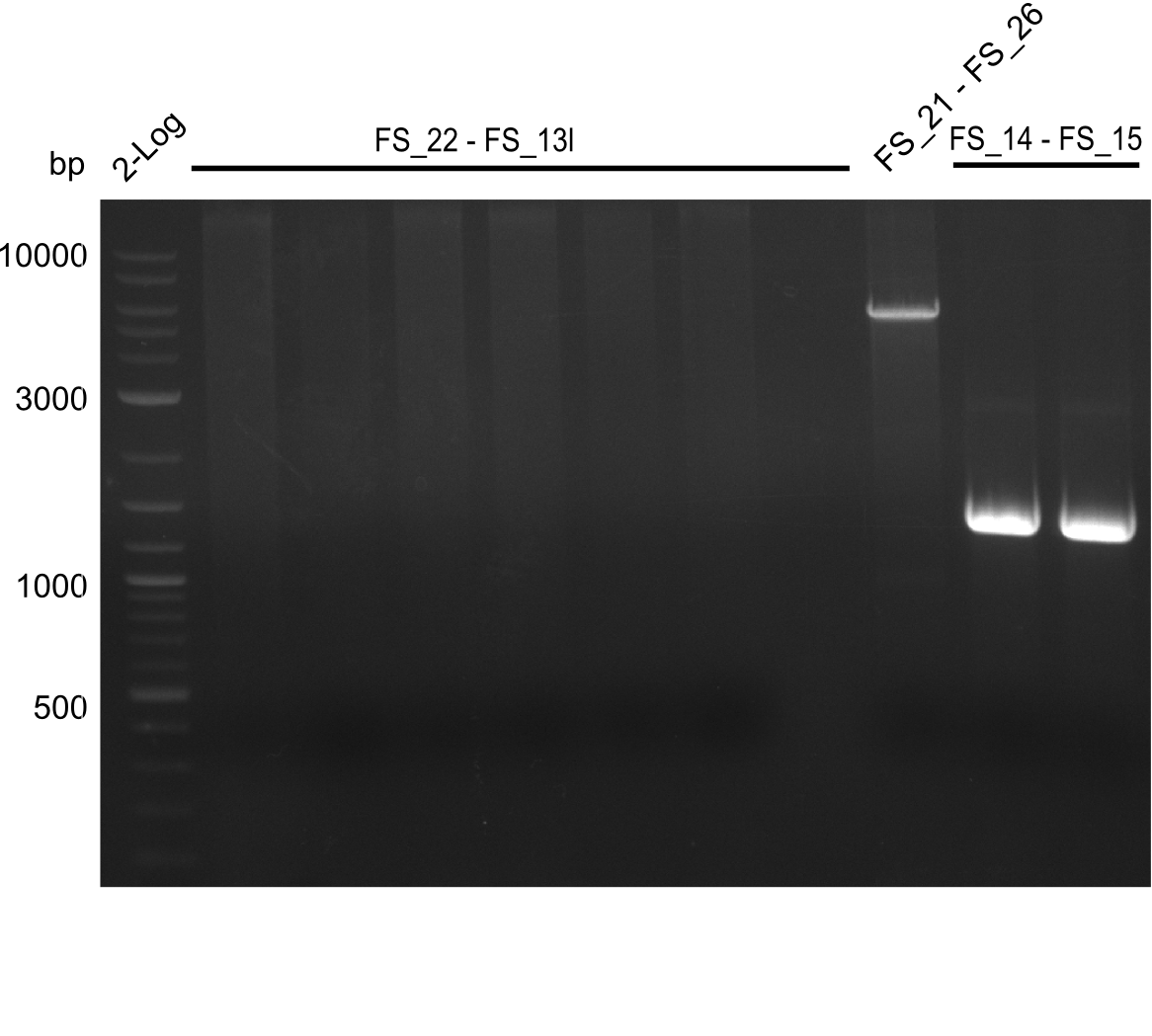
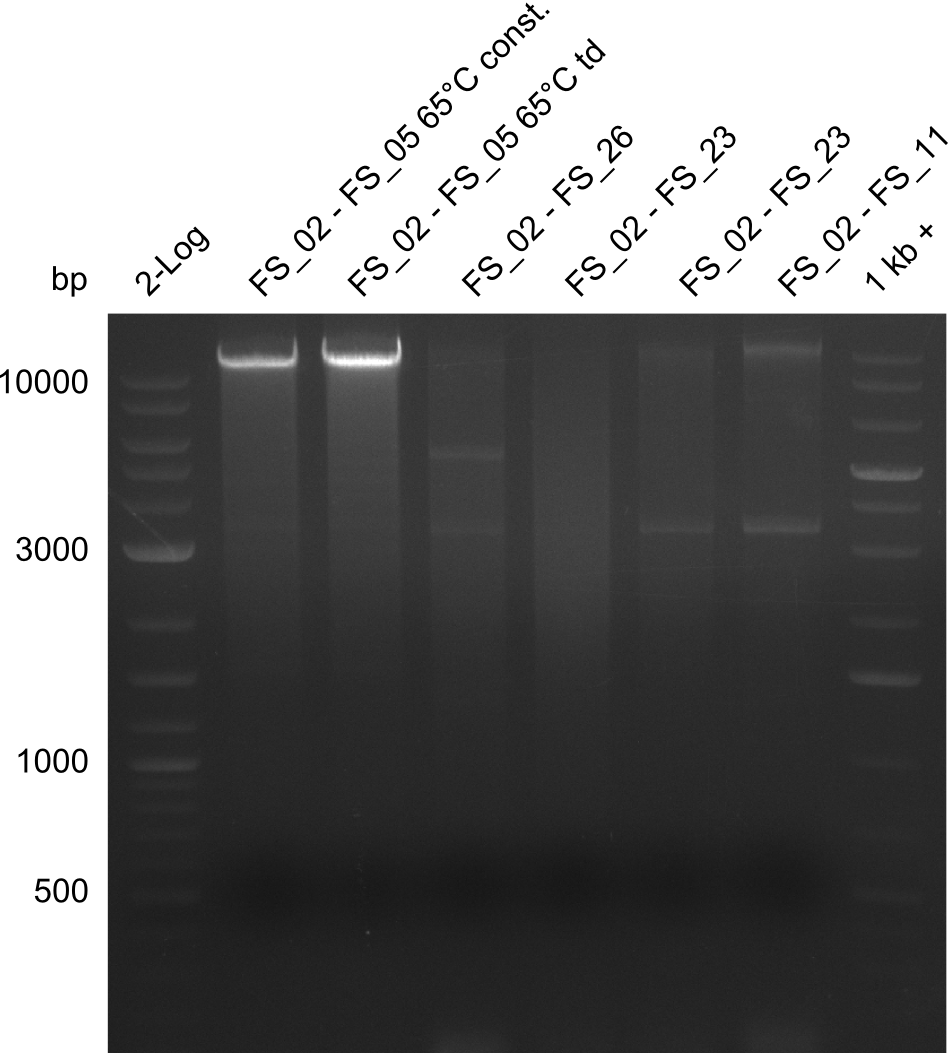
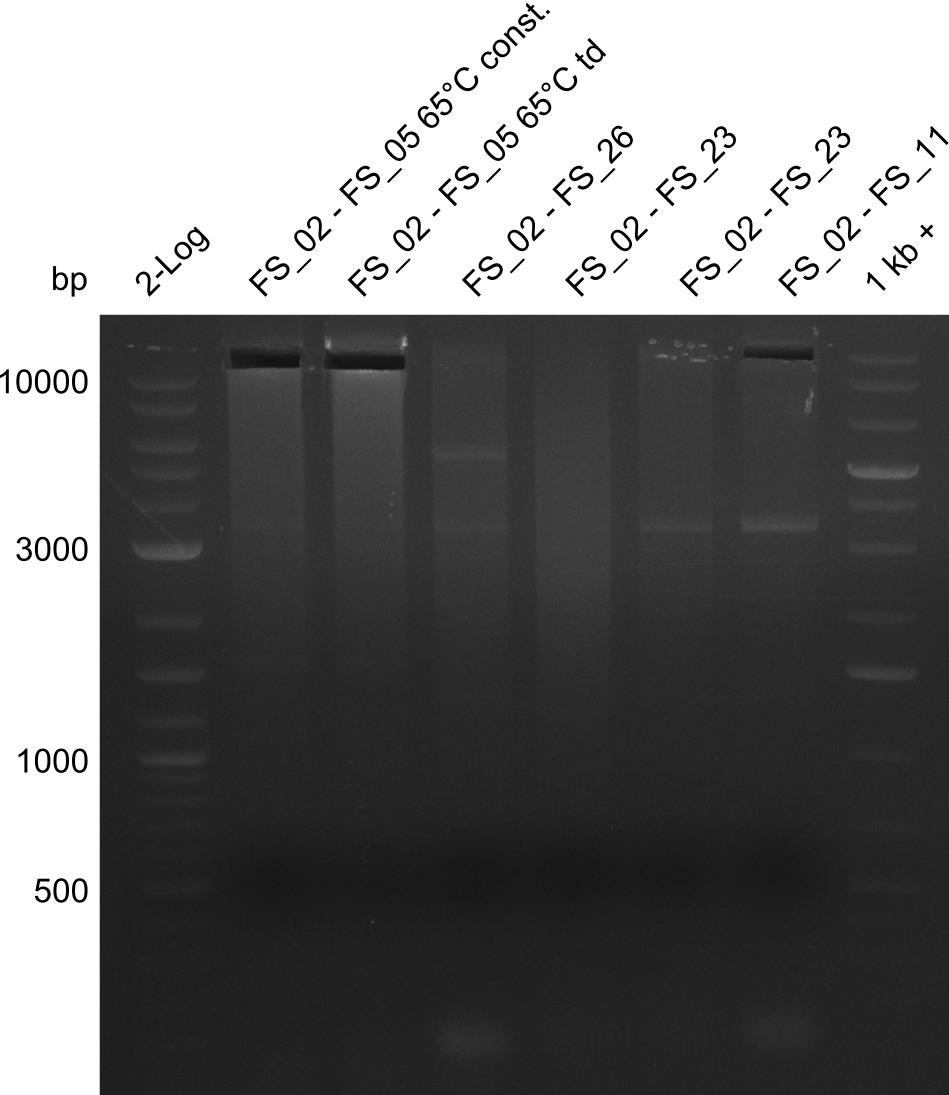
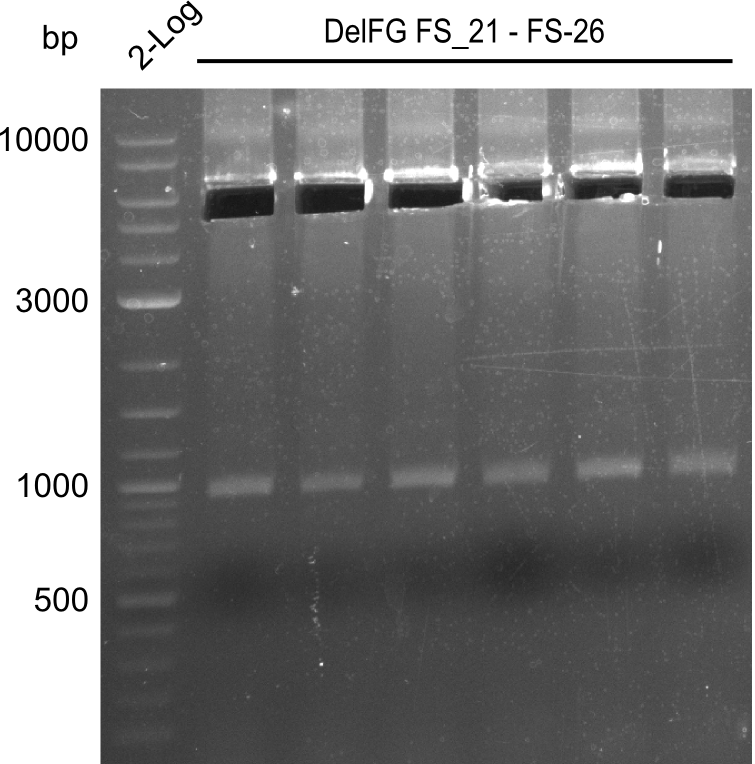
Amplification of Del Genes from D. acidovorans DSM-39 genome
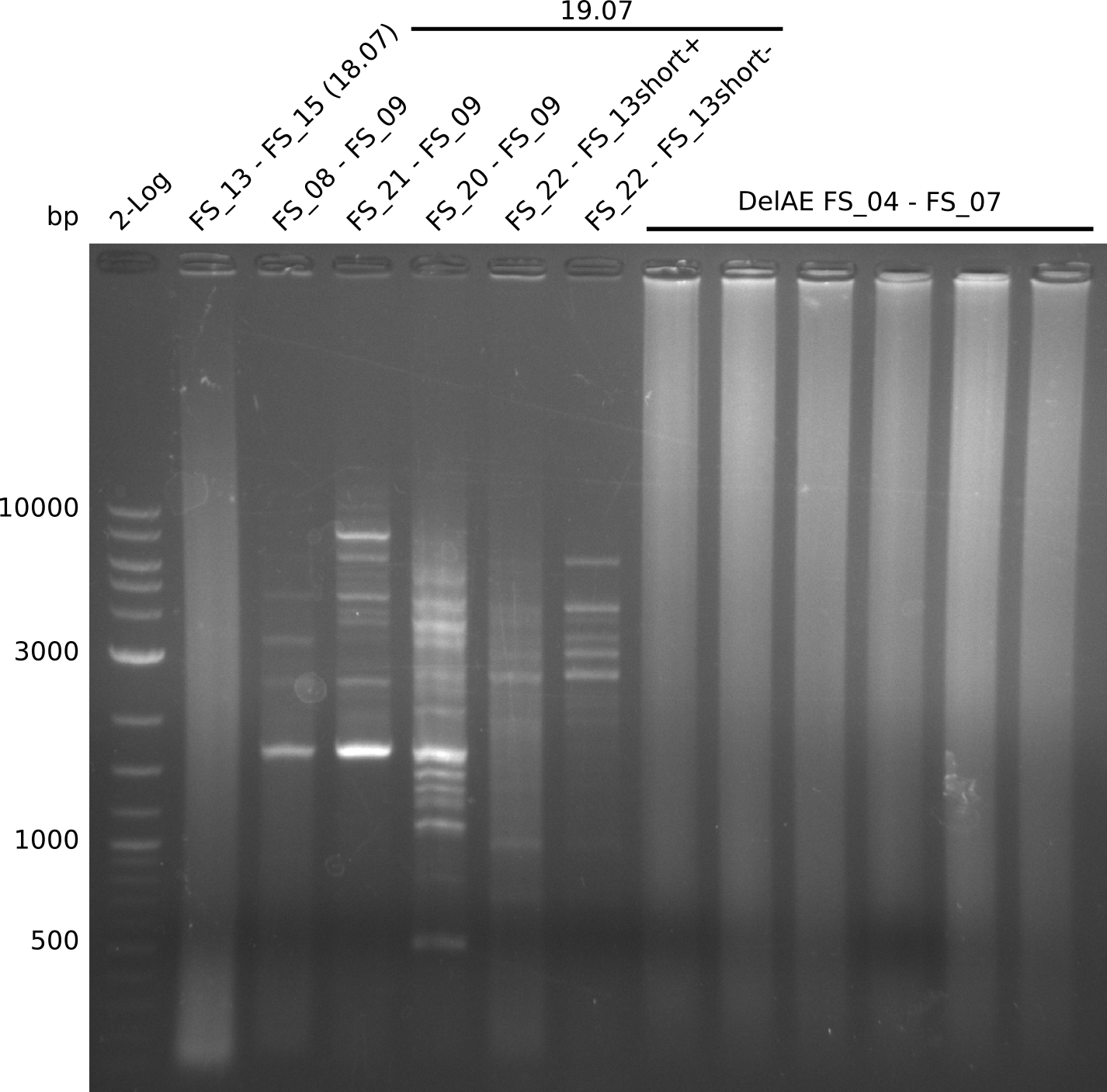
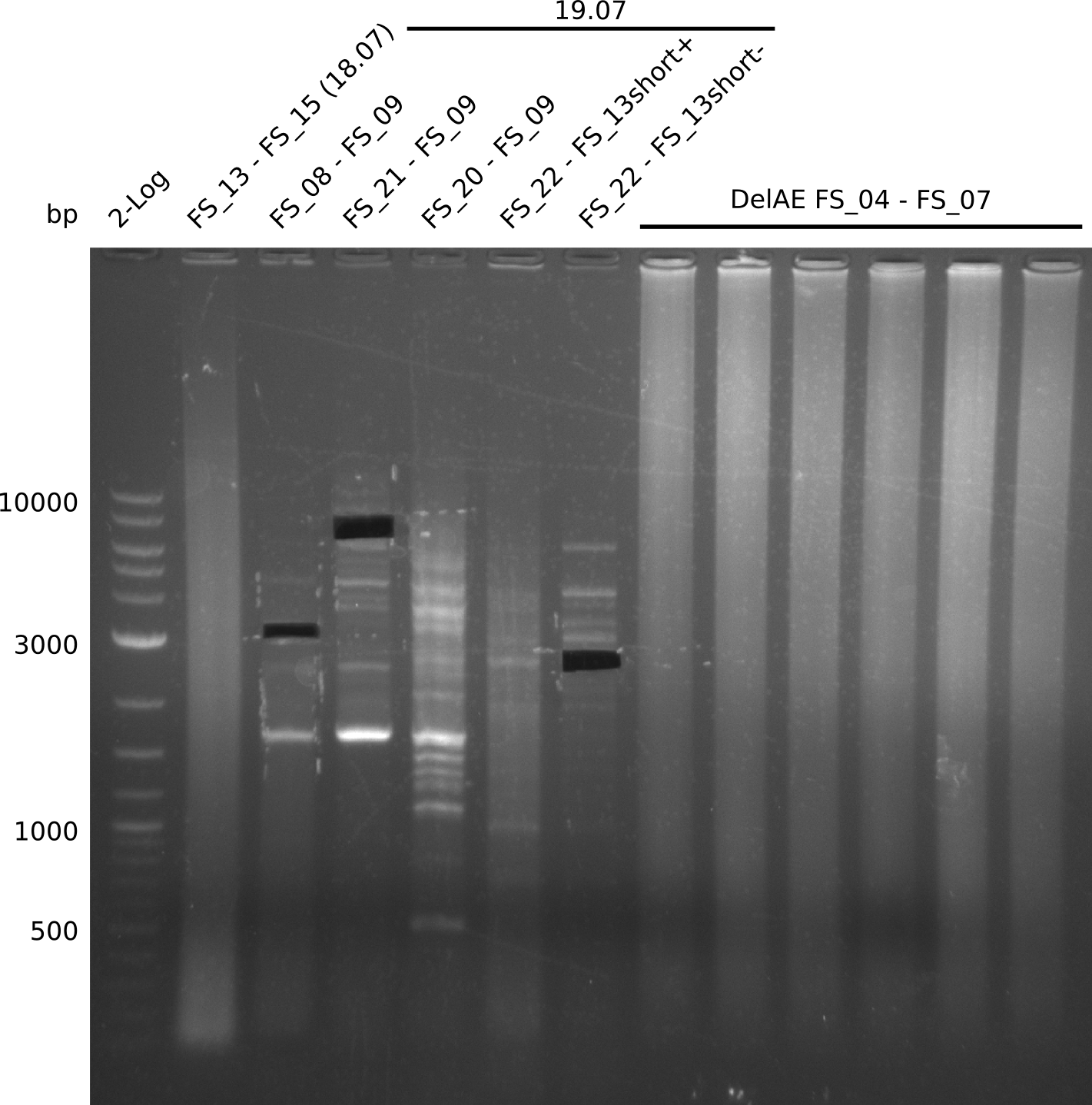
Goals
This week we will try to amplify the sequence region of DelF-G with several combinations of primers including both, the intial primers and the recently ordered ones beeing FS_20, FS_21, FS_22, FS_23, FS_24 and FS_26. Furthermore we will continue opimtizing our protocol for the amplification of DelO-P. To evaluate whether the very weak bands on the corresponding agarose gels represent the intended amplicons, we decided to carry out re PCR and test restriction digests.
Results
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | Primer FS_02 and FS_03 | ||
| DelE-G | Primer FS_04 and FS_07 | |||
| DelF-G | Primer FS_20 and FS_07 | |||
| Primer FS_20 and FS_09 | ||||
| Primer FS_21 and FS_09 | ||||
| DelG | Primer FS_08 and FS_09 | |||
| Primer FS_08 and FS_11_short | ||||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13 | ||
| Primer FS_22 and FS_13_short | ||||
| DelL-P | Primer FS_13_short and FS_15_short | |||
| Re-PCRs | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelE DelF DelG | DelE-G | Primer FS_04 and FS_07 | ||
| DelF-G | Primer FS_06 and FS_09 | |||
| Primer FS_21 and FS_09 | ||||
| DelG | Primer FS_08 and FS_09 | |||
| Primer FS_08 and FS_11_short | ||||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13 | ||
19-07-2013
Amplificaction from FS_02 to FS_03; 5.3 kb
- Reaction
2x ~50 µL
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2.5 |
| FS_03: (1/10) | 2.5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 19 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE did not work since a different cycler was used
20-07-2013
Amplification from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2.5 |
| FS_03: (1/10) | 2.5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2,5 |
| dd H2O | 19 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE worked but a smear occured, therefore bands were cut out carefully and only used for a test restriction digest
15-07-2013
Amplification from FS_04 to FS_07, 11.1 kb
3x 20µl of reaction mix
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 2 |
| FS_07: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelEG worked as expected
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
21-07-2013
Re-PCR of DelEG (FS_04 to FS_07; 11.1 kb; 05-07-2013)
- Reaction
| what | µl |
|---|---|
| Fragment FS_04 to FS_07 amplified 15-07-2013 | 1 |
| FS_04: (1/10) | 2 |
| FS_07: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:00 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Re-PCR of DelEG in order to increase specifity did not work
16-07-2013
Amplification from FS_08 to FS_11_short; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2 |
| FS_11_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 24 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Conditions II
| Biorad C1000 Touch Block B | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
- Conditions III
| Biometra T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Only a small band with conditions II was visible
- There were no bands with the other two
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
18-07-2013
Amplification from FS_08 to FS_11_short; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2 |
| FS_11_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad C1000 Touch | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 64 ↓ 0.5 | 5 | |
| 72 | 2:10 min | |
| 18 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:10 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelG did not work, no product was detectable
- PCR will be repeated at higher temperature to decrease formation of secondary structures and thereby improve binding of primers to the desired sequence
Re-Amplification from FS_08 to FS_11_short; 6.5 kb; 09-07-2013)
- Reaction
| what | µl |
|---|---|
| DelG (09-07-2013) | 4 |
| FS_08 (1/10) | 2 |
| FS_11_short (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 1 |
- Conditions II
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 16 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 10min |
| 1 | 4 | inf |
Results:
- Amplification of DelG was not sucessful, the Re-PCR did not yield the desired product
- forward Primer will be reused but reverse primer changed, in order to obtain a shorter amplicon and another strategy for the Gibson Assembly
19-07-2013
Amplification from FS_08 to FS_09; 3.3 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 6min |
| 1 | 12 | inf |
Results:
- Amplification of DelG did not lead to the desired product, a small band of the desired size was visible and cut for validation and Re-PCR
- Re-PCR will be run
21-07-2013
Re-PCR from FS_08 to FS_09; 3.3 kb; 19-07-2013
- Reaction
| what | µl |
|---|---|
| Fragment FS_08 to FS_09 (19-07-2013) | 1 |
| FS_08: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 6min |
| 1 | 12 | inf |
Results:
- Re-Amplification of DelG did not work
- initial amplification will be repeated at lower annealing temperature
18-07-2013
Amplification from FS_13s to FS_15s; 6.4 kb
- Reaction of DelLP
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_13_short: (1/10) | 2 |
| FS_15_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions of Del LP
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- The second try to amplify DelL-P did not work. There were no bands visible on the gel.
- It does not make sense to try this again.
17-07-2013
Amplification from FS_22 to FS_13(s); 2.7 kb
4 reactions (with long and short primer FS13 and with conditionI and conditionII)
- Reaction of DelOP
| what | µl |
|---|---|
| D.acidovorans | 1 |
| FS_22: (1/10) | 2 |
| FS_13 (long or short): (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I of DelOP
| Biorad C1000 Touch Block B | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:00 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:00 min | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
- Conditions II of DelOP
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:00 min | |
| 1 | 72 | 5:00 min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP failed again
- PCR will be repeated, annealing will be carried out 65°C (touchdown) as primer binding seems not to occure at high temperatues as the ones tested in the last amplification attempts
19-07-2013
Amplification from FS_22 to FS_13s; 2.7 kb
--> reaction mixture with and without DMSO
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 4/5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP failed again, unexpected bands as well as a smear occured
- Annealing temperature will be further decreased, to investigate if amplification of the intended product occurs at lower temperatures
20-07-2013
Amplification from FS_22 to FS_13s; 2.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 4 |
- Conditions
| Biorad C1000 Touch Block B | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 60 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP did not work, only unintented products as well as a slight smear occured
- Reaction will be carried out again at lower annealing temperature to allow primer binding to the intended sequences
21-07-2013
Amplification from FS_22 to FS_13s; 2.7 kb
4x 20µl (with, without DMSO; 60touchdown, 72 twostep)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 4/5 |
- Conditions I
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 60 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
- Conditions II
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 72 | 1:00 min | |
| 1 | 72 | 5:00 min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP led to inconclusive results, a very thin band of the intended length as well as several other bands and a smear occured
- Band were cut out and DNA purified using QIAquick Gel Extraction Kit to verify amplicon in a re-PCR
Re-PCR of DelOP (FS_22 to FS_13s; 2.7 kb; 19-07-2013)
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short (19-07-2013) | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | - |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated as re-pcr from the previously obtained sample
22-07-2013
Re-PCR of DelOP (FS_22 to FS_13; 2.7 kb; 19-07-2013)
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short 19-07-2013) | 1 |
| FS_22: (1/10) | 2 |
| FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Re-PCR did lead ot a band at the expected size but also a smear and several unexpected bands
- Re-PCR will be repeated
24-07-2013
Re-PCR of DelOP (FS_22 to FS_13; 2.7 kb; 19-07-2013)
3x20µl
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short (19-07-2013) | 1 |
| FS_22: (1/10) | 2 |
| FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Re-PCR of DelOP did not work, no bands were visible
- Re-PCR will be repeated
15-07-2013
Re-Amplification from FS_06 to FS_09; 8.5 kb; 13-07-2013)
- Reaction
| what | µl |
|---|---|
| DelEG (FS_06-FS_09; 13-07-2013) | 1 |
| FS_06: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
Cycler incubation room right
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 3:00 min | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work
- two unecpected bands appeared like in the previous trials
19-07-2013
Amplification from FS_20 to FS_09; 8.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:50 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:50 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work with FS_20 to FS_09
- as PCR worked with a different set of Primers beeing FS_21 to FS_09, see below, the PCR for this primer combination will be optimized instead of using FS_20 to FS_09
Amplification from FS_21 to FS_09; 8.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
-->New cycler (not two block)
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:50 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:50 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification worked with FS_21 to FS_09 but not with FS_20 to FS_09
- band was cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be repeated to get rid of side product at about 1.5 kb as well as smear, therefore annealing temperature will be increased
20-07-2013
Amplification from FS_21 to FS_09; 8.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 2:50 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:50 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work only an uneqxpected band a smear occured, but the intended product was not detectable
- other primer combinations might be tried or PCR has to be repeated with the previous conditions not leading to an optimal product quality
21-07-2013
Amplification from FS_20 to FS_07; 5.2 kb
2x 20µl (with, without DMSO)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 2 |
| FS_07: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 4/5 |
- Conditions
| Biorad C1000 Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:00 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work perfectly, neither with nor without 5% DMSO, nevertheless band of expected size was cut out carefully to be used for a Re-PCR
Re-PCR from FS_21 to FS_09; 8.1kb; 19-07-2013)
- Reaction
| what | µl |
|---|---|
| Gel extracted fragments FS_21 to FS_09 (19-07-2013) | 2 |
| FS_21: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 3 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:50 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:50 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work with primers FS_21 to FS_09
- PCR will be repeated with different primers
Amplification and purification of Del Genes from DSM-39
Goals
As our the amplifications of of the sequence region DelF-G did not progress last week, we decided to focus on this region in week 13. All combinations of initial and newly ordered primers will be tried with various PCR protocols. Furthermore, we tried to get higher amounts of the fragments for which protocols have already been established in order to have enough of each product to carry out suitable test restriction digests.
Results
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | Primer FS_02 and FS_03 | ||
| Primer FS_02 and FS_24 | ||||
| DelA-F | Primer FS_02 and FS_05 | |||
| DelA-G | Primer FS_02 and FS_26 | |||
| Primer FS_02 and FS_23 | ||||
| DelE | Primer FS_04 and FS_24 | |||
| DelE-G | Primer FS_04 and FS_09 | |||
| DelF-G | Primer FS_06 and FS_07 | |||
| Primer FS_20 and FS_07 | ||||
| Primer FS_20 and FS_23 | ||||
| Primer FS_20 and FS_11_short | ||||
| Primer FS_21 and FS_07 | ||||
| Primer FS_21 and FS_11_short | ||||
| Primer FS_21 and FS_23 | ||||
| Primer FS_20 and FS_09 | ||||
| Primer FS_21 and FS_09 | ||||
| DelG | Primer FS_08 and FS_11_short | |||
| Primer FS_08 and FS_23 | ||||
| DelO DelP | DelO-P | Primer FS_22 and FS_13 | ||
| Primer FS_22 and FS_13_short | ||||
| Re-PCRs | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelG | DelG | Primer FS_10 and FS_23 | ||
| DelO DelP | DelO-P | Primer FS_22 and FS_13 | ||
| Test restriction digests of PCR amplified fragments | ||||
|---|---|---|---|---|
| Fragment | Primer | Digestion enzyme | Expected bands [bp] | |
| DelA-E | Primer FS_02 and FS_03 | EcoRI-HF | 3054, 2260 | |
| DelA-F | Primer FS_02 and FS_05 | EcoRI-HF | 4624, 4354, 2260 | |
| DelE-G | Primer FS_04 and FS_07 | PvuI-HF | 6187, 4917 | |
| DelF-G | Primer FS_20 and FS_07 | XmaI | 4307, 879 | |
| Primer FS_20 and FS_07 | ClaI | 2420, 1519, 1247 | ||
| DelG | Primer FS_08 and FS_23 | BglII | 3291, 1936, 1303 | |
| DelO-P | Primer FS_22 and FS_13_short | EcoRI-HF | 1883, 960 | |
26-07-2013
Restriction digest of fragment FS_02 to FS_03; 5.3 kb; 08-07-2013 with EcoRI-HF
Incubation at 37°C for 1 h 45 min
| what | µL |
|---|---|
| FS_02 to FS_03 (08-07-2013) | 15 |
| EcorRI-HF | 0.5 |
| Buffer CutSmart | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 3054, 2260 |
Results:
- restriction digest of DelAE did not work, since incubation time might have been to short
27-07-2013
Amplificaction from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2.5 |
| FS_03: (1/10) | 2.5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 19 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 12 min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE didnt work out, only smear occured
- repeat PCR with better cycler
Amplificaction from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1.5/1 |
| FS_02: (1/10) | 2.5/5 |
| FS_03: (1/10) | 2.5/5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 19/14 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 12 min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE worked with both 200 and 400 nM of Primers, nevertheless amplification was more specific with the higher primer concentration
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
Amplification I from FS_02 to FS_24; 7.1 kb
4 reactions, 2 with 200nM Primers and 2 with 400nM Primers (both concentrations for each condition)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 4/2 |
| FS_24: (1/10) | 4/2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:50 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3:50 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:50 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAE worked with 200 nM primer concentration at an annealing temperature of 68°C and 400 nM at an annealing temperature of 65°C, the product obtained at 65°C was more specific
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
Amplification II from FS_02 to FS_24; 7.1 kb
2 reactions, 68°C Touchdown with 200nM Primers and 65°C constant with 400nM Primers
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2/4 |
| FS_24: (1/10) | 2/4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 5:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 5:40 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 5:40 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification did not work, neither with 200nM and 68°C touchdown, nor with 400nM and 65°C constant.
- Repeat amplfication with different conditions as primers did not bind very effectively
Amplification III from FS_02 to FS_24; 7.1 kb
2 reactions, 66°C Touchdown with 200nM Primers and 60°C constant with 400nM Primers
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2/4 |
| FS_24: (1/10) | 2/4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 5:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 5:40 min | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 5:40 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification from DelAE (7.1 kbp) failed again
- stick to the old strategy and use previously obtained fragments with different other fragments for gibson assembly.
29-07-2013
Restriction digest of FS_02 to FS_03; 5.3 kb;(27-07-2013; II) with BglII
Incubation at 37°C for about 3 h
| what | µL |
|---|---|
| FS_02 to FS_03 (27-07-2013; II) | 15 |
| BglII | 1 |
| Buffer 3.1 | 2 |
| dd H2O | 2 |
Expected fragment lengths: 2,146 kb; 1,862 kb; 1,306 kb
Results:
- Restriction digest shows the expected product sizes
- indicator for correct amplicon but to be sure, PCR product will be prepared for single read sequencing by GATC
26-07-2013
Restriction digest of FS_02 to FS_05; 11.2 kb; 04-07-2013 with EcoRI-HF
Incubation at 37°C for 1hour 45min
| what | µL |
|---|---|
| FS_02 to FS_05 (04-07-2013) | 15 |
| EcoRI-HF | 0.5 |
| Buffer CutSmart | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 4624, 4354, 2260 |
Results:
- restriction digest did not work as expected
- test restriction will be repeat with higher amount of enzyme and longer incubation time
27-07-2013
Amplification of FS_02 to FS_05; 11.2 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2.5 |
| FS_05: (1/10) | 2.5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0,5 | 5 | |
| 72 | 3:00 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:00 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAE worked, but a slight smear as well as several unexpected bands occured
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit but PCR has to be further optimized in order to improve product quality
27-07-2013
Amplification from FS_02 to FS_26; 16.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 3:50 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
Results:
- Amplification of DelAG did not work, unspecific band at approximately 3.5 kbp occured
Amplification 1 from FS_02 to FS_23; 22.8 kb
4 reactions, 2 with 200nM Primers and 2 with 400nM Primers (both concentrations for each condition)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 4/2 |
| FS_23: (1/10) | 4/2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 6:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 6:40 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 6:40 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAG did not work, several unspecific bands occured
Amplification 2 from FS_02 to FS_23; 22.8 kb
4 reactions, 2 with 200nM Primers and 2 with 400nM Primers (both concentrations for each condition)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 4/2 |
| FS_23: (1/10) | 4/2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad MyCycler, Biometra TProfessional Basic and Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 6:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
Results:
- Amplification of DelAG did not work, independent of the cycler only unspecific bands at 4.0 and 5.2 kbp occured
27-07-2013
Amplification from DelE (FS_04 to FS_24; 1.6 kb)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3:00 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:00 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification did not work.
24-07-2013
Restriction digest of fragment FS_04 to FS_07; 11.1 kb with PvuI-HF
Incubation at 37°C for 45 min
| what | µl |
|---|---|
| FS_04 to FS_07 (14-07-2013 and 15-07-2013) | 15 |
| PvuI-HF | 0.8 |
| Buffer CutSmart | 2 |
| dd H2O | 2.8 |
| Expected fragment lengths [bp] | 6187, 4917 |
Results:
- restriction digest did not work
- digest will be repeated with newly amplified and purified DelEG
28-07-2013
Amplification from FS_04 to FS_09 ; 14.4 kb
2 reactions with conditions I and II
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_04: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 4:40 | |
| 1 | 72 | 13 min |
| 1 | 10 | inf |
- Conditions II
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 4:40 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 4:40 | |
| 1 | 72 | 13 min |
| 1 | 10 | inf |
Amplification from FS_26 to FS_07
This amplification did not make sense, two reverse Primer were used. We mixed up Primer FS_24 with Primer FS_26.
2 reactions with conditions I and II
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_26: (1/10) | 2 |
| FS_07: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 13 min |
| 1 | 10 | inf |
- Conditions II
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
Results:
- Amplification of DelEG did not work, neither with annealing at a constant temperature of 65°C nor with a touchdown starting from 68°C
- later on it was discovered, that primers had been mixed up
Amplification from FS_26 to FS_24
This amplification did not make sense, two reverse Primer were used.
2 reactions with conditions I and II
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_26: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 13 min |
| 1 | 10 | inf |
- Conditions II
| Biorad C1000 Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
Results:
- Amplification of DelEG did not work, neither with annealing at a constant temperature of 65°C nor with a touchdown starting from 68°C
- later on it was discovered, that primers had been mixed up
22-07-2013
Re-PCR from FS_10 to FS_23; 3.3 kb; 13-07-2013
- Reaction
| what | µl |
|---|---|
| Fragment FS_10 to FS_11_short (13-07-2013) | 1 |
| FS_10: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
--> First cycles with wrong program
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:10 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:10 min | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Results:
- Re-Amplification of DelG lead to the expected fragment but also to a smear, therefore fragment will not be used for Gibson Assembly but restriction digests to validate the product.
Amplification from FS_08 to FS_23; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 4/- |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG worked, leading to the expected product as well one other unexpected band.
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
Amplification from FS_08 to FS_11s; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2 |
| FS_11_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 14 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 16 | 98 | 1 |
| 60 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
23-07-2013
Amplification from FS_08 to FS_23; 6.5 kb
4x 20µl
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_08: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- Gel to proof quality of gel extraction was run
26-07-2013
Restriction digest of Fragment FS_08 to FS_23; 6.5 kb; 23-07-2013 with BglII
Incubation at 37°C for 1hour 45min
| what | µl |
|---|---|
| FS_08 to FS_23 cut1 (23-07-2013) | 15 |
| BglII | 0.5 |
| Buffer 3.1 | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 3291, 1936, 1303 |
Results:
- Restriction digest of DelG (FS_08 to FS_23) was successful though one unexpected band (presumably carry over) appeared
- Construct will be further validated by sequencing before potential Gibson Assembly
25-07-2013
Re-PCR I from FS_22 to FS_13; 2.7 kb; 19-07-2013
3x20µl
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short (19-07-2013) | 1 |
| FS_22: (1/10) | 4 |
| FS_13_long: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| dd H2O | 1 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP led to a smear and several unexpected bands
- Re-PCR will be repeated
Re-PCR I from FS_22 to FS_13; 2.7 kb; 19-07-2013
3x20µl
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short (19-07-2013) | 1 |
| FS_22: (1/10) | 2 |
| FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- a smear as well as a band of the expeceted size occured
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- DNA will be used for restriction digests to validate amplicon
26-07-2013
Restriction digest of fragment FS_22 to FS_13s; 2.7 kb; 21-07-2013 with EcoRI-HF
Incubation at 37°C for 1h 45min
| what | µl |
|---|---|
| FS_22 to FS_13short (21-07-2013) | 20 |
| EcorRI-HF | 0.8 |
| CutSmart | 2.5 |
| dd H2O | 2.5 |
| Expected fragment lengths [bp] | 1883, 960 |
Results:
- Restriction digest did not work, the amplified fragment was either not cut or the fragments are not visible due to a very small amount of DNA
- Re-PCR will be repeated to obtain the higher amounts of DNA required for the restriction digest
30-07-2013
Re-PCR from FS_22 to FS_13; 2.7 kb; 19-07-2013
3x20µl
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short (19-07-2013) | 1 |
| FS_22: (1/10) | 2 |
| FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated with different primer concentrations to estimate whether primer dimes might be the reason for insufficient amplification
Restriction digest from FS_22 to FS_13; 2.7 kb; 25-07-2013 with EcoRI-HF
Incubation at 37°C for
| what | µl |
|---|---|
| FS_22 to FS_13long(25-07-2013) | 17 |
| EcoRI | 1 |
| Buffer CutSmart | 2 |
Amplification from FS_22 to FS_13s; 2.7 kb
2x20µl
- Reaction
| what | µl |
|---|---|
| D.acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 4 |
| FS_13_short: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| dd H2O | 1 |
20µl
- Reaction
| what | µl |
|---|---|
| D.acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP resulted in a small band atthe desired lenght, but also a smear and several unexpected bands
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- amplicon will be used for restriction digest to validate the construct
Amplification from FS_22 to FS_13(s/l); 2.7 kb
- Reaction
| what | µl |
|---|---|
| D.acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short/FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 4/5 |
- Conditions
| Biorad C1000 Touch | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 70 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
22-07-2013
Amplification from FS_20/FS_21 to FS_09; 8.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20/FS_21: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 2:20 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:20 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work
- other primers/combinations of primers will be used
- somehow getting annoyed by this part of D. Acidovorans
Amplification from FS_20/FS_21 to FS_11_short; 11.6 kb
4x 20µl (70 touchdown, 65 touchdown)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20 or FS_21: (1/10) | 2 |
| FS_11_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
- Conditions II of Del FG
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work neither with a touchdown PCR starting at an annealing temperature of 65°C nor 70°C
Amplification from FS_20/FS_21 to FS_23; 11.6 kb
4x 20µl (70 touchdown, 65 touchdown)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20 or FS_21: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
- Conditions II
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
Results:
- Neither the amplification with the primers FS_20 to FS_23 or FS_21 to FS_23 did work. Another primer combination has to be tried.
24-07-2013
Amplification of DelFG (FS_06 to FS_07; 5.2 kb)
- Reaction I
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
Amplification from FS_20 to FS_07; 5.2 kb
- Reaction II
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
Amplification from FS_21 to FS_07; 5.2kb
- Reaction III
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions for reactions I - III
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- The amplification of FS_06 to FS_07 did not work. No bands were visible
- The amplification of FS_21 to FS_07 led to several bands, but none of these was the intended product
- The amplfication of FS_20 to FS_07 also led to several bands, one band at the right height was observed. Consequently the specificity of the PCR will be increased by a higher annealing temperature
Amplification from FS_20 to FS_07; 5.2 kb
2x20µl (one with conditions I, other one with conditions II)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions I
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- nethertheless bands were cut out and DNA purified using QIAquick Gel Extraction Kit for restriction digest
Amplification from FS_20 to FS_07; 5.2 kb
4x20µl
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- nethertheless bands were cut out and DNA purified using QIAquick Gel Extraction Kit for restriction digest
25-07-2013
Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; 23-07-2013 and 24-07-2013 with XmaI
Incubation at 37°C for 45 min
| what | µl |
|---|---|
| FS_20 to FS_07 (23-07-2013 and 24-07-2013) | 15 |
| XmaI | 0.8 |
| Buffer CutSmart | 2 |
| dd H2O | 2.2 |
| Expected fragment lengths [bp] | 4307, 879 |
Results:
- restriction digest of DelFG did not work, only very slight bands were visible
- digest will be repeated with higher amount of DNA and enzyme to improve analysis on the gel
26-07-2013
Amplification from FS_20 to FS_07; 5.2 kb
- Reaction I
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 70 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- Annealing temperature will be increased further, to optimize primer specifity
Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; 23-07-2013 and 23-07-2013) with ClaI
Incubation at 37°C for 45 min
| what | µl |
|---|---|
| FS_20 to FS_07 (23-07-2013 and 23-07-2013) | 25 |
| ClaI | 1 |
| Buffer CutSmart | 3 |
| dd H2O | 1 |
| Expected fragment lengths [bp] | 2743, 1519, 1208 |
Results:
- restriction digest of Del FG did not lead to the expected results
- as no DNA was visible in the restriction digest, experiment will be repeated with a higher amount of DNA
Amplification from FS_06 to FS_07; 5.2 kb
4 x 20µL
- Reaction I
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 72 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 70 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- nethertheless bands were cut out and DNA purified using QIAquick Gel Extraction Kit for restriction digest
- Does anyone know, why we are constantly repeating this totally deficient PCR?
Amplification from FS_6/FS_20/FS_21 to FS_24 (PRIMER FS_24 MIXED UP!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06/FS_20/FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:20 | |
| 1 | 72 | 7 min |
| 1 | 10 | inf |
Results:
- no PCR product occured since the wrong primers were used
28-07-2013
Amplification from FS_21 to FS_24; (PRIMER 24 was MIXED UP!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
Results:
- no PCR product occured since the wrong primers were used
Amplification and purification of Del Genes from D. acidovorans DSM-39 genome
Goals
Our goal still remains to amplifiy the missing fragments from the Delftibactin cluster of D. Acidovorans. However, since we want to validate all fragments before we use them in a Gibson Assembly, and since last weeks restriction digests were either negative or inconclusive, these digests will be repeated.
By the end of this week we will dicuss the further procedure and decide whether we again need to change our cloning strategy or not. Additionally a nested PCR of the sequence region surrounding DelOP might still be an option to provide specific template suitable for a PCR with our intended Gibson primers.
Results
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-F | Primer FS_02 and FS_05 | ||
| DelA-G | Primer FS_02 and FS_26 | |||
| DelF-G | Primer FS_06 and FS_26 | |||
| Primer FS_20 and FS_26 | ||||
| Primer FS_21 and FS_26 | ||||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13 | ||
| Primer FS_22 and FS_13_short | ||||
| Test restriction digests of PCR amplified fragments | ||||
|---|---|---|---|---|
| Fragment | Primer | Digestion enzyme | Expected bands [bp] | |
| DelA-E | Primer FS_02 and FS_03 | BglII | 2146, 1862, 1306 | |
| DelA-F | Primer FS_02 and FS_05 | BglII | 7229, 2146, 1862 | |
| DelF-G | Primer FS_21 and FS_26 | XmaI | 4268, 1202 | |
| DelG | Primer FS_08 and FS_23 | ClaI | 3291, 1936, 1303 | |
| DelO-P | Primer FS_22 and FS_13 | EcoRI-HF | 1883, 960 | |
29-07-2013
Restriction digest of fragment FS_02 to FS_03 (5.3 kb; 27-07-2013; II) with BglII
Incubation at 37°C for about 3 h
| what | µL |
|---|---|
| FS_02 to FS_03 (27-07-2013; II) | 15 |
| BglII | 1 |
| Buffer 3.1 | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 2146, 1862, 1306 |
Results:
- Restriction digest shows the expected product sizes
- indicator for correct amplicon but to be sure, PCR product will be prepared for single read sequencing by GATC
29-07-2013
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 27-07-2013 with BglII
Incubation at 37°C for about 3 hours
| what | µL |
|---|---|
| FS_02 to FS_05 (27-07-2013) | 15 |
| BglII | 1 |
| Buffer 3.1 | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 7229, 2146, 1862 |
Results:
- Test restriction digest worked out as predicted
30-07-2013
Amplification from FS_02 to FS_05; 11.2 kb
2 reactions 200nM/400nM Primers
- Reaction
| what | µL 2nd PCR |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02 (1/10) | 5/2.5 |
| FS_05 (1/10) | 5/2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 11.5/16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification did not work properly, as there were several unspecific bands. However we still had product.
30-07-2013
Amplification from FS_02 to FS_26; 16.7 kb
4 reactions, 68°C Touchdown and 65°C constant with 200nM and 400nM Primers
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2/4 |
| FS_26: (1/10) | 2/4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 5:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 5:40 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 5:40 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAG did not work, neither annealing at 65°C constant nor a touchdown starting from 68°C led to the desired product
29-07-2013
Restriction digest of fragment FS_08 to FS_23; 6.5 kb; 23-07-2013 with ClaI
Incubation at 37°C for about 3 hours
| what | µl |
|---|---|
| FS_08 to FS_23 (2) (23-07-2013) | 15 |
| ClaI | 1 |
| Buffer CutSmart | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 3291, 1936, 1303 |
Results:
- Restriction digest of DelG (FS_08 to FS_23) was successful all predicted bands are clearly visible
30-07-2013
Re-PCR of DelOP FS_22 to FS_13; 2.7 kb; 19-07-2013
3x20µl
- Reaction
| what | µl |
|---|---|
| Fragment FS_22 to FS_13_short (19-07-2013) | 1 |
| FS_22: (1/10) | 2 |
| FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated with different primer concentrations to estimate whether primer dimes might be the reason for insufficient amplification
Restriction digest of fragment FS_22 to FS_13; 2.7 kb; 25-07-2013 with EcoRI-HF
Incubation at 37°C for
| what | µl |
|---|---|
| FS_22 to FS_13long(25-07-2013) | 17 |
| EcoRI | 1 |
| Buffer CutSmart | 2 |
| Expected fragment lengths [bp] | 1883, 960 |
Amplification from FS_22 to FS_13s; 2.7 kb
2x20µl
- Reaction
| what | µl |
|---|---|
| D.acidovorans | 1 |
| FS_22: (1/10) | 4 |
| FS_13_short: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| dd H2O | 1 |
20µl
- Reaction
| what | µl |
|---|---|
| D.acidovorans | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP resulted in a small band atthe desired lenght, but also a smear and several unexpected bands
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- amplicon will be used for restriction digest to validate the construct
Amplification from FS_22 to FS_13(s/l); 2.7 kb
- Reaction
| what | µl |
|---|---|
| D.acidovorans | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short/FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1/- |
| dd H2O | 4/5 |
- Conditions
| Biorad C1000 Touch | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 70 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
01-08-2013
Amplification from FS_22 to FS_13(s); 2.7 kb
- Reaction
| what | µl |
|---|---|
| D.acidovorans DSM-39 | 1 |
| FS_22: (1/10) | 2 |
| FS_13_short/FS_13_long: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 92 | 5 | |
| 91 | 5 | |
| 90 | 5 | |
| 89 | 5 | |
| 88 | 5 | |
| 87 | 5 | |
| 86 | 5 | |
| 85 | 5 | |
| 84 | 5 | |
| 82 | 5 | |
| 80 | 5 | |
| 78 | 5 | |
| 76 | 5 | |
| 74 | 5 | |
| 72 | 5 | |
| 70 | 5 | |
| 68 | 5 | |
| 66 | 5 | |
| 64 | 5 | |
| 62 | 5 | |
| 60 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification of DelOP did not work with stepwise cooling to the intended annealing temperature of 60°C
07-08-2013
Amplification from FS_22 to FS_13; 2.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_22: (1/10) | 2 |
| FS_13: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Amplification from FS_22 to FS_13; 2.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_22: (1/10) | 2 |
| FS_13: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP led to a band of the desired size as well as a smear and several different sideproducts
- Band was cut out and DNA purified using QIAquick Gel Extraction Kit to be restriction digested for validation of the PCR product
- Amplification will be repeated at lower temperature to obtain more product
29-07-2013
Amplification from FS_21 to FS_24 ; (WRONG PRIMER!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
Amplification from FS_21 to FS_24; (WRONG PRIMER!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
Amplification from FS_21 to FS_24; (WRONG PRIMER!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
Amplification from FS_06/FS_20/FS_21 to FS_26 ; 5.5 kb
3x20µl
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06 or FS_20 or FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 64 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
- Furthermore, amplification with FS_21 to FS_26 led to the intended product and satisfying specifity
- Amplification will be repeated at the same annealing temperature to obtain the amount of PCR product required for Gibson Assembly
- Amplfication will be repeated at lower annealing temperature to increase the yield
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
30-07-2013
Amplification from FS_21 to FS_26 ; 5.5 kb
3x20µl with conditions I, 2x20µl with conditions II
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 64 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- restriction digest with XmaI will be conducted to validate the PCR product
31-07-2013
Restriction digest of fragment FS_21 to FS_26 (5.5 kb; 29-07-2013) with XmaI
Incubation at 37°C for about 3 hours
| what | µl |
|---|---|
| FS_21 to FS_26 (29-07-2013) | 17 |
| XmaI | 1 |
| Buffer CutSmart | 2 |
| Expected fragment lengths [bp] | 4268, 1202 |
Results:
- Restriction digest of DelFG with XmaI did not lead to the expected result
- digest will be carried out with a different enzyme, as the used one was outdated and digest might therefore not be very reliable
02-08-2013
Restriction digest of fragment FS_21 to FS_26 (5.5 kb; 30-07-2013) with ClaI
Incubation at 37°C for about 3 hours
| what | µl |
|---|---|
| FS_21 to FS_26 (30-07-2013) | ~15 |
| ClaI | 1 |
| Buffer CutSmart | 2 |
| Expected fragment lengths [bp] | 2743, 1519, 1208 |
Results:
- Restriction digest of DelFG did not display any product
- digest will be repeated with higher amount of DNA after new amplification from the genome of D. Acidovorans
07-08-2013
Amplification from FS_21 to FS_26 ; 5.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful, gel displays highly specific product of convincing yield
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- restriction digest with ClaI will be conducted to validate PCR product
Strategy
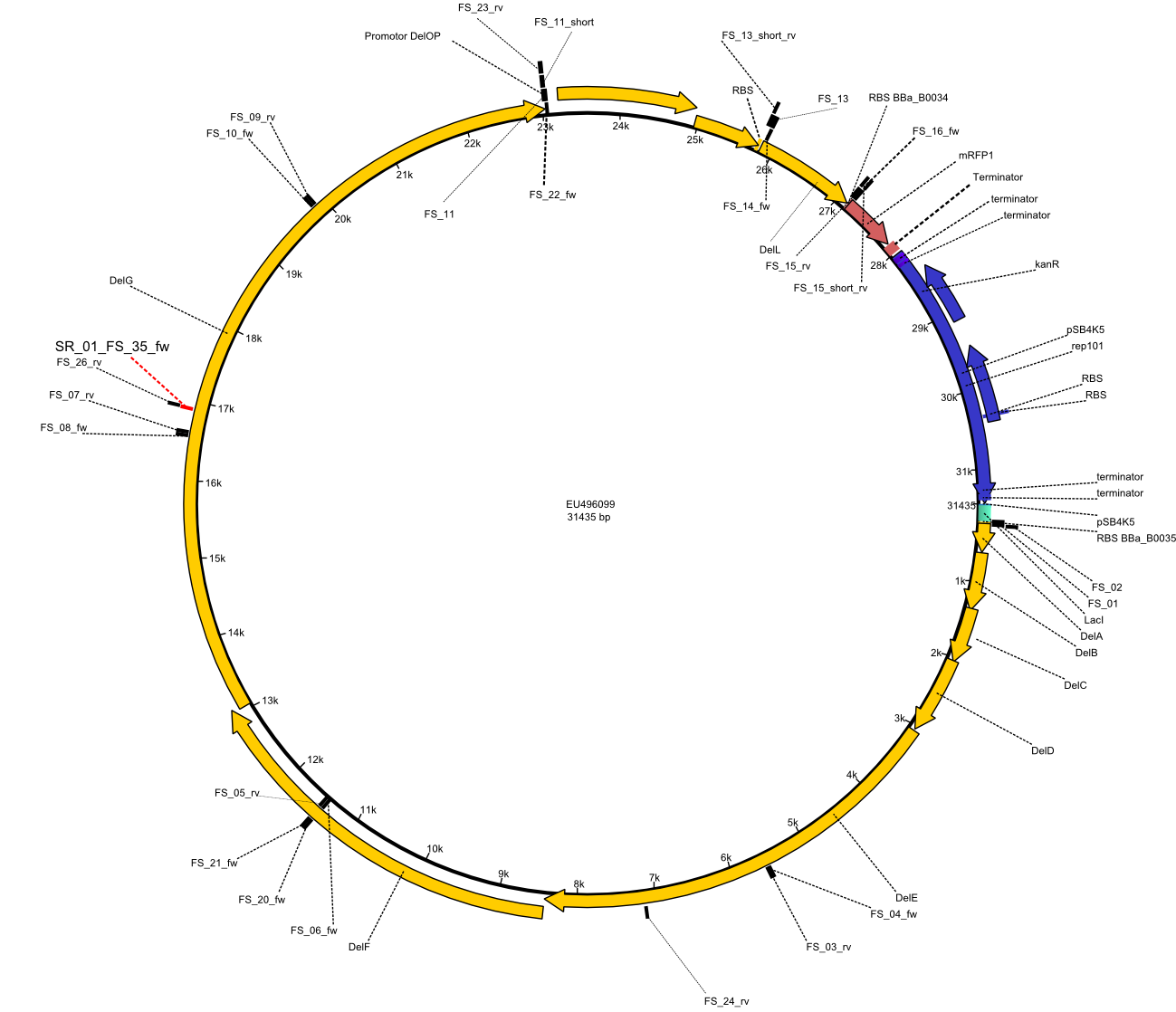
As already indicated last week, we decided to order another primer for the sequence region of DelF-G, beeing the primer FS_35_SR_01. This primer shows significantly better secondary structures and therefore annealing should be better. Furthermore we ordered various primers suitable for a nested PCR of DelOP.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_35: DelG_fw | 2013-08-05 | Amplification of DelG from Delftia acidovorans Gibson Primer | CATGCAGATCATCGTGGATGAATTC |
Cultivation of D. acidovorans substrain SPH-1
Single read sequencing of several PCR products obtained so far was carried out by GATC. We aligned the so obtained sequences to the SPH-1 reference genome. (6.8 Mbp, [http://www.ncbi.nlm.nih.gov/genome/659 NCBI Genome]). Our alignments revealed significant differences between the two D. acidovorans strains: DSM-39 which we used as PCR template and SPH-1 based on which we have designed all PCR primers. As result of this sequence analysis, the strain D. acidovorans SPH-1 was ordered in order to have a suitable template for the primers we already designed and ordered so far. We hoped, that haveing the right strain at hand now, our PCRs should work better and we could hopefully proceed with the Gibson assembly soon. Lyohilized D. Acidovorans SPH-1 cells obtained from DSMZ were reactivated with Reactivation_medium and cultivated in Acidovorax complex medium according to the recoomondations by the DSMZ. Subsequently, liquid culture were inocculated (for glycerol stocks) and bacteria were also spread onto ACM-agar plates. Then we repeated our PCRs either by doing colony-PCR from colonies on the ACM-agar plates or by using 1µL of glycerol stock as template.
Amplification of Del Genes from D. acidovorans SPH-1 genome
Goals
Since we wanted to ensure that our amplicons fit the intended sequence and avoid any mutations resulting from primer missmatches all PCRs were repeated (even the previously successful ones), now using D. Acidovorans SPH-1 as template. Established PCR protocols will be repeated with the new strain as template and adopted if necessary. For the DelF-G fragment with which we have had problems before, we will try whether PCRs work better with the new template or the recently ordered primer FS_35/SR_01. The amplification of DelO-P will be repeated with the intially ordered Primers FS_22 and FS_13 to test whether the changed template also improves these PCRs. If it turns out, that primers are still not able to bind to the intended template, the abovementioned nested PCR strategy will be conducted as primers FS_27 to FS_33 also arrived this week.
Results
| PCRs from D.acidovorans SPH-1 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | Primer FS_02 and FS_03 | ||
| DelA-F | Primer FS_02 and FS_05 | |||
| DelA-G | Primer FS_02 and FS_23 | |||
| Primer FS_02 and FS_11 | ||||
| Primer FS_02 and FS_26 | ||||
| DelF-G | Primer FS_21 and FS_26 | |||
| DelG | Primer FS_08 and FS_23 | |||
| Primer SR_01 and FS_23 | ||||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13 | ||
| Primer FS_22 and FS_13_short | ||||
| Primer FS_22 and FS_27 | ||||
| Primer FS_33 and FS_13_short | ||||
| Primer FS_33 and FS_27 | ||||
| Primer FS_31 and FS_29 | ||||
| Primer FS_31 and FS_30 | ||||
| Primer FS_32 and FS_29 | ||||
| Primer FS_32 and FS_30 | ||||
| Primer FS_22 and FS_28 | ||||
| Primer FS_22 and FS_29 | ||||
| DelL | Primer FS_14 and FS_15 | |||
| pSB4K5 | pSB4K5 | Primer FS_01 and FS_16 | ||
| Test restriction digests of PCR amplified fragments | ||||
|---|---|---|---|---|
| Fragment | Primer | Digestion enzyme | Expected bands | |
| DelF-G | Primer FS_21 and FS_26 | ClaI | 2743, 1519, 1208 | |
| BglII | 5500 | |||
| DelG | Primer FS_08 and FS_23 | ClaI | 3415, 3115 | |
| DelO-P | Primer FS_22 and FS_13 | EcoRI-HF | 1883, 960 | |
07-08-2013
Sequencing Results
We send the following samples (amplified fragments of D. acidovorans DSM-39 and backbone pSB4K5 from the partsregistry) to GATC for sequencing:
- AE (FS_02-FS_03) with Primer FS_02
- AE (FS_02-FS_03) with Primer FS_03
08-08-2013
Amplification from FS_02 to FS_03; 5.3kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02: (1/10) | 4 |
| FS_03: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:30 | |
| 1 | 72 | 7min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE was repeated successfully with the new strain SPH-1
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- if fragment is used in Gibson assembly the amplification has to be repeated to increase the amount of product
07-08-2013
Amplification I from FS_02 to FS_05; 11.2 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02 (1/10) | 2.5 |
| FS_05 (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAF did not work
- Repeat amplification in case any errors were made
Amplification II from FS_02 to FS_05; 11.2 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02 (1/10) | 2.5 |
| FS_05 (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAF did not work as no product appeared, therefore conditions of PCR have to be optimized
- as extremely long amplicons in the past were usually easier to amplify from agar plate (colony PCR) the same conditions will be tried on the colony as template
08-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans SPH-1 (from agarplate) | 1 |
| FS_02 (1/10) | 2.5 |
| FS_05 (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAF did not work as no product appeared, therefore conditions of PCR have to be optimized
- PCR will be repeated with glycerol stock as template
09-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| what | µL 2nd PCR |
|---|---|
| D. acidovorans SPH-1 (glycerol stock) | 1 |
| FS_02 (1/10) | 2.5 |
| FS_05 (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3 min | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF did not work as no product appeared, therefore conditions of PCR have to be optimized
- PCR will be repeated with a higher annealing temperature as primers might not have bound due to secondary structes
- PCR will be repeated with lower annealing temperatue as primers might not have bound due to high temperatures
10-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| Reagent | DelAF | |
|---|---|---|
| Template | D.acidovorans SPH-1 colony | |
| Primer fw | 2 µL FS_02 | 4 µL FS_02 |
| Primer rev | 2 µL FS_05 | 4 µL FS_05 |
| Phusion flash Ready Mix | 10 µL | |
| dd H2O | 1 µL | - |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 3:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF did not work at a temperature of 58°C constant
- as primers might not have been bound at this low temperatue but neither bound at a temperature of 68°C (touchdown or constant), PCR will be repeated at a temperature of 65°C as touchdown and at constant annealing
Amplification II + III from FS_02 to FS_05; 11.2 kb
- Reaction
| Reagent | DelAF II + III | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4.5 µL FS_02 | |||||||||
| Primer rev | 4.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
- Conditions
| Biorad MyCycler | ||||||
|---|---|---|---|---|---|---|
| Cycles | temperature [°C] DelAF II | Time | Cycles | temperature [°C] DelAF III | Time | |
| 1 | 98 | 10 s | 1 | 98 | 10 s | |
| 30 | 98 | 1 s | 12 | 98 | 1 s | |
| 65 ↓ 0.5 | 5 s | |||||
| 65 | 5 s | 72 | 3:15 min | |||
| 18 | 98 | 1 s | ||||
| 72 | 3:15 min | 66 | 5 s | |||
| 72 | 3:15 min | |||||
| 1 | 72 | 10 min | 1 | 72 | 10 min | |
| 1 | 10 | inf | 1 | 10 | inf | |
Results:
- Amplification of DelAF from the new strain finally worked, both conditions (65°C constant and 65°C touchdown) led to specific product but the touchdown PCR resulted in a smear
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be repeated with a slightly lower temperature with constant annealing to increase yield but avoid the smear which appeared due to the low annealing temperatures used in the last cycles of the touchdown PCR.
11-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| Reagent | DelAF |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.5 µL FS_02 |
| Primer rev | 4.5 µL FS_05 |
| Phusion Ready Mix | 10 µL |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 62 | 5 | |
| 72 | 3:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked out though again a smear and several other bands appeared
- bands were cut out very precise to avoid carry over of any unwanted amplicon and DNA purified using QIAquick Gel Extraction Kit
- gradient PCR will be carried out to determine optimal annealing temperature of primers and thereby obtain product of higher specificity suitable for gibson assembly
07-08-2013
Amplification from FS_02 to FS_26; 16.7 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 3:50 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
Results:
- Amplification did not work as expected, nethertheless band was cut out for further amplification with a Re-PCR or test restriction digest.
08-08-2013
Amplification from FS_02 to FS_23; 22.8 kb
- Reaction I
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02: (1/10) | 4 |
| FS_23: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Reaction II
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02: (1/10) | 4 |
| FS_13short: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 6:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
10-08-2013
Amplification from FS_02 to FS_26; 16.7 kb
Reaction
| Reagent | DelAG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4.5 µl FS_02 | |||||||||
| Primer rev | 4.5 µl FS_26 | |||||||||
| DMSO | 1 µl | |||||||||
| Phusion Ready Mix | 10 µl | |||||||||
Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 5:20 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 5:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAG did not work no product was detectable
- PCR will be repeated with different Primers
Amplification from FS_02 to FS_11_short; 22.8 kb and from FS_02 to FS_23; 22.8 kb
- Reaction
| Reagent | DelAG | |
|---|---|---|
| Template | D.acidovorans SPH-1 colony | |
| Primer fw | 4.5 µl FS_02 | 4.5 µl FS_02 |
| Primer rev | 4.5 µl FS_11_short | 4.5 µl FS_23 |
| DMSO | 1 µl | |
| Phusion Ready Mix | 10 µl | |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 7:20 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 7:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAG did not work no product was detectable, therefore a Re-PCR using the amplification of DelAG from 10-08-13 will be conducted
11-08-2013
Re-PCR from FS_02 to FS_11; 22.8 kb; 10-08-2013
- Reaction
| Reagent | DelAG |
|---|---|
| Template | PCR-Product FS02-FS11s from 10-08-2013 |
| Primer fw | 4.5 µl FS_02 |
| Primer rev | 4.5 µl FS_11 |
| Phusion Ready Mix | 10 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 7:20 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 7:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAG did not work, therefore the template used for the Re-PCR presumably was not the desired amplicon
08-08-2013
Amplification from FS_04 to FS_05; 6 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_04: (1/10) | 4 |
| FS_05: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:30 | |
| 1 | 72 | 7min |
| 1 | 12 | inf |
Results:
- Amplification of DelEF worked, though several side products and a smear occured
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
07-08-2013
Amplification from FS_08 to FS_23; 6.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_08: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| dd H2O | 4 |
| DMSO | 1 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
Amplification from SR_01 to FS_23; 6.2 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| SR_01: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
08-08-2013
Restriction digest of fragment SR_01 to FS_23; 6.2 kb; 07-08-2013 with BglII
Incubation at room temperature for about 4h and at 37°C for 1 hours
| what | µl |
|---|---|
| SR_01 to FS_23 (07-08-2013) | 20 |
| BglII | 1 |
| Buffer 3.1 | 3 |
| dd H2O | 6 |
| Expected fragment lengths [bp] | 3415, 3115 |
Results:
- Restriction digest of DelG (SR_01 to FS_23) worked out as predicted, PCR-fragment will be sequenced before potential Gibson Assembly
Concentration measurement
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelG | SR01-FS23 | 07-08-2013 | 34.9 ng/µl |
| DelG | FS08-FS23 | 07-08-2013 | 25.6 ng/µl |
09-08-2013
Amplification from SR_01 to FS_23; 6.2 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| SR_01: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG did not work
- Amplification will be repeated as the protocol was established and carried out successfully several times before, so a pipetting error might be the reason of the failure
08-08-2013
Amplification from FS_22 to FS_13, 2.7 kb
- Reaktion
| what | µl |
|---|---|
| D. acidovorans SPH-1 (colony-pcr) | 1 |
| FS_22: (1/10) | 4 |
| FS_13long: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 50 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated at a higher annealing temperature of 65°C to increase primer binding
Restriction digest of fragment FS_22 to FS_13; 2.7 kb; 07-08-2013) with EcoRI-HF
Incubation at Room temperature for about 4h and at 37°C for 1 hours
| what | µl |
|---|---|
| SR_22 to FS_13 (07-08-2013) | 20 |
| EcoRI-HF | 1 |
| Buffer CutSmart | 3 |
| dd H2O | 6 |
| Expected fragment lengths [bp] | 1883, 960 |
Results:
- Restriction digest did not work, amplicon was not cut and total amount of DNA was probably too low
- Restriction digest will be repeated as soon as PCR of DelOP works reliable and reproducable with higher amounts of DNA
Concentration measurement OP (07-08-2013)
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelOP | FS22-FS13 | 07-08-2013 | 6.1 ng/µl |
09-08-2013
Amplification I from FS_22 to FS_13, 2.7 kb
- Reaktion
| what | µl |
|---|---|
| D. acidovorans SPH-1 (colony-pcr) | 1 |
| FS_22: (1/10) | 4 |
| FS_13long: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated at a lower annealing temperature of 55°C in case primers were not able to bind at high temperatures
Amplification II from FS_22 to FS_13; 2.7 kb
wrong Primers were used
- Reaktion
| what | µl |
|---|---|
| D. acidovorans SPH-1 (colony-pcr) | 1 |
| FS_22: (1/10) | 4 |
| FS_13long: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work as the wrong primers were used
- Amplification will be repeated with different amounts of DMSO
Amplification III from FS_22 to FS_13; 2.7 kb
3 samples, one with, two without DMSO (one with much template, one with less template)
- Reaktion
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| FS_22: (1/10) | 4 |
| FS_13long: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 0/1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work, neither with lower amount of template, nor with higher, furthermore the DMSO concentration did not influence the PCR for the given annealing temperature
- PCR will be repeated with recently ordered new Primers which bind outside the intended template in the Del-Cluster of D. acidovorans SPH-1 in order to obtain a specific template suited for a nested PCR of DelOP with Primers FS_22 and FS_14l
10-08-2013
Amplification I, nested PCR different Primers; 2.7 kb
- Reaction
| Reagent | DelOP | ||||
|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | ||||
| Primer fw | 4 µl FS_32 | 4 µl FS_32 | 4 µl FS_32 | 4 µl FS_32 | 4 µl FS_32 |
| Primer rev | 4 µl FS_13l | 4 µl FS_27 | 4 µl FS_28 | 4 µl FS_29 | 4 µl FS_30 |
| Phusion Ready Mix | 10 µl | ||||
| dd H2O | 1 µl | ||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated as touchdown PCR with annealing starting at 68°C and different primer combinations to the template needed for the nested PCR of DelOP with primers FS_22 to FS_13l
Amplification II, nested PCR with different primers; 2.7 kb
- Reaction
| Reagent | DelOP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4 µl FS_22 | 4 µl FS_22 | 4 µl FS_33 | 4 µl FS_33 | 4 µl FS_31 | 4 µl FS_31 | 4 µl FS_32 | 4 µl FS_32 | 4 µl FS_22 | 4 µl FS_22 |
| Primer rev | 4 µl FS_13s | 4 µl FS_27 | 4 µl FS_13s | 4 µl FS_27 | 4 µl FS_29 | 4 µl FS_30 | 4 µl FS_29 | 4 µl FS_30 | 4 µl FS_28 | 4 µl FS_29 |
| DMSO | 1 µl | |||||||||
| Phusion Ready Mix | 10 µl | |||||||||
| dd H2O | 1 µl | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelOP worked with the Primer combinations FS22-FS13short, FS_22-FS_27, FS_33-FS_13short, FS_33-FS_27, FS_32-FS_30 and FS_22-FS28
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- all the obtained PCR products will be used as specific templates for a nested PCR using primers FS_22-FS_13long
Amplification III from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4 µl FS_22 | |||||||||
| Primer rev | 4 µl FS_13l | |||||||||
| DMSO | 1 µl | |||||||||
| Phusion Ready Mix | 10 µl | |||||||||
| dd H2O | 1 µl | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 1:00 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work
- hope and wait for the nested PCR
11-08-2013
Nested PCR of DelOP-Fragments from 10-08-2013
- Reaction
| Reagent | DelOP | ||||||
|---|---|---|---|---|---|---|---|
| Template | FS22-13s | FS31-30 | FS33-27 | FS22-28 | FS22-13s | FS31-30 | FS33-29 |
| Primer fw | 4.5 µl FS_22 | ||||||
| Primer rev | 4.5 µl FS_13l | ||||||
| DMSO | 1 µl | ||||||
| Phusion Ready Mix | 10 µl | ||||||
- Conditions
| Biometra TProfessional Basic | |||||
|---|---|---|---|---|---|
| Cycles | temperature [°C] Template FS22-13s, FS31-30, FS33-27, FS22-28 | Time | Cycles | temperature [°C] Template FS22-13s, FS31-30, FS33-29, Time | |
| 1 | 98 | 10 s | 1 | 98 | 10 s |
| 30 | 98 | 1 s | 12 | 98 | 1 s |
| 68 ↓ 0.5 | 5 s | ||||
| 58 | 5 s | 72 | 1:00 min | ||
| 18 | 98 | 1 s | |||
| 72 | 1:00 min | 66 | 5 s | ||
| 72 | 1:00 min | ||||
| 1 | 72 | 5 min | 1 | 72 | 5 min |
| 1 | 10 | inf | 1 | 10 | inf |
Results:
- Nested PCR of DelOP did not work, though concentrations of the used template were fine as checked previously
- PCR will be with an annealing temperature of 55°C with the template obtained from the primer combination of FS_22-FS_13short as nested PCR
07-08-2013
Results Sequencing
Send the following samples (amplified fragments of D. acidovorans DSM-39 and backbone pSB4K5 from the partsregistry) to GATC for sequencing:
- DelL (FS_14 to FS_15) with Primer FS_14
- DelL (FS_14 to FS_15) with Primer FS_15
Amplification of DelL (FS_14 to FS_15; 1.4 kb)
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_14: (1/10) | 2.5 |
| FS_15: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 42 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 42 | |
| 1 | 72 | 12 min |
| 1 | 4 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
08-08-2013
Concentration measurement of DelL
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelL | FS14-FS15 | 07-08-2013 | 51 ng/µl |
09-08-2013
Amplification of DelL (FS_14 to FS_15; 1.4 kb)
- Reaction
| what | µl |
|---|---|
| D. acidovorans | 1 |
| FS_14: (1/10) | 2.5 |
| FS_15: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 42 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 42 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
07-08-2013
Results Sequencing
Send the backbone pSB4K5 from the partsregistry to GATC for sequencing:
- pSB4K5 with Primer FS_01
- pSB4K5 with Primer FS_16
Results:
- The sequence is the same as expected and indicated in the partsregistry.
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(2x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 98 | 1 |
| 60 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
08-08-2013
Concentration measurement
The concentration of the gel purified fragment was measured using a NanoDrop Instrument.
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| pSB4K5 | FS16-FS01 | 07-08-2013 | 55.9 ng/µl |
08-08-2013
Restriction digest of fragment from FS_21 to FS_26; 5.5 kb; 29-07-2013) with ClaI
Incubation at Room temperature for about 4h and at 37°C for 1 hours
| what | µl |
|---|---|
| FS_21 to FS_26 (29-07-2013) | 10 |
| ClaI | 1 |
| Buffer CutSmart | 1.5 |
| dd H2O | 2.5 |
| Expected fragment lengths [bp] | 2743, 1519, 1208 |
Results:
- restriction digest of DelFG led to the expected fragment sizes, therefore restriction digest, implies amplification of the correct fragment
- PCR product will be further validated by single read sequencing before Gibson Assembly
Concentration measurement DelFG
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelFG | FS21-FS26 | 07-08-2013 | 24.3 ng/µl |
09-08-2013
Amplification from FS_21 to FS_26; 5.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans SPH-1 (colony-pcr) | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
Strategy
Most PCRs have worked!! Yeah- finally!!! Thus, all the fragments needed for Gibson assembly are now available, however yields are still suboptimal for some of the fragments amplified. Furthermore, we designed screening primers for our final construct pFSN. These primers will be used to analyze our transformed bacteria for presence of the complete DelRest construct pFSN by colony-PCR.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_47_screening_BB_AF_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGGCCGATTCATTAATGC |
| FS_48_screening_BB_AF_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TAACGGTATCGGTATCGCTTTG |
| FS_49_screening_AFI_AFII_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTTCTCTGGAAGATGGATAC |
| FS_50_screening_AFI_AFII_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGACGAAAAAGCCGACCAC |
| FS_51_screening_AF_FG(21-26)_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGGATATCGACTGGACTGCCTG |
| FS_52_screening_AF_FG(21-26)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGCACCACATCGACGAAACGG |
| FS_53_screening_FG(21-26)_G_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTACGGCCTATCACATCAGCG |
| FS_54_screening_FG(21-26)_G_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACCTGGGTGTTCACGAAAAAGCC |
| FS_55_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTATCTCTACATGCATCGCTAC |
| FS_56_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | AGGACATTTTCCGCACCCCG |
| FS_57_screening_G_OP_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GCTGGCGTTTTCCATAAG |
| FS_58_screening_OP_L_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACAACTTCCAGCACAGCCTGTTC |
| FS_59_screening_OP_L_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CGTTGAAGATTTCGTTGACG |
| FS_60_screening_L_BB_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CATCTTCAAGGTGTTCTATGAAC |
| FS_61_screening_L_BB(with_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CAGTTTAACTTTGTAGATGAAC |
| NK_01_FS_62_screening_L_BB(without_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTCACCGACAAACAACAGATAAAACG |
Amplification and purification of Del Genes from SPH-1
Goals
As we were able to establish amplification protocols for most of our desired amplicons by the end of last week, we will repeat these PCRs to obtain the required amounts and concentrations. Furthermore we will optimize the PCR conditions of DelO-P by using higher annealing temperatures, hopefully inhibiting secondary structe formation of the primers. Afterwards we will assemble our construct, transform it into E.Coli DH10B and screen for (hopefully many) positive clones using the recently ordered screening primers.
Results
| PCRs from D.acidovorans SPH-1 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-F | Primer FS_02 and FS_05 | ||
| DelA-G | Primer FS_02 and FS_11 | |||
| Primer FS_02 and FS_11_short | ||||
| DelF-G | Primer FS_21 and FS_07 | |||
| Primer FS_21 and FS_26 | ||||
| DelG | Primer SR_01 and FS_23 | |||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13_short | ||
| Primer FS_22 and FS_13 | ||||
| Test restriction digests of PCR amplified fragments | ||||
|---|---|---|---|---|
| Fragment | Primer | Digestion enzyme | Expected bands | |
| DelO-P | Primer FS_22 and FS_13 | EcoRI-HF | 1883, 960 | |
12-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4.5 µL FS_02 | |||||||||
| Primer rev | 4.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 67.5 - 65.0 (ΔT = 0.5) ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 65.5 - 63.0 (ΔT = 0.5) | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked well, gradient displays an optimal annealing temperature of 65.5°C, which will be used for further amplifications
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
13-08-2013
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 12-08-2013 with EcoRI-HF
Incubation at 37°C for 2 hours
| what | µL |
|---|---|
| FS_02 to FS_05 (12-08-2013) | 20 |
| EcoRI-HF | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 1.5 |
Expected fragment sizes: 2.26kbp; 4.62kbp; 4.32kbp
Results:
- Weak bands of about 4.6kbp visible, as well as on of about 2.6kbp
- digest will be repeated using higher concentrations of DNA to clearify results of restriction digest
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
1 sample contains 2µL of DMSO
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4.5 µL FS_02 | |||||||||
| Primer rev | 4.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65.5 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 63.5 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked though a slight smear occured, therefore PCR will be repeated on the more precise Biometra TProfessional Basic cylcer again
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
14-08-2013
Concentration measurement (FS_02 to FS_05; 11.2 kb)
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelAF | FS02-FS05 | 11-08-2013 | ~10 ng/µL |
| DelAF | FS02-FS05 | 12-08-2013 | 0 ng/µL |
| DelAF | FS02-FS05 | 12-08-2013 | 0 ng/µL |
Results:
- concentrations are not sufficient for gibson assembly
- PCR will be repeated and gel slices of different reactions will be pooled for one gel extraction using QIAquick Gel Extraction Kit to obtain the concentrations needed for gibson assembly
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 11-08-2013 with EcoRI-HF
Incubation at 37°C for 2 hours 45min
| what | µL |
|---|---|
| FS_02 to FS_05 (11-08-2013) | 18 |
| EcoRI-HF | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 3.5 |
Expected fragment sizes: 2.26kbp; 4.62kbp; 4.32kbp
Results:
- One band of about 5kbp and one of 2.5kbp
- no clear result, digest will be repeated with another enzyme (ClaI), as the enzyme used was beyond expiration date
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 10-08-2013 with ClaI
Incubation at 37°C for 2 hours
| what | µL |
|---|---|
| FS_02 to FS_05 (10-08-2013) | 20 |
| ClaI | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 1.5 |
Expected fragment sizes: 6.9kbp, 4.3kbp
Results:
- restriction digest displays the expected fragments, therefore amplification of the desired fragment can be assumed
- PCR product will be prepared for sequencing by GATC to proof amplification of the desired DNA sequence before Gibson Assembly
17-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction of DelAF
6x20 µL
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 2.5 µL FS_02 | |||||||||
| Primer rev | 2.5 µL FS_05 | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
| DMSO | 1 µL | |||||||||
| dd H2O | 4 µL | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65.5 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 63.5 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked
- bands were cut out, pooled and DNA purified using QIAquick Gel Extraction Kit to obtain concentrations needed for gibson assembly
18-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction of DelAF
6x20 µL
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 2.5 µL FS_02 | |||||||||
| Primer rev | 2.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
| dd H2O | 4 µL | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65.5 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 63.5 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
12-08-2013
Amplification from FS_02 to FS_11; 22.8 kb
- Reaction
| Reagent | DelAG |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.5 µl FS_02 |
| Primer rev | 4.5 µl FS_11 |
| DMSO | 1 µl |
| Phusion Ready Mix | 10 µl |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 72.0 - 67.0 (ΔT = 0.5) ↓ 0.5 | 5 | |
| 72 | 7:20 | |
| 18 | 98 | 1 |
| 70.0 - 65.0 (ΔT = 0.5) | 5 | |
| 72 | 7:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- none of the amplifications led to yields of DNA sufficient for Gibson Assembly, therefore the used combination of primers seems not to be sufficient for amplification of 22.0 kbp
13-08-2013
Amplification from FS_02 to FS_11_short; 22.8 kb
- Reaction
| Reagent | DelAG |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.0 µl FS_02 |
| Primer rev | 4.0 µl FS_11_short |
| DMSO | 2 µl |
| Phusion Ready Mix | 10 µl |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 72.0 - 67.0 (ΔT = 0.5) ↓ 0.5 | 5 | |
| 72 | 7:20 | |
| 18 | 98 | 1 |
| 70.0 - 65.0 (ΔT = 0.5) | 5 | |
| 72 | 7:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
15-08-2013
Amplification from SR_01 to FS_23; 6.2 kb
- Reaction (5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| SR_01: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- Bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- Amplification will be repeated to increase the concentration for the Gibson Assembly
16-08-2013
Concentration measurement
the fragment was gel purified
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelG | SR01-FS23 | 15-08-2013 | 52.6 ng/µl |
| DelG after nucleotide removal | SR01-FS23 | 15-08-2013 | 30 ng/µl |
Amplification from SR_01 to FS_23; 6.2 kb
- Reaction (5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| SR_01: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- Bands were cut out and DNA purified using QIAquick Gel Extraction Kit
12-08-2013
Amplification I from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP | |
|---|---|---|
| Template | D.acidovorans SPH-1 colony | 1µl FS_22-FS_13_short 10-08 |
| Primer fw | 2 µl FS_22 | |
| Primer rev | 2 µl FS_13 | |
| Phusion Ready Mix | 10 µl | |
| dd H2O | 5 µl | |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated at very high annealing temperatures to ensure that primers with very complex secondary structure such as the Gibson-Primer FS_13long are able to bind
Amplification II and III from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP |
|---|---|
| Template | 1µl FS_22-FS_13_short 10.08 |
| Primer fw | 4 µl FS_22 |
| Primer rev | 4 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
- Conditions
| Biorad MyCycler | ||||||
|---|---|---|---|---|---|---|
| Cycles | temperature [°C] DelOP II | Time | Cycles | temperature [°C] DelOP III | Time | |
| 1 | 98 | 10 s | 1 | 98 | 10 s | |
| 30 | 98 | 1 s | 12 | 98 | 1 s | |
| 74.0 - 70.0 (ΔT = 2.0) ↓ 0.5 | 5 s | |||||
| 72 | 1:00 min | 72 | 3:15 min | |||
| 18 | 98 | 1 s | ||||
| 66 | 5 s | |||||
| 68 / 70 / 72 | 1:00 min | |||||
| 1 | 72 | 10 min | 1 | 72 | 10 min | |
| 1 | 10 | inf | 1 | 10 | inf | |
Results:
- Amplification of DelOP was successful, 2-step PCR with FS_22-FS_13s as template led to expected bands as well as a slight smear and an unintended product
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be repeated to obtain more sample
Re-PCR from FS_22 to FS_13; 2.7 kb; 10-08-2013)
3x 20µl
- Reaction
| Reagent | DelOP |
|---|---|
| Template | 1µl FS_22-FS_13_short 10-08-2013 |
| Primer fw | 4 µl FS_22 |
| Primer rev | 4 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:05 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successfull
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- gradient PCR with annealing at 74°C, 72°C and 70°C will be carried out to increase yield
Amplification V from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.5 µl FS_22 |
| Primer rev | 4.5 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 75.0 - 72.0 (ΔT = 0.5) | 1:05 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successfull
- The gradient PCR shows that amplification worked best with an annealing temperature of 72.3°C, interestingly this annealing temperature was used for the remaining mastermix of the gradient PCR which included more DMSO than the other samples as mixing of DMSO often is not perfect and therefore the very last sample is of higher DMSO concentration
- Therefore PCR will be repeated with an annealing temperature of 72.3°C and 10% DMSO
13-08-2013
Restriction digest of fragment FS_22 to FS_13; 2.7 kb; 12-08-2013) with EcoRI-HF
Incubation at 37°C for 2 hours
| what | µl |
|---|---|
| FS_22 to FS_13 (12-08-2013) | 20 |
| EcoRI-HF | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 1.5 |
| Expected fragment lengths [bp] | 1883, 960 |
Results:
- restriction digest shows the expected bands, therefore validation if PCR amplicon is sufficient for Gibson Assembly
Amplification from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4 µl FS_22 |
| Primer rev | 4 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1/2 µl |
| dd H2O | 1/- µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:05 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successfull with 10% DMSO leading to one specific band of intended size
- PCR will be repeated with 10% DMSO to obtain the amount of product which is necessary for Gibson Assembly and test restriction digest
14-08-2013
Concentration measurement DelOP
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelOP | FS22-FS13 | 12-08-2013 | 11 ng/µl |
17-08-2013
Amplification from FS_22 to FS_13; 2.7 kb
6x20 µl
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 2.5 µl FS_22 |
| Primer rev | 2.5 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 2 µl |
| dd H2O | 3 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
18-08-2013
Amplification from FS_22 to FS_13; 2.7 kb
6x20 µl
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 2.5 µl FS_22 |
| Primer rev | 2.5 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 2 µl |
| dd H2O | 3 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
15-08-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(2x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
Restriction digest of pSB4K5 (FS_01 to FS_16; 4.2 kb; 15-08-2013) with DpnI
Incubation at 37°C for about 6 hours
| what | µl |
|---|---|
| FS_16 to FS_01 (15-08-2013) | 30 |
| DpnI | 2 |
| CutSmart Buffer | 4 |
| dd H2O | 4 |
Results:
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
16-08-2013
Concentration measurement
The concentration of the gel purified, and DpnI digested fragment was measured using a NanoDrop Instrument.
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| pSB4K5 DpnI digested | FS01-FS16 | 15-08-2013 | 30 ng/µl |
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(3x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
17-08-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(3x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
Restriction digest of pSB4K5 (FS_01 to FS_16; 4.2 kb; 17-08-2013) with DpnI
Incubation at 37°C for about 6 hours
| what | µl |
|---|---|
| FS_16 to FS_01 (17-08-2013) | 30 |
| DpnI | 2 |
| CutSmart Buffer | 4 |
| dd H2O | 4 |
Results:
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
18-08-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(3x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Restriction digest of pSB4K5 (FS_01 to FS_16; 4.2 kb; 18-08-2013) with DpnI
Incubation at 37°C for about 6 hours
| what | µl |
|---|---|
| FS_16 to FS_01 (18-08-2013) | 30 |
| DpnI | 2 |
| CutSmart Buffer | 4 |
| dd H2O | 4 |
Results:
- Digested fragment was run on a gel.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
13-08-2013
Amplification from FS_21 to FS_07; 5.2 kb
- Reaction
| Reagent | µl |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4 µl FS_21 |
| Primer rev | 4 µl FS_07 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
| dd H2O | 1 µl |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 2:00 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG worked, but amount of DNA was insufficient
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be carried out with another cylcer which by experience of the last amplifications led to higher product yields.
15-08-2013
Amplification from FS_21 to FS_26 ; 5.5 kb
- Reaction
(5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- obtained DNA will consequently be used for Gibson Assembly
- PCR will be repeated to get higher amounts of DNA and increase concentration by purifying several gel slices using the same QIAquick spin column
16-08-2013
Concentration measurement DelFG
all fragments were gel purified, FG were additionally purified with the nucleotide removal kit
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelFG | FS21-FS26 | 29-07-2013 | 61.4 ng/µl |
| DelFG after nucleotide removal | FS21-FS26 | 29-07-2013 | 30 ng/µl |
Amplification from FS_21 to FS_26 ; 5.5 kb
- Reaction
(5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- obtained DNA will consequently be used for Gibson Assembly
15-08-2013
Gibson Assembly and electroporation
Assembly of our fragments
Those gel-purified fragments were added for the Gibson assembly of our construct pFSN.
| Fragment | Primer | Date | Concentration (ng/µl) | Volume (µl) |
|---|---|---|---|---|
| pSB4K5 | FS_01 - FS_16 | 14-08-13 | 13 | 1.35 |
| AF | FS_02 - FS_05 | 13-08-13 | 49.75 | 2.65 |
| FG | FS_21 - FS_26 | 07-08-13 | 24.5 | 2.69 |
| G | FS_35_SR_01 - FS_23 | 08-08-13 | 34.9 | 2.1 |
| OP | FS_22 - FS_13 | 13-08-13 | 36.3 | 0.89 |
| L | FS_14 - FS_15 | 07-08-13 | 51 | 0.31 |
| Gibson MasterMix NEB | 2x | 10 | ||
- Gibson Assembly according to protocols of NEB, 1h at 50°C
Gibson backbone (pSB4K5)
The backbone pSB4K5 was also incubated with the Gibson mastermix. We will use this as one negative control in the electroporation to test the efficiency.
| Fragment | Date | Concentration (ng/µl) | Volume (µl) |
|---|---|---|---|
| pSB4K5 | 14-08-13 | 13 | 1.35 |
| H2O | 8.65 | ||
| Gibson MasterMix NEB | 2x | 10 | |
- Gibson Assembly according to protocols of NEB, 1h at 50°C
Electroporation
The following samples were electroporated
| mixture | volume used | cells | |
|---|---|---|---|
| 5 µl Gibson Assembly of pFSN construct | 10 µl H2O | 1 µl | 50 µl DH10β old |
| 5 µl Gibson Assembly of pFSN construct | 10 µl H2O | 14 µl | 50 µl DH10β old |
| 5 µl Gibson Assembly of Backbone (control) | 10 µl H2O | 1 µl | 50 µl DH10β old |
| pSB4K5 14-08-13 | 1.35 µl | 50 µl DH10β old | |
| pSB6A1 Hanna [69.57 ng/µl] 1:10 | 1 µl | 50 µl DH10β Fanny new | |
- Cells were transdered into 400 µl SOC-medium (NEB) after electroporation
- incubation and growth of cells for 1 hour at 37°C
- cells were centrifuged for 3 min at 6000rpm, supernatant discarded and cells resuspended in remaining SOC-medium
- cells were plated on agar plates containing kanamycine, 10µl on one plate, remaining µl on another plate for electroporation of the construct pFSN and on agar plates containing ampicillin for electroporation of the control backbone pSB6A1
20130816ElectroporationA.JPG
1 µl of diluted Gibson electroporated; 10 µl of E.coli plated |
20130816ElectroporationB.jpg
1 µl of diluted Gibson electroporated; Rest of E.coli plated |
20130816ElectroporationC.jpg
14 µl of diluted Gibson electroporated; 10 µl of E.coli plated |
20130816ElectroporationD.jpg
14 µl of diluted Gibson electroporated; Rest of E.coli plated |
P1010689.JPG
1 µl of diluted Gibson (backbone control) electroporated; 10 µl of E.coli plated |
P1010690.JPG
1 µl of diluted Gibson (backbone control) electroporated; Rest of E.coli plated |
P1010692.JPG
1 µl of backbone control electroporated; 10 µl of E.coli plated |
P1010693.JPG
1 µl of backbone control electroporated; Rest of E.coli plated |
P1010704.JPG
Test of electrocompetence DH10β with 1 µl pSB6A1; 10 µl of E.coli plated |
P1010705.JPG
Test of electrocompetence DH10β with 1 µl pSB6A1; Rest of E.coli plated |
16-08-2013
Colony PCRs
As a lot of colonies grew we screened if DelL and pSB4K5 are present.
Different probes:
- A: 1 µl of diluted Gibson electroporated; 10 µl of E.coli plated
- B: 1 µl of diluted Gibson electroporated; Rest of E.coli plated
- C: 14 µl of diluted Gibson electroporated; 10 µl of E.coli plated
- D: 14 µl of diluted Gibson electroporated; Rest of E.coli plated
- Only red colonies were chosen for screening
- The expected amplicon size is 1.5 kb
- Reaction mixture
| Reagent | A1, A2 | B1-B5 | C1-C3 | D1-D40 |
|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | 2 red colonies A (A1, A2) | 5 red colonies B (B1-B5) | 3 red colonies C (C1-C3) | 40 red colonies D (D1-D40) |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl |
- PCR Conditions
--> T100
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 12 | 95 | 1:00 |
| 66 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 95 | 1:00 |
| 63 | 5 | |
| 72 | 1:30 min | |
| 1 | 12 | inf |
Result:
- None of the screened colonies led to the correct amplicon, therefore the correct assembly has not worked in any of the white colonies
- screening will be repeated, with white colonies as template, as bacteria including the correct 32 kbp backbone might not be capable of expressing mRFP anymore due to the large insert between lac promotor and mRFP
17-08-2013
Colony PCRs
Different probes:
- A 1 µl of diluted Gibson Assembly electroporated; 10 µl of E.coli plated
- B 1 µl of diluted Gibson Assembly electroporated; remaining E.coli plated
- C 14 µl of diluted Gibson Assembly electroporated; 10 µl of E.coli plated
- D 14 µl of diluted Gibson Assembly electroporated; remaining E.coli plated
- Only white colonies where chosen for screening
- The expected amplicon size is 1.5 kb
- Reaction mixture
| Reagent | B1w | C1w | D1w-D10w |
|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl |
| Colonies | 1 white colony B (B1w) | 1 white colony C (C1w) | 10 white colonies D (D1w-D10w) |
| DreamTaq | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl |
- PCR Conditions
--> T100
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 12 | 95 | 1:00 |
| 66 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 95 | 1:00 |
| 63 | 5 | |
| 72 | 1:30 min | |
| 1 | 12 | inf |
Result:
- Colony D8w shows expected amplicon of 1.5kbp
- The other colonies were screened negative.
- D8w will be transfered to liquid culture to repeat screening PCR as well as for further validation beeing screening for the other insert fragments as well as test restriction digest
- Furthermore another electorporation will be carried out to yield more clones positive for screening with the Primers VR and FS_14, therefore the remaining 10µl of the Gibson assembly will be purified using isopropanol precipitation
Electroporation
We electroporated the Gibson assembly (15-08) again to increase the chance of a positive clone. We purified it by isopropanol purification.
| mixture | volume used | Cells | |
|---|---|---|---|
| Assembled fragments (15-08) isopropanol precipitated | 15 µl | 50 µl DH10β | |
- Cells were transdered into 400 µl SOC-medium (NEB) after electroporation
- incubation and growth of cells for 1 hour at 37°C
- cells were centrifuged for 3 min at 6000rpm, supernatant discarded and cells resuspended in remaining SOC-medium
- cells were plated on agar plates containing kanamycine, 10µl on one plate, remaining µl on another plate for electroporation of the construct pFSN
P1010701.JPG
15 µl of Gibson 15-08 (isopropanol precipitated) electroporated; 10 µl of E.coli plated |
P1010702.JPG
15 µl of Gibson 15-08 (isopropanol precipitated) electroporated; Rest of E.coli plated |
18-08-2013
Colony PCRs
Different probes:
- D: 14 µl of diluted Gibson Assembly electroporated; remaining E.coli plated
- E: 1 µl isopropanol precipitated Gibson Assembly (15-08) ; 10 µl of E.coli plated
- F: 1 µl isopropanol precipitated Gibson Assembly (15-08); remaining E.coli plated
- The expected amplicon size is 1.5 kb
- 3 colonies were sceened in one PCR
- Reaction mixture
| Reagent | D8w | D11w-D24w | E1w-E19w | F1w-F20w | E1r-E10 | F1r-F20r |
|---|---|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | Colony D8w liquid culture | 24 white colonies D (D1w-D24w) | 19 white colonies E (E1w-E19w) | 30 white colonies F (F1w-F20w) | 10 red colonies E (E1r-E10r) | 20 red colonies F (F1r-F20r) |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
- PCR Conditions
--> T100
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 12 | 95 | 1:00 |
| 66 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 95 | 1:00 |
| 63 | 5 | |
| 72 | 1:30 min | |
| 1 | 12 | inf |
Result:
- colony D8w again showed the expected amplicon in the colony PCR and will therefore be further analyzed for the other fragments as well as restriction digested and partially sequenced as soon as screening primers arive
- Furthermore colonies E16w and F25w are screened positive for the fragment DelL
Validation of pFSN
We screened for more colonies with pFSN but we didn't find any more positive clones.
Furthermore we did some tests for the validation of pFSN in colony D8w.
Results
| Validation of pFSN in clone D8w | ||
|---|---|---|
| Method | Details method | Result |
| Screening for inserted fragments | Fragments pSB4K5 and DelA-F | |
| Fragments DelA-F and DelF-G | ||
| Restriction Digest | EcoRI | |
| BamHI | ||
| BglII | ||
| Sequencing gibson overlaps | Primer FS_13_short | |
| Primer FS_22 | ||
| Primer VFII | ||
| Primer VR | ||
There is a mutation at the beginning of the mRFP cassette. This explains why the colonies do not get red. All other tests for valiation were positive. Consequently colony D8w includes the right plasmid. Because we cannot use mRFP as expression control we will do SDS-PAGE instead to prove that the proteins are expressed.
19-08-2013
Concentration measurement
The concentration of the gel purified, and DpnI digested fragments was measured using a NanoDrop Instrument.
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| pSB4K5 DpnI digested | FS_01-FS_16 | 17-08-2013 | 101.2 ng/µl |
| pSB4K5 DpnI digested | FS_01-FS_16 | 18-08-2013 | 159.2 ng/µl |
19-08-2013
Colony PCRs for testing ligation of pSB4K5 and DelL
- Reaction mixture
| Reagent | E13w | E14w | E15w | F25w | F26w | F27w |
|---|---|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | Colony E13w liquid culture | Colony E14w liquid culture | Colony E15w liquid culture | Colony F25w liquid culture | Colony F26w liquid culture | Colony F27w liquid culture |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
Result:
- Colony F26w was screened positive, as it shows the expected band at 1.4kbp, the others were screened negative
- 8 ml of colony D8w and colony D26w (pFSN-1) were midi-preped according to protocol 'Preparation of large plasmids'.
Colony PCRs for testing ligation of pSB4K5 and DelAF, as well as DelAF and DelFG
- Reaction mixture
| Reagent | D8w | D8w | F26w | F26w |
|---|---|---|---|---|
| Primer_fw (1/10) | 2 µl FS_47 | 2 µl FS_51 | 2 µl FS_47 | 2 µl FS_51 |
| Primer_rv (1/10) | 2 µl FS_48 | 2 µl FS_52 | 2 µl FS_48 | 2 µl FS_52 |
| Colonies | Colony D8w liquid culture | Colony D8w liquid culture | Colony F26w liquid culture | Colony F26w liquid culture |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl |
| Expected band | 547 bp | 724 bp | 547 bp | 724 bp |
Results:
- The screening was positive for clone D8w with both primer combinations.
- Thus this clone contains the backbone pSB4K5, the genes DelA, DelB, DelC, DelD, DelE, DelF, DelL and at least a part of the gene DelG.
- For colony F26w the screenings were negativ.
Restriction digest of pFSN (31.435 kbp)
Incubation at 37°C for about 6 hours
| Reagent | µl | ||
|---|---|---|---|
| pFSN | 1 | ||
| EcoRI | BamH1 | BglII | 2 |
| CutSmart Buffer | 0.5 | ||
| dd H2O | 1.5 | ||
| Expected fragment lengths [bp] EcoRI | 9798, 7856, 5276, 4624, 2529, 1212 | ||
| Expected fragment lengths [bp] BamHI | 23860, 7435 | ||
| Expected fragment lengths [bp] BglII | 14223, 11919, 3291, 1892 | ||
Results:
- All digests displayed the expected bands and thus were positive.
- Therefore the pFSN plasmid is very likely our desired construct.
20-08-2013
Sequencing
- The construct pFSN was isolated from overnight culture using the 'Preparation of large plasmids' protocol.
- Single read sequencing was carried out by GATC Biotech
- obtained .abi files were analyzed with UniPro Ugene, part of sequence showing convenient chromatogram was chosen for alignment using ClustalOmega
Sequence-Files from GATC Biotech
Alignments
Results:
- Sequencing of the final construct pFSN for all internal Gibson sites validated the intended sequence
- Sequence of mRFP shows mutations within the primer site, therefore it can be concluded that mRFP is not functional
- Colonies of D8w will be further analyzed by FACs to investigate whether mRFP is functional or not
- Additionaly, SDS-page of clone D8w will be conducted to check for expression of the inserted Del genes
- Concluding the results of colony PCRs, restriction digests and sequencing it can be concluded, that we successfully amplified 28 kbp of genomic DNA from our donator organism Delftia acidovorans SPH-1 distributed from the DSMZ, ligated the thereby obtained 6 fragments including the standard biobrick backbone pSB4K5 sucessfully in the intended order and consequently transformed a plasmid of 32 kbp in total into E. Coli using electroporation
21-08-2013
- Another 8 ml of colony D8w (pFSN-1) were midi-preped according to protocol 'Preparation of large plasmids'.
Validation of pFSN
This week we will analyze our positive D8w clone for expression of our pFSN plasmid. Therefore a SDS page will be carried out
Results
This week we carried out an SDS-PAGE with our clone D8w containing the pFSN plasmid. We used E.coli DH10ß as control. For our transformed E.coli a clear band at about 190 kDa was visible. This corresponds to the molecular weight of DelE (192,473 kDa). Consequently we showed that DelE is recombinantly expressed. Futhermore one could sense a very weak band at the height of about360 kDa. However this band could not be captured on an image. Consequently the samples have to be applied on a gel in a higher about again.
30-08-2013
SDS-PAGE
Lysis of cells
- 1 ml over night cultures in LB of electrocompetent DH10ß, DH10ß with pSB4K5 and DH10ß with pFSN
- meausured ODs (DH10ß= 1,32; DH10ß+pSB4K5= 1,28; DH10ß+pFSN= 1,12)
- centrifugation at 13000 rpm for 10 min
- discarded supernatant
- resuspended in 66 µl(DH10ß), 64 µl(DH10ß+pSB4K5) and DH10ß+pFSN= 56 µl 1x loading buffer (40% ddH2O, 12.5% TrisHCl pH=7.5, 20% Glycerol (50%), 20% SDS (10%), 5% β-Mercaptoethanol, 2.5% Bromophenol blue)
- boiled samples for 10 min at 98°C
Electrophoresis
- Applied 2.5 µl and 10 µl of each sample on SDS-polyacrylamide gel
- Let it run for 1 h at 180 V
Staining and destaining procedure
- Gel was stained in Coomassie Brilliant Blue for 1 h on shaker
- Destaining with destaining buffer (50% ddH2O, 40% Methanol, 10% Acetic acid) until bands were visible
Results:
- One band at a height of about 190 kDa was visible in DH10ß+pFSN, but not in the controls. As the protein DelE has a molecular weight of 192.52 kDa this band is probably DelE.
- A very weak band could be detected well above the 260 kDa band of the ladder. We suspect that this might be DelG, as it has a molecular weight of 357.33 kDa, however the band was too weak to capture it in a picture. Thus a bigger amount of the samples have to be applied on a SDS-PAGE again.
Validation of pFSN
SDS-PAGE will be repeated to analyze expression of Del genes from our plasmid pFSN
Results
This week we appiled the samples of week 18 on an SDS-PAGE again, as last time we were able to detect DelE, but only saw a band at the height of DelG which was so weak that it could not be captured on a photograph. Therefore we applied more of the samples. The results were positiv. A band could be detected at about 360 kDa was visible. The band of DelE was present again as well. Therefore we were able to prove that DelG is also expressed. As the genes DelA, DelB, DelC, DelD and DelF are regulated by the same promotor as DelE and DelG we assume that these proteins are expressed likewise.
02-09-2013
SDS-PAGE of Samples 30-08-2013
Electrophoresis
- Applied 15 µl and 25 µl of the sample DH10ß+pSB4K5 and DH10ß+pFSN 30-08-2013 on a SDS-polyacrylamide gel again.
- Let it run for 1 h at 180 V
Staining and destaining procedure
- Gel was stained in Coomassie Brilliant Blue for 1 h on shaker
- Destaining with destaining buffer (50% ddH2O, 40% Methanol, 10% Acetic acid) until bands were visible
Results:
- Bands of about 190 kDa and 350 kDa were visible in the sample DH10ß+pFSN but not DH10ß+pSB4K5. These bands correlate to the proteins DelE (192.52 kDa) and DelG (357.33 kDa). The other proteins which should be expressed by our plasmid could not be detected as there is much background of the proteins of the cells at these heights.
- Still, as the genes DelA, DelB, DelC, DelD and DelF are closer to the promotor as DelG, we suspect that these genes are expressed as well.
 "
"