Team:Heidelberg/Tyrocidine
From 2013.igem.org
(Difference between revisions)
| (24 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
} | } | ||
.carousel-inner { | .carousel-inner { | ||
| - | margin-top: | + | margin-top:15%; |
} | } | ||
| Line 16: | Line 16: | ||
<div class="container"> | <div class="container"> | ||
<!--Project Description--> | <!--Project Description--> | ||
| - | <div style=" | + | <div> |
| - | + | <h1><span style="font-size:180%;color:#800000;">Novel NRPS</span> <span style="font-size:180%;color:#666666;">&</span> <span style="font-size:180%;color:#0B2161;">Indigoidine-Tag.</span><span class="text-muted" style="font-family:Arial, sans-serif; font-size:100%"> Exploring Modularity of NRPS by Shuffling Modules.</span></h1> | |
| - | + | ||
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col-sm- | + | <div class="col-sm-12 col-md-6"> |
<!--Months--> | <!--Months--> | ||
| - | <ul class="pagination" style="margin-bottom:2%; margin-left: | + | <ul class="pagination" style="margin-bottom:2%; margin-left:17%;"> |
<!--<button type="button" class="btn btn-default">May</button> | <!--<button type="button" class="btn btn-default">May</button> | ||
| Line 160: | Line 160: | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
<b>Module Shuffling</b><br/> | <b>Module Shuffling</b><br/> | ||
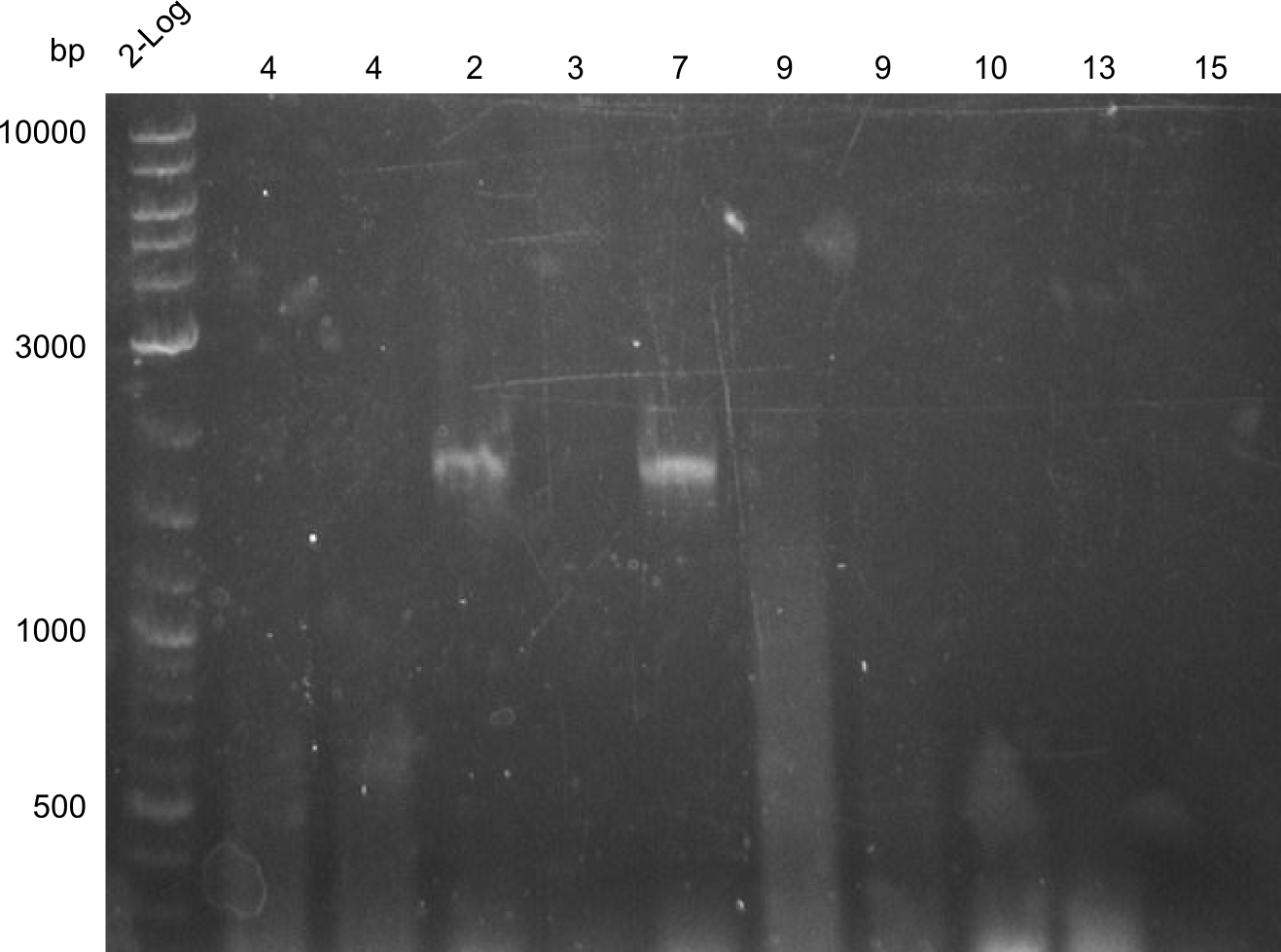
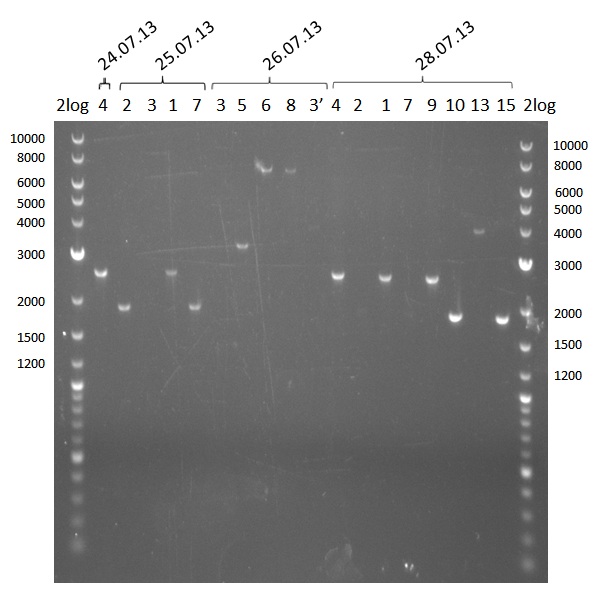
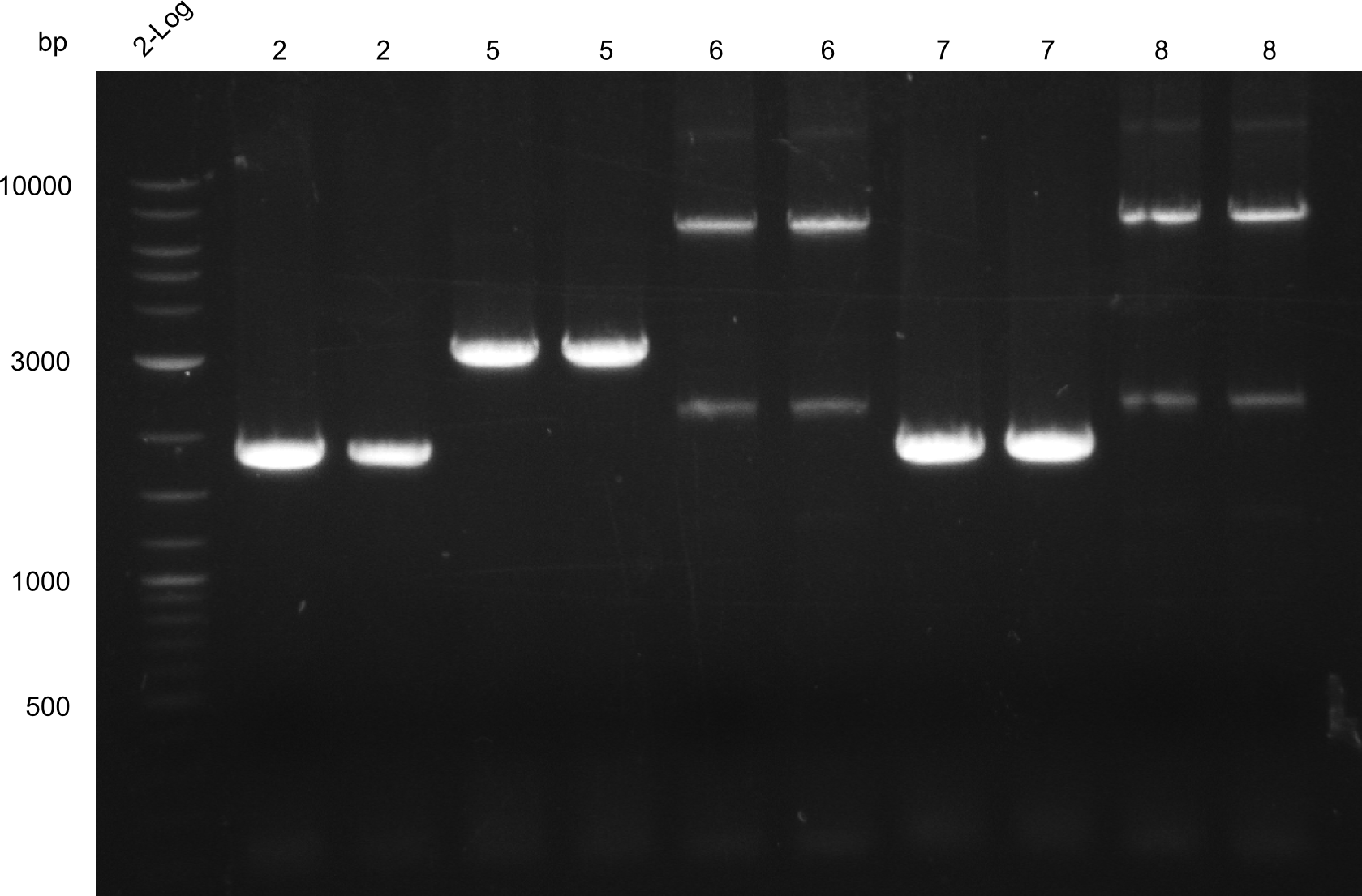
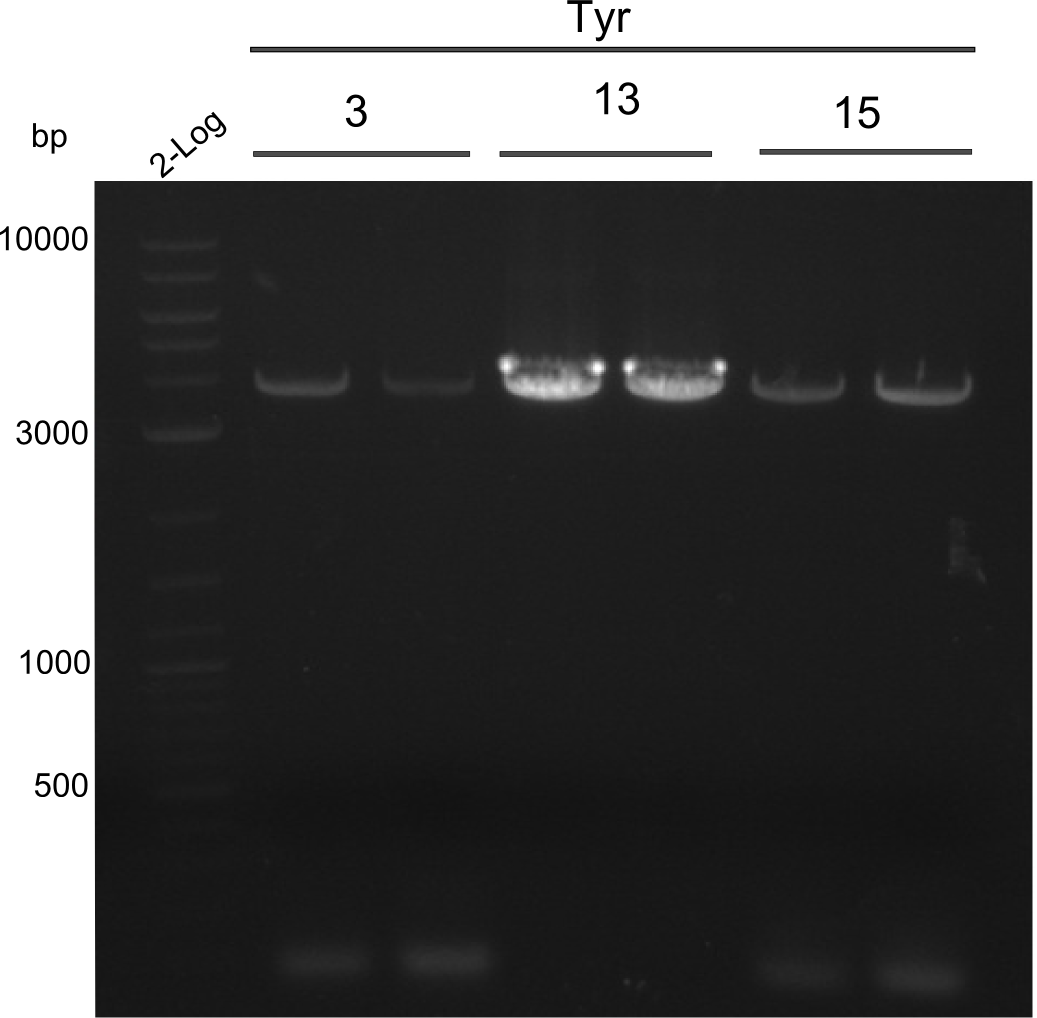
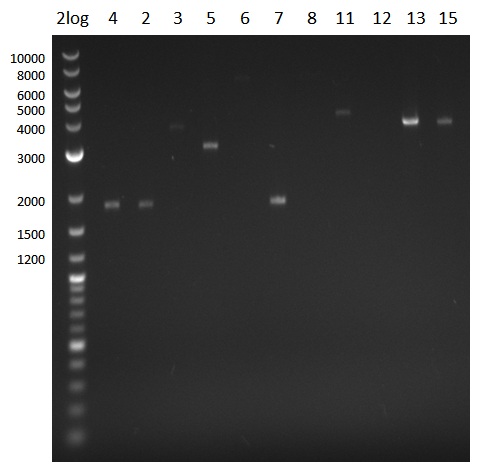
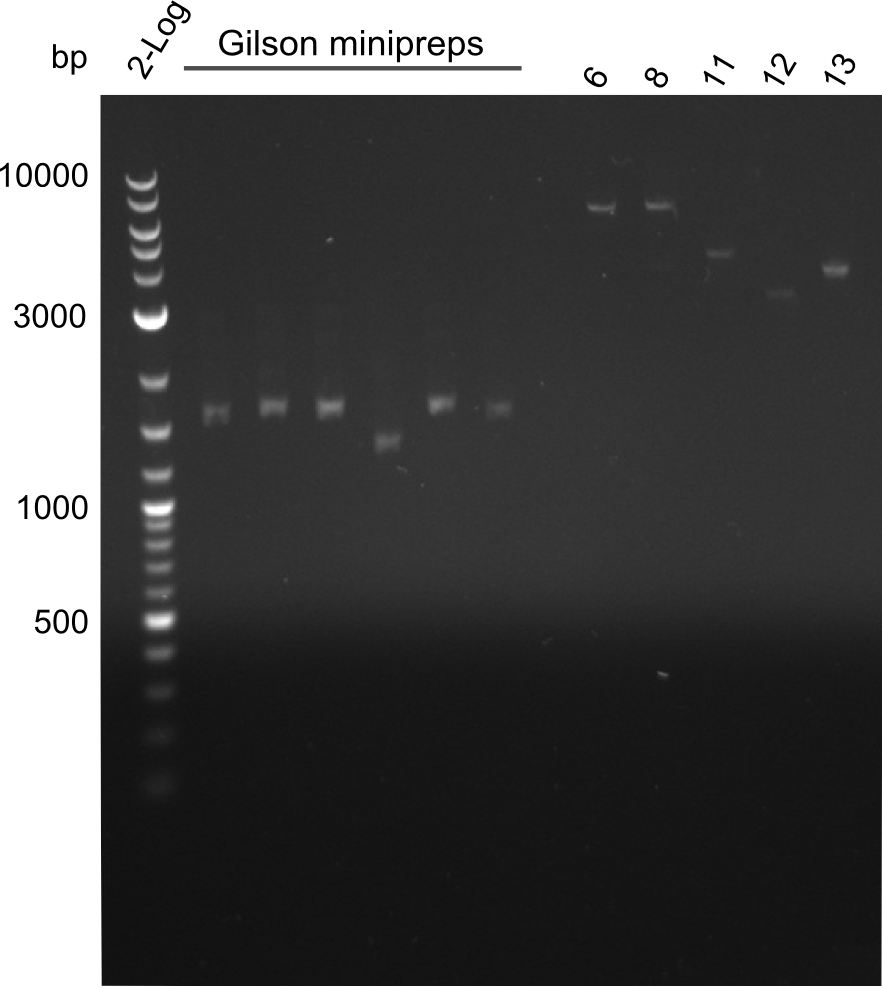
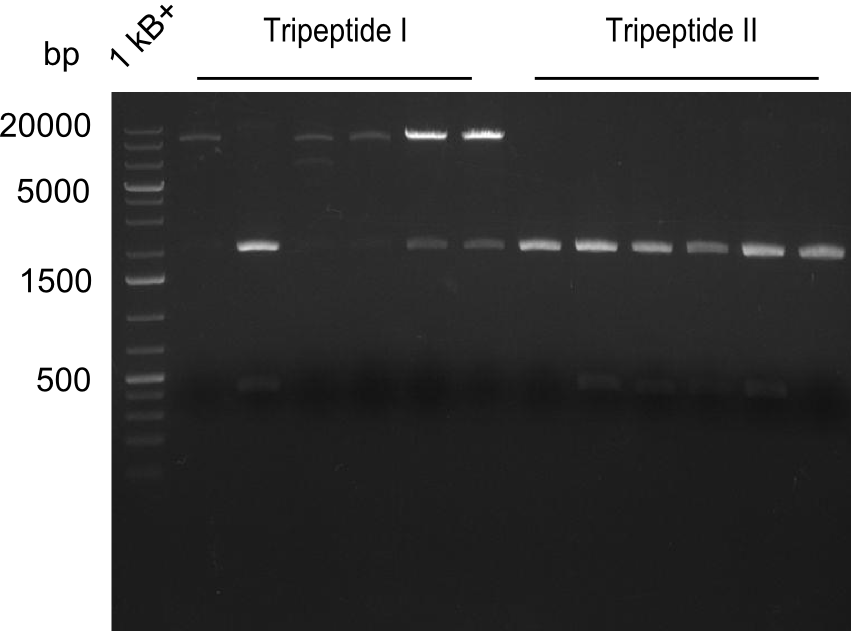
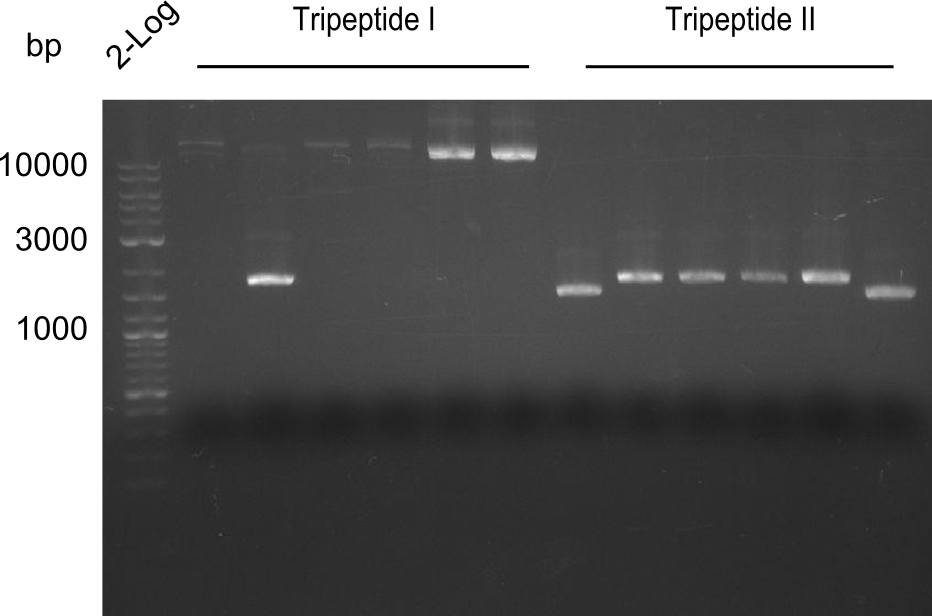
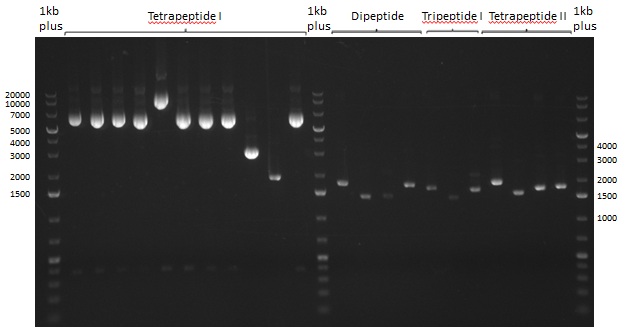
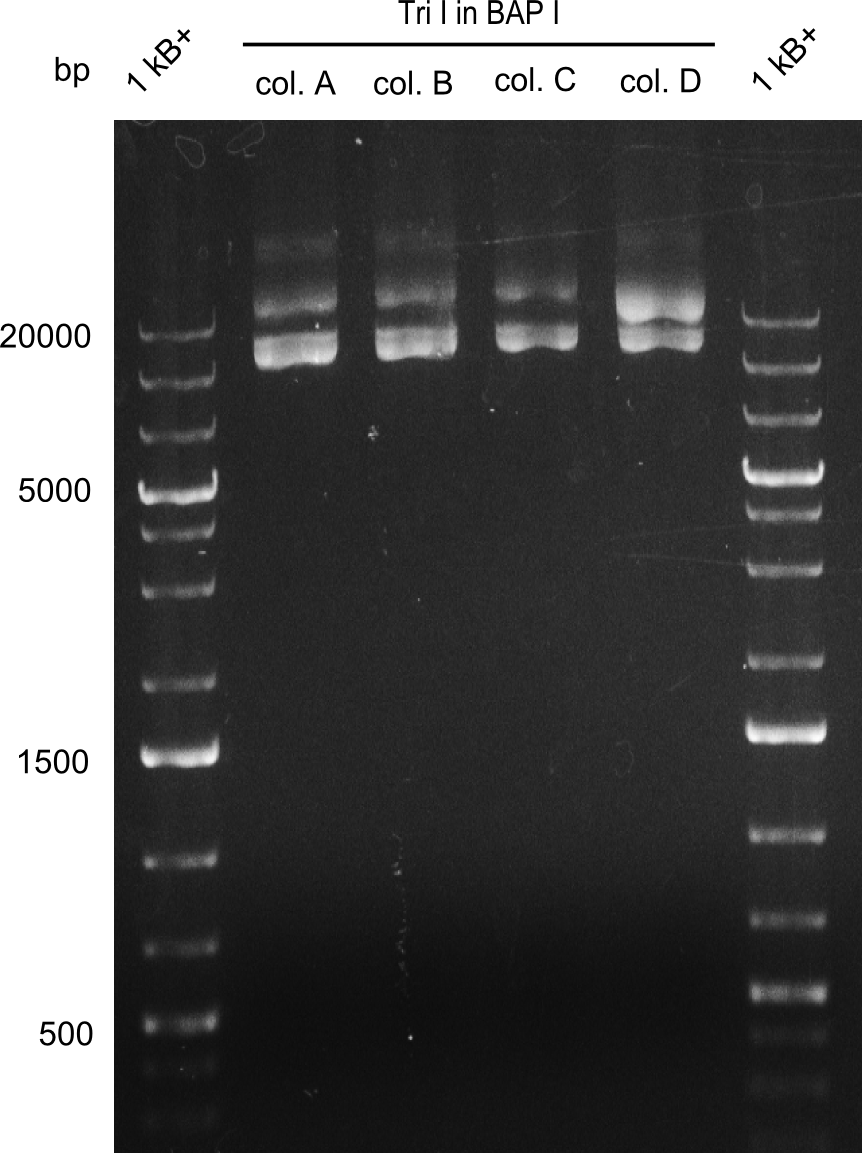
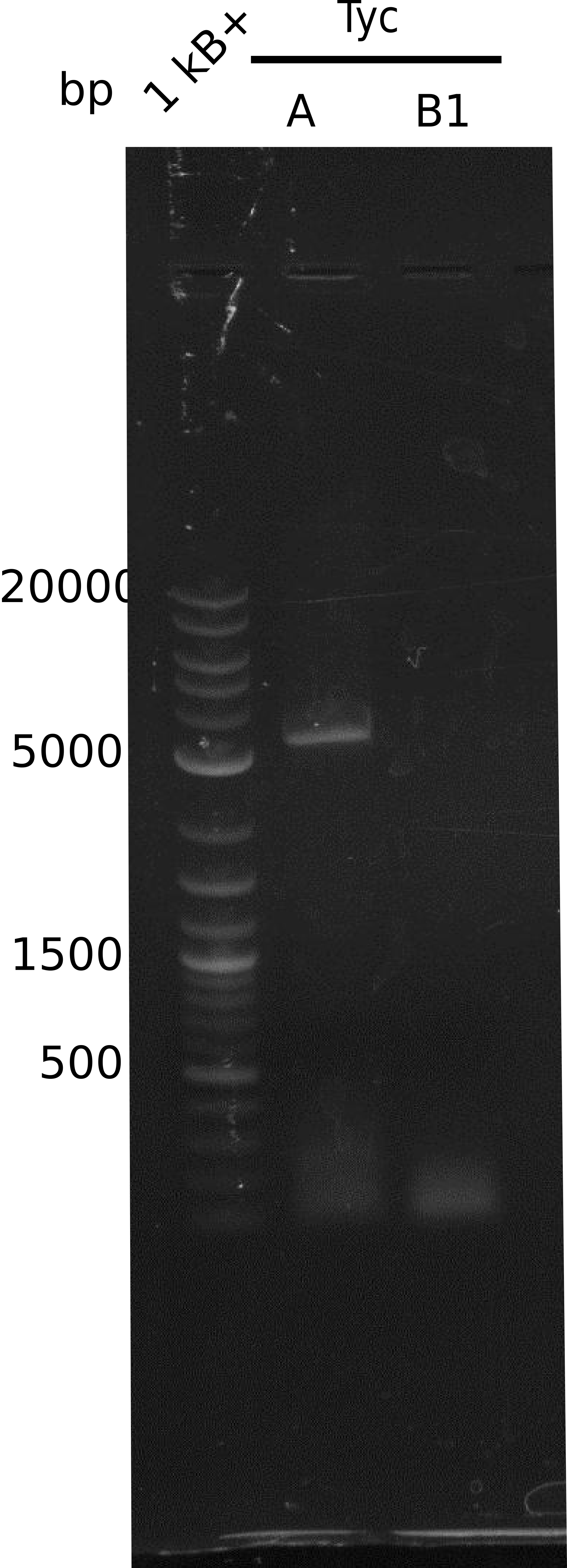
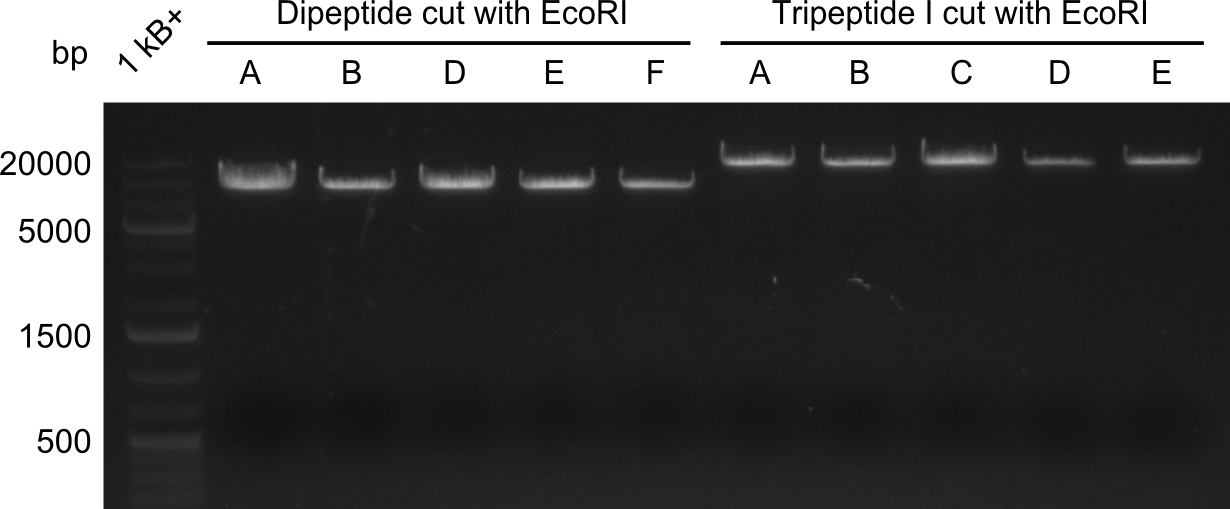
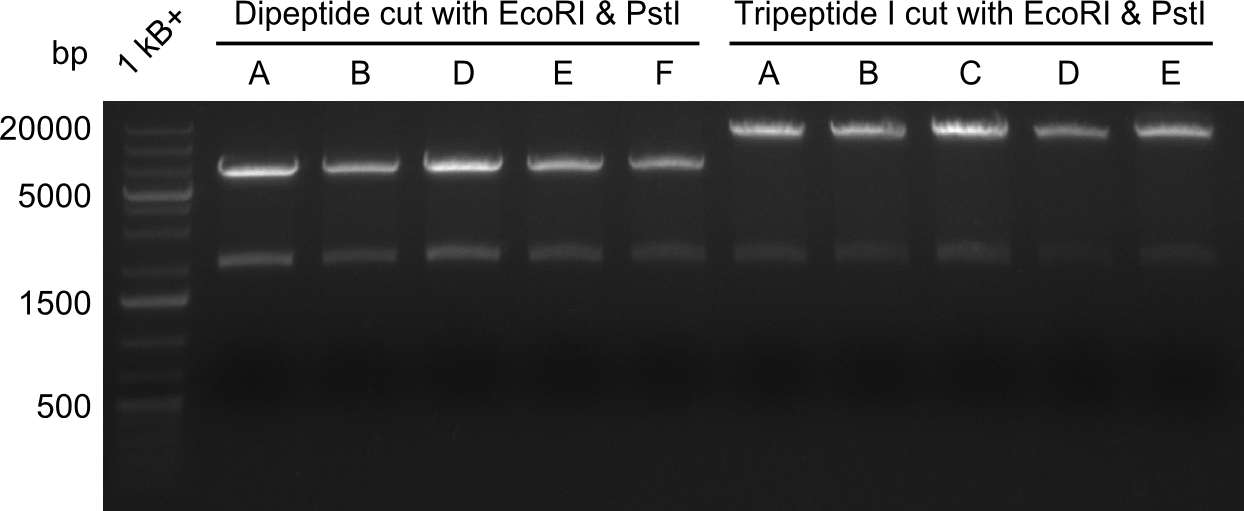
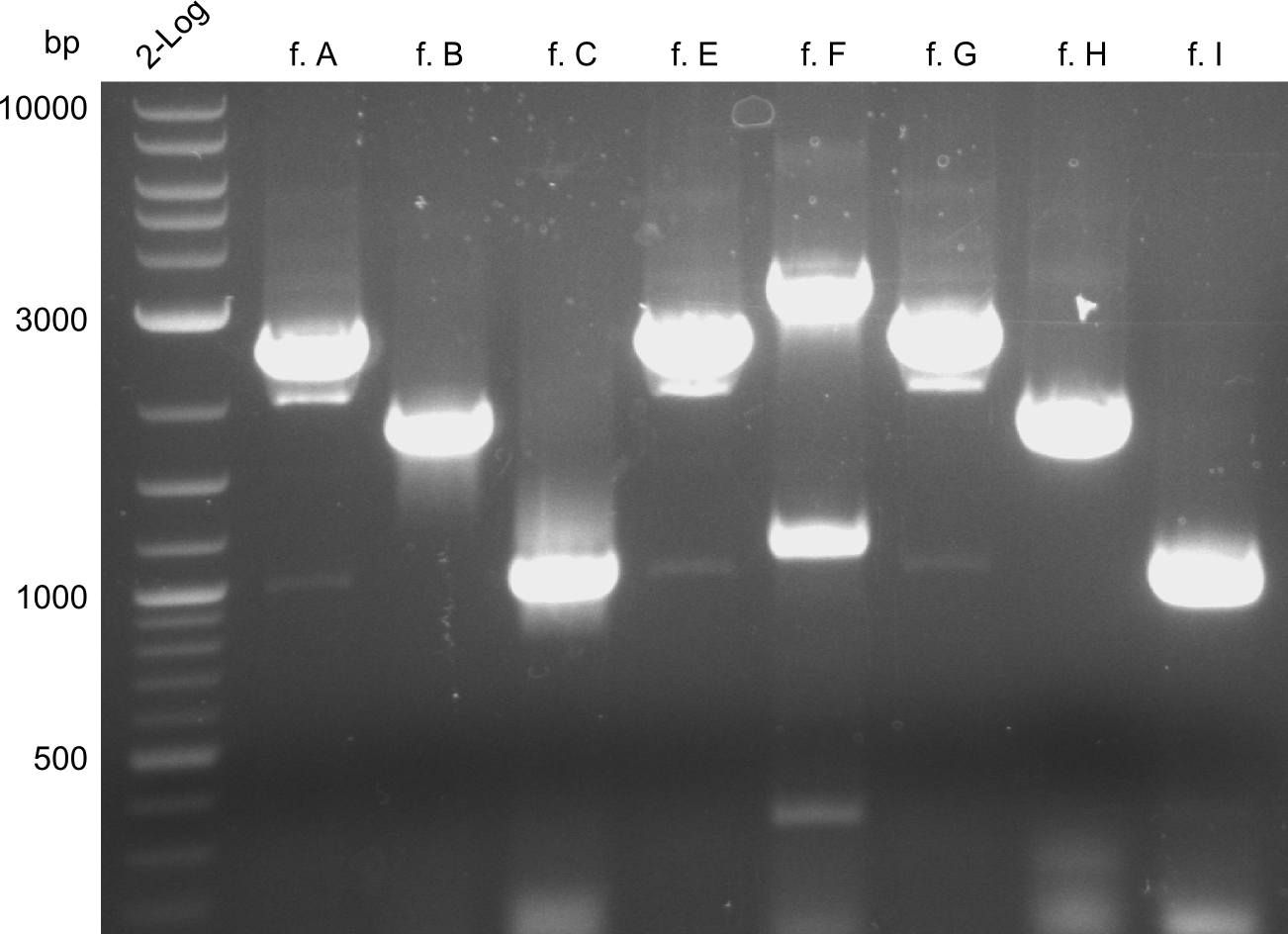
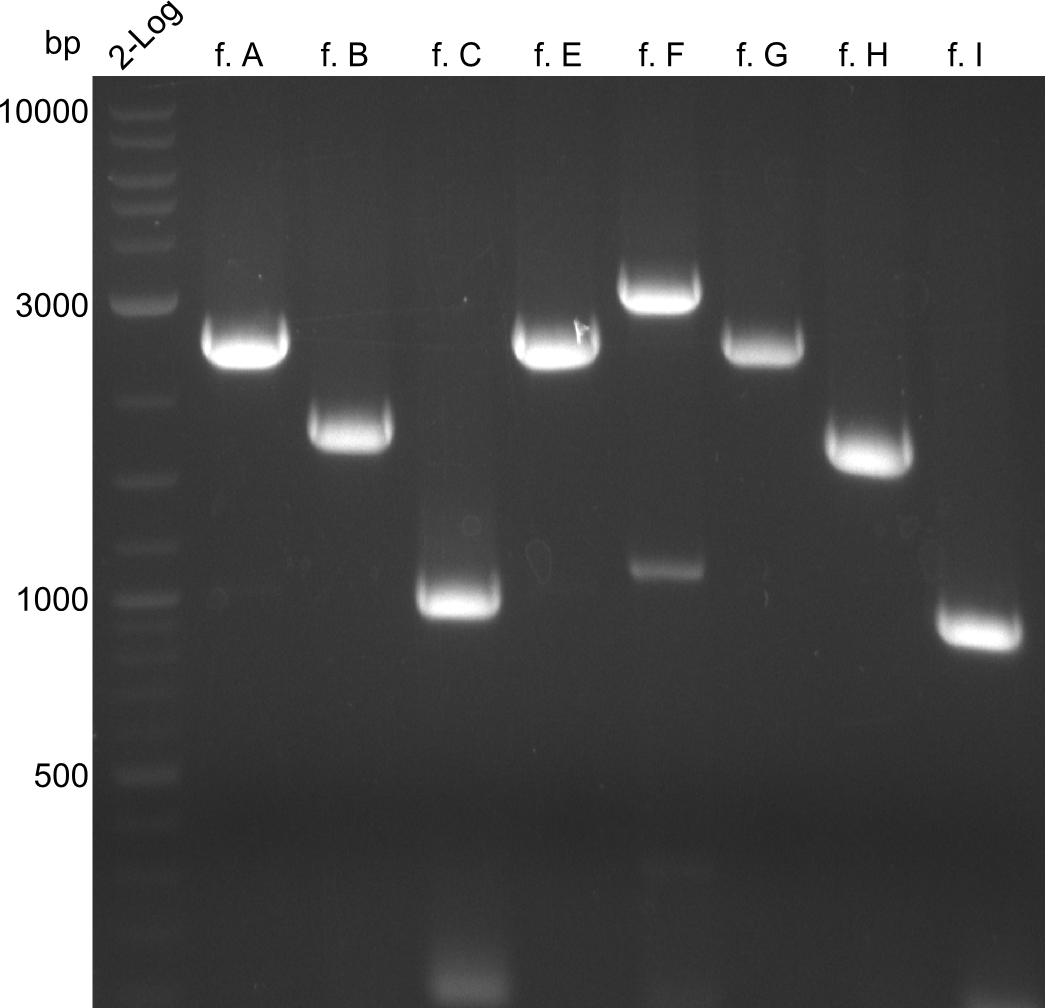
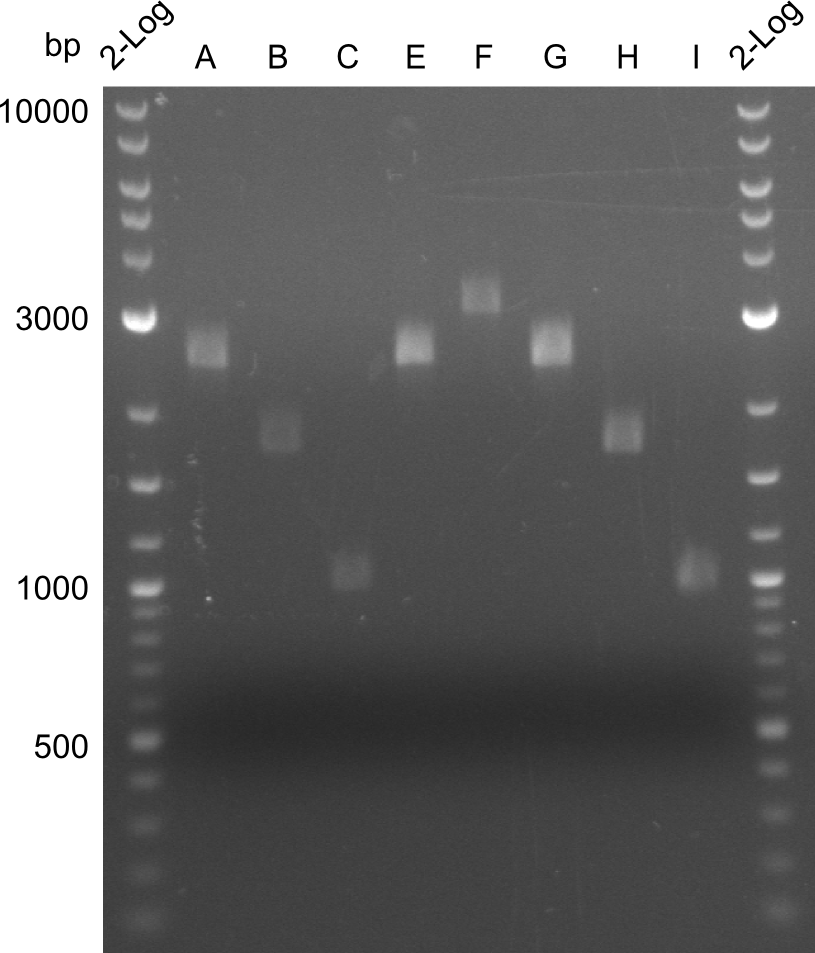
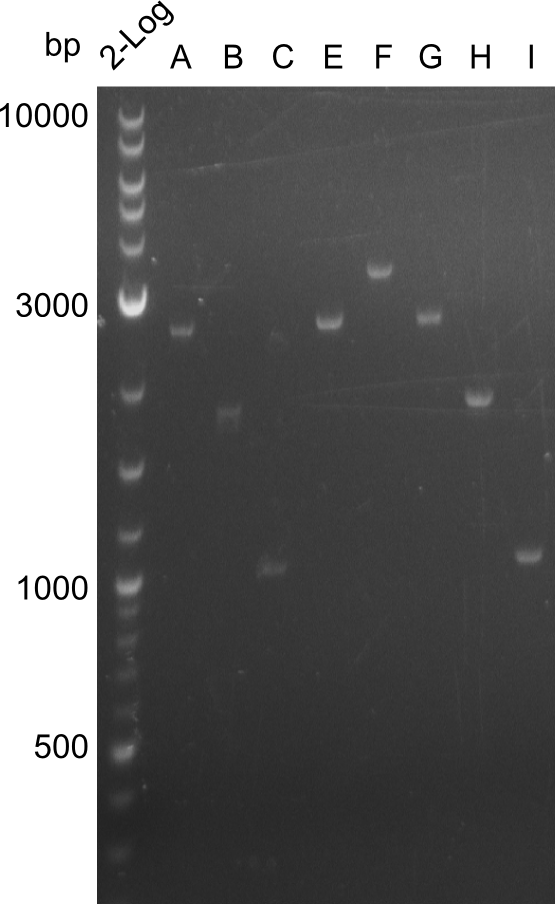
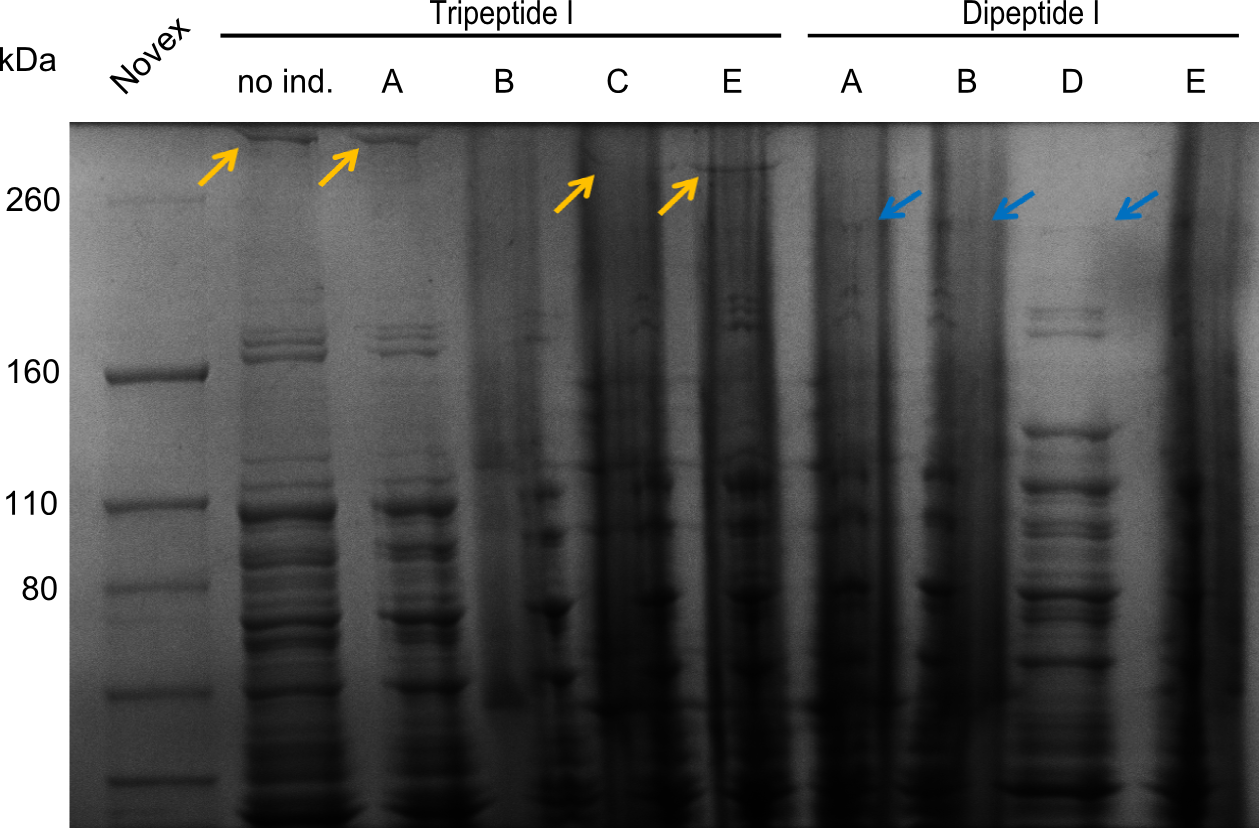
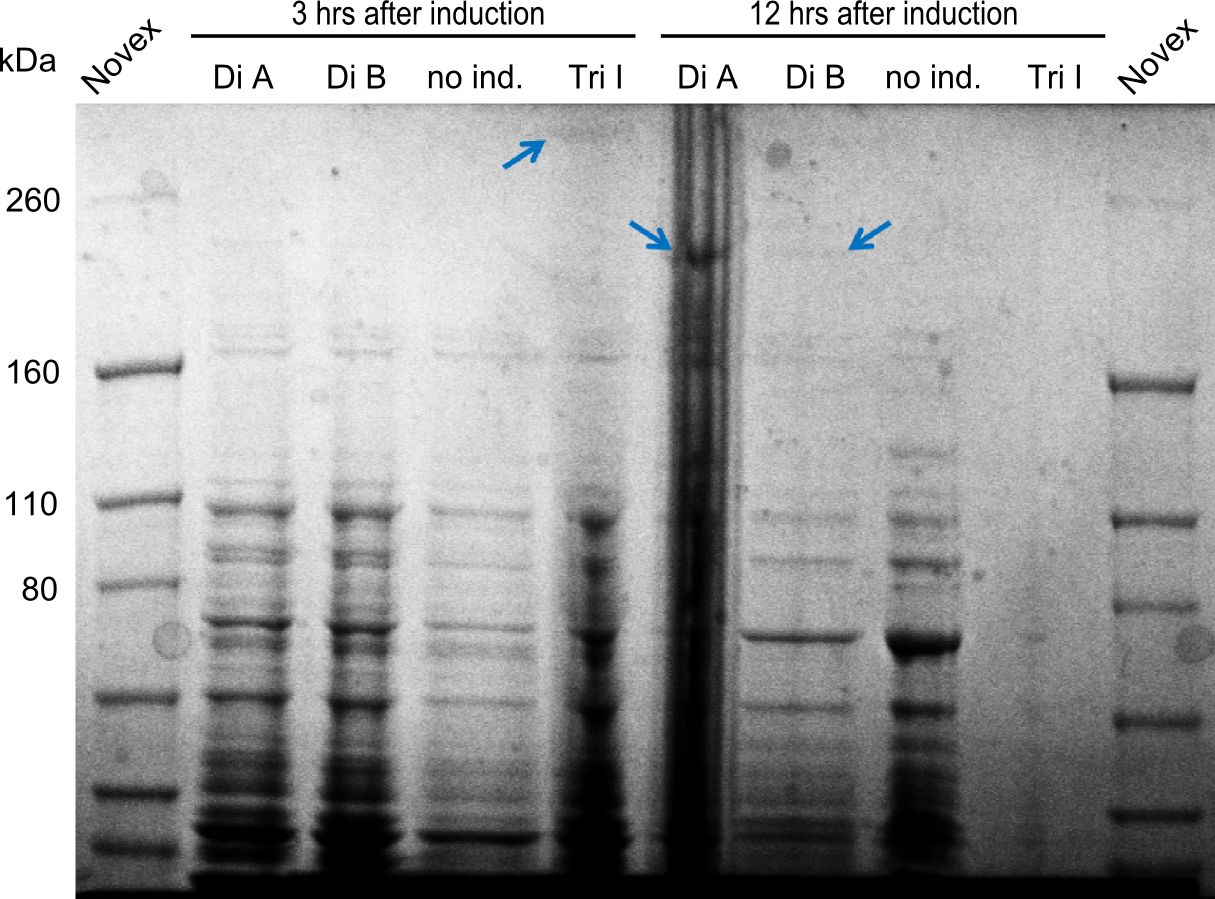
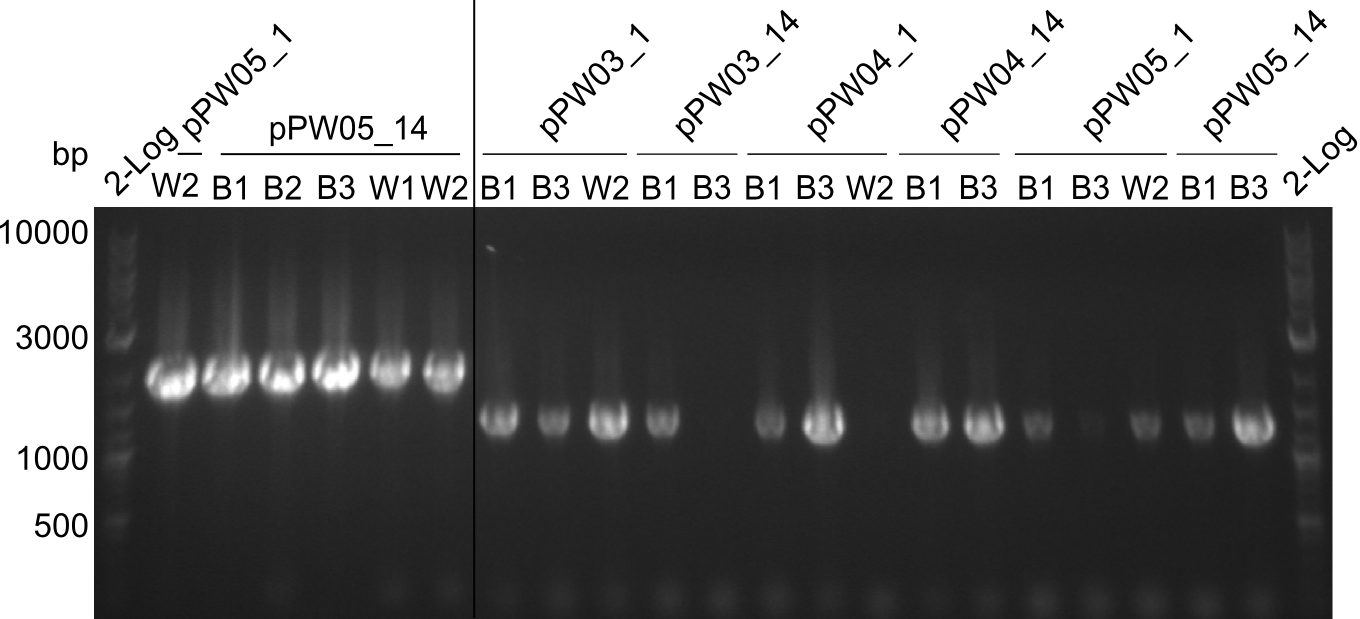
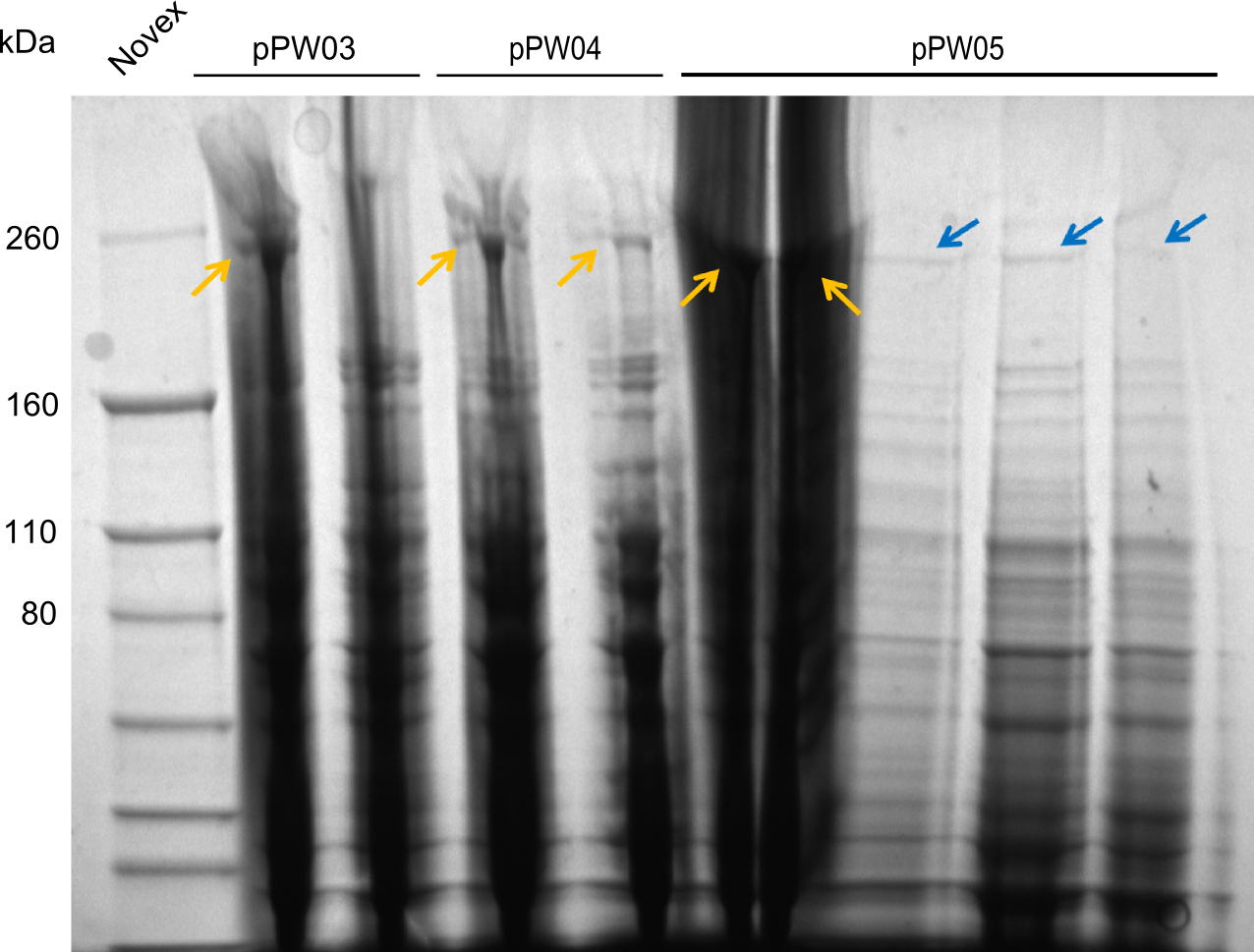
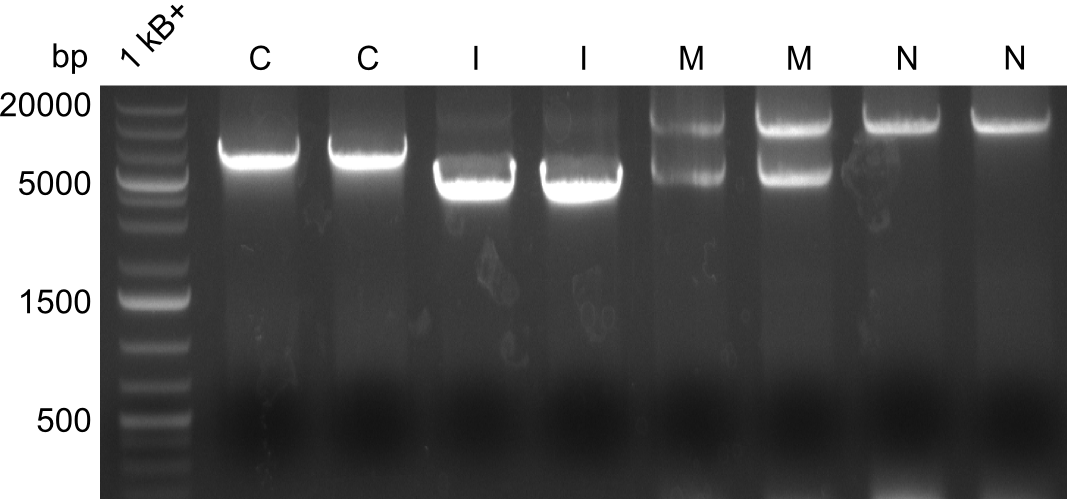
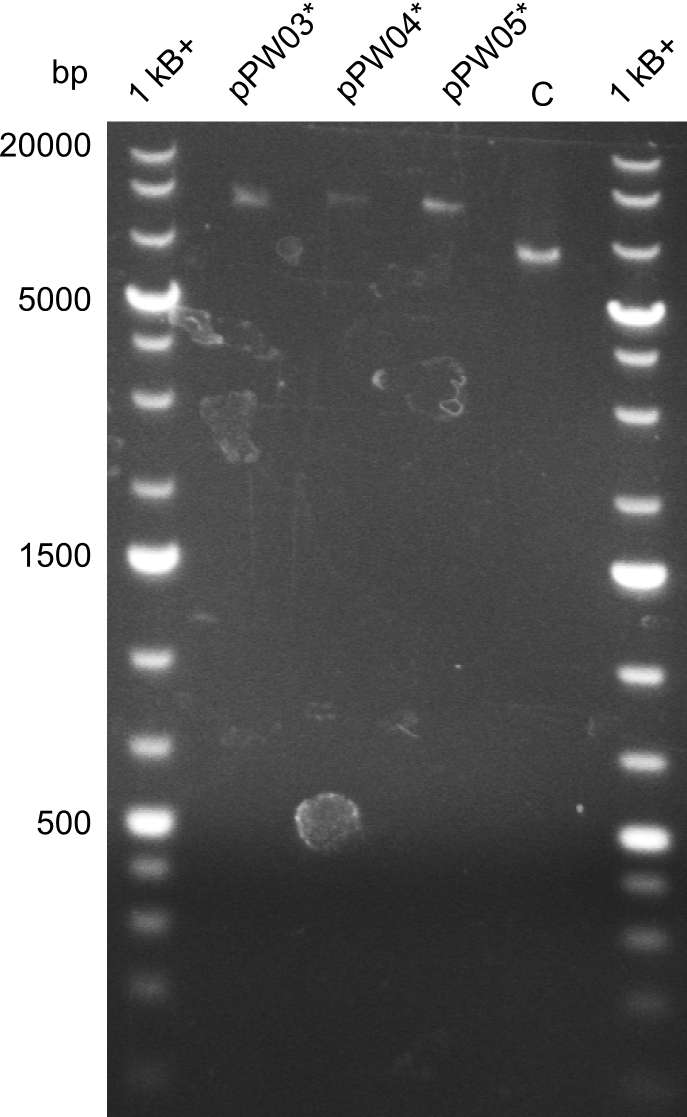
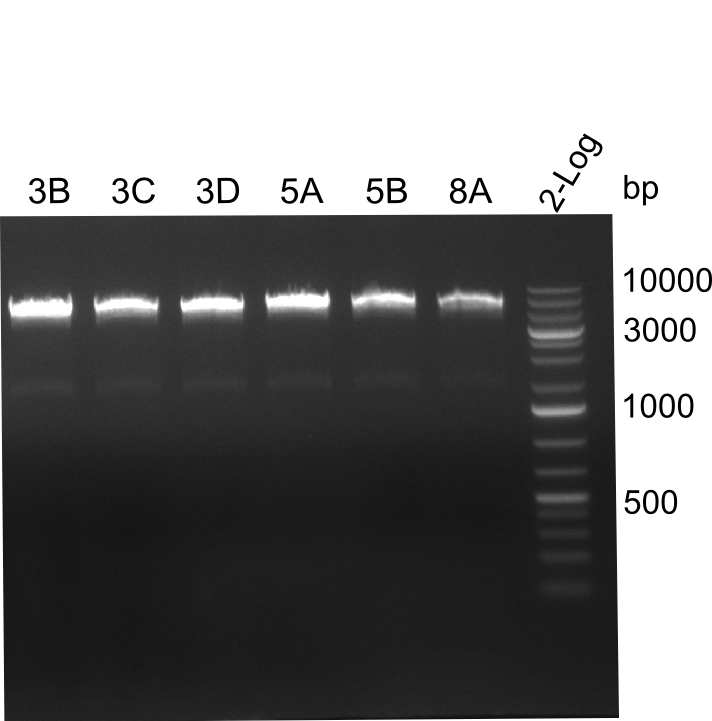
| - | We started the week with a SDS-PAGE analyzing the expression capacity of several clones for the Dipeptide- and Tripeptide-I-NRPS. It showed a positive band for the Dipeptide synthetase (212 kDa) and an inconclusive band for the Tripeptide-I synthetase (381 kDa), because the top band of our ladder was at 260 kDa | + | We started the week with a SDS-PAGE analyzing the expression capacity of several clones for the Dipeptide- and Tripeptide-I-NRPS. It showed a positive band for the Dipeptide synthetase (212 kDa) and an inconclusive band for the Tripeptide-I synthetase (381 kDa), because the top band of our ladder was at 260 kDa. We interpreted this as a positive result for Tripeptide-I-synthetase as well. |
<br/><br/> | <br/><br/> | ||
<b>Interspecies <br/>Module Shuffling</b><br/> | <b>Interspecies <br/>Module Shuffling</b><br/> | ||
| Line 166: | Line 166: | ||
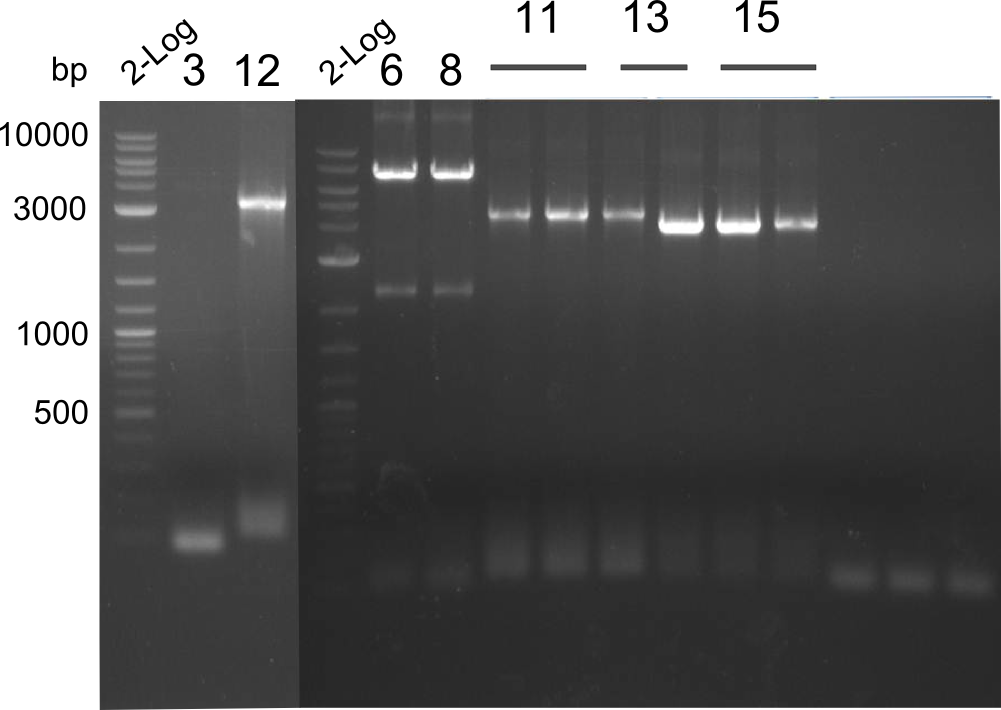
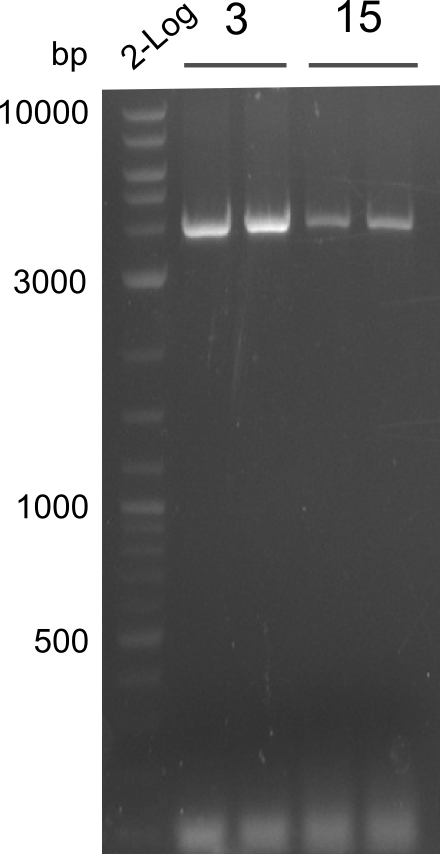
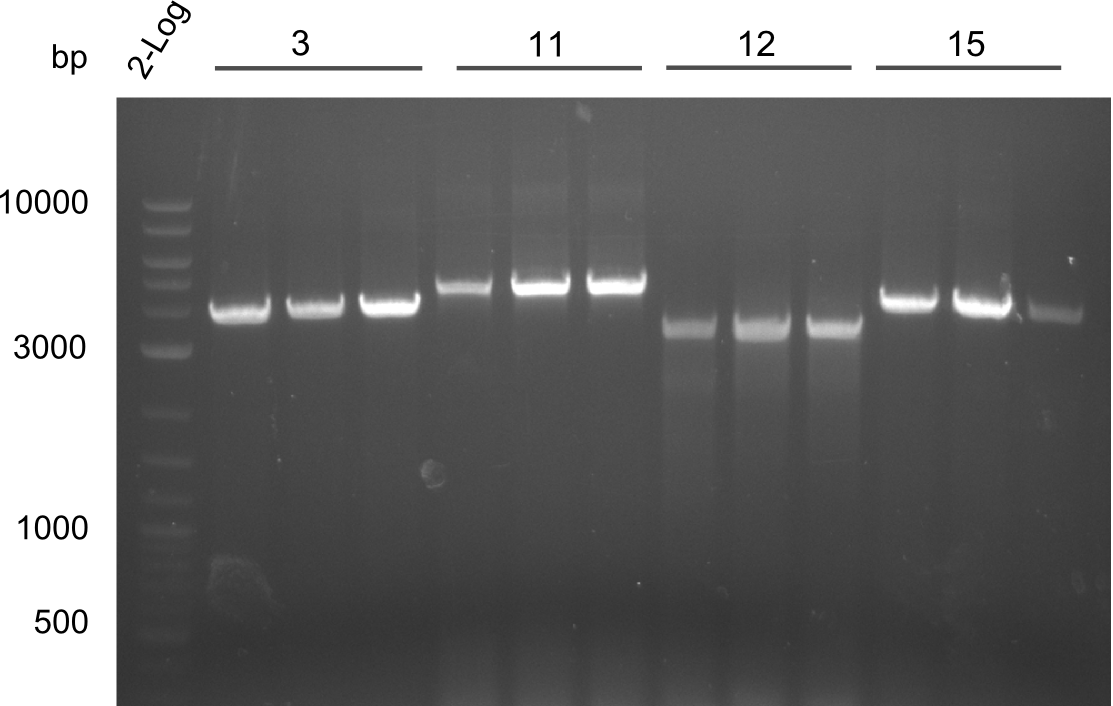
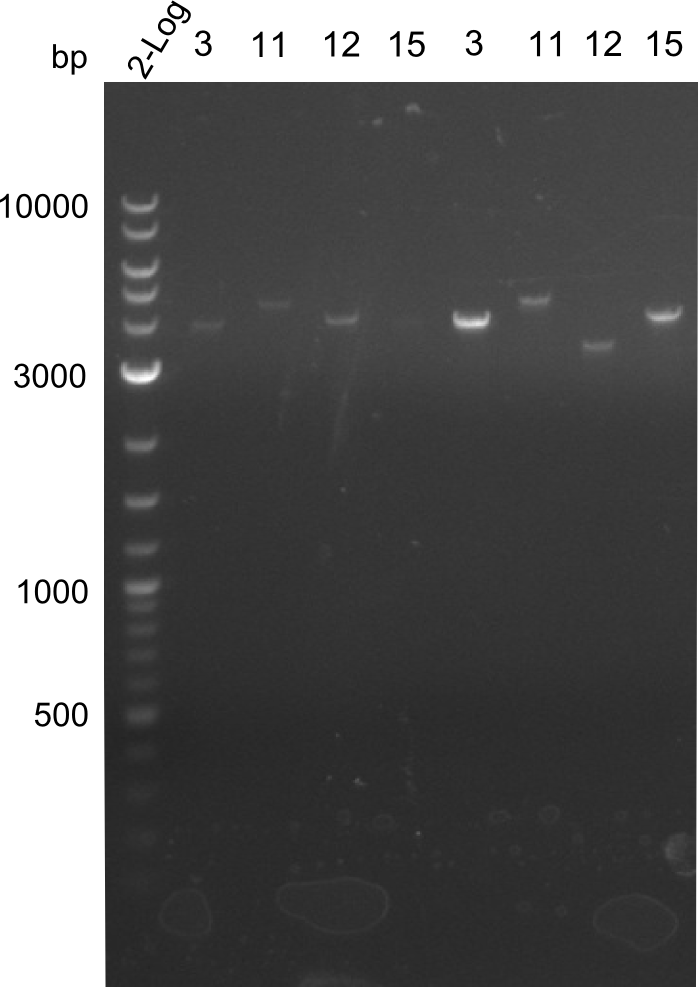
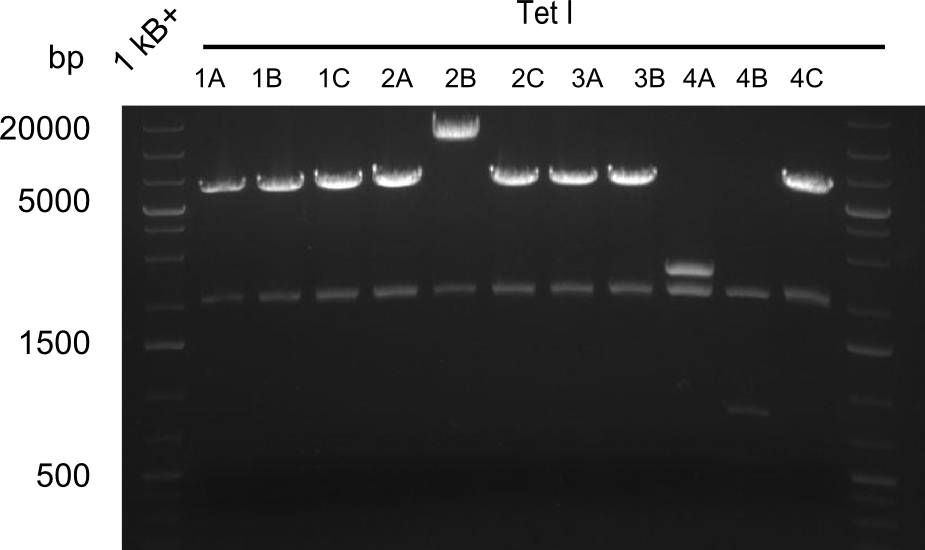
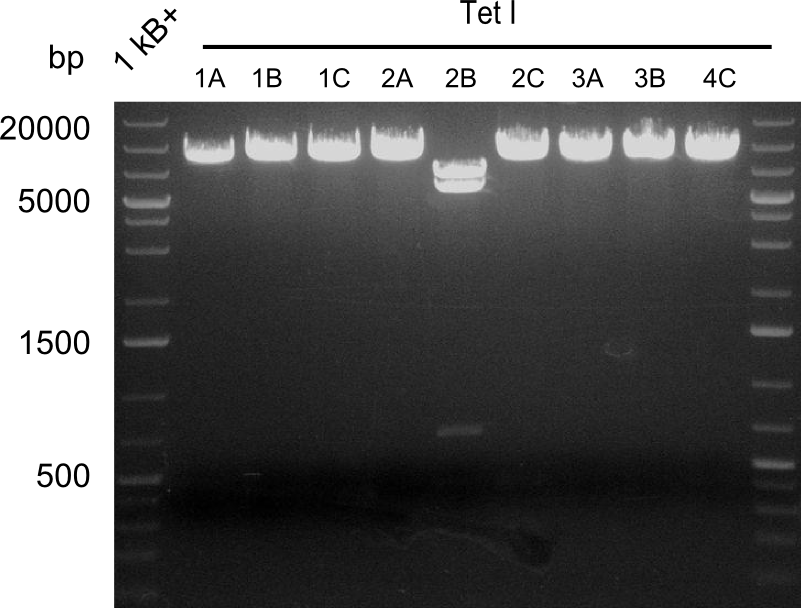
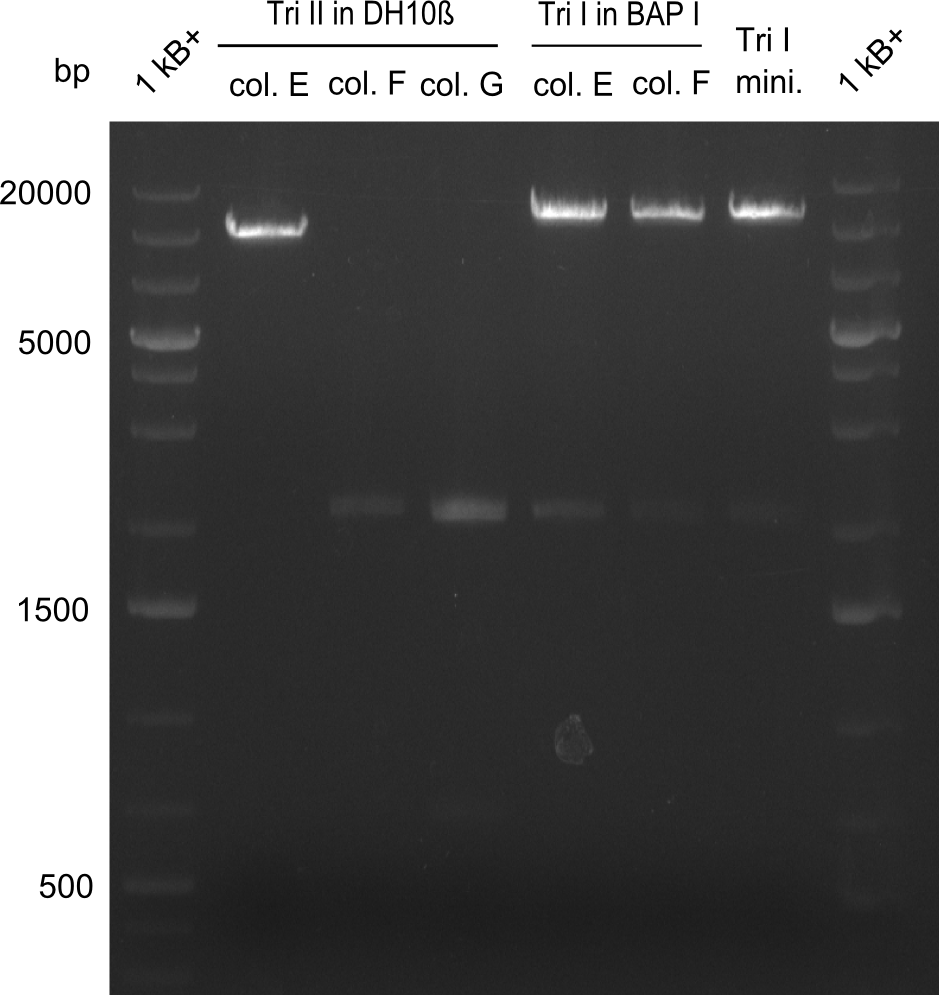
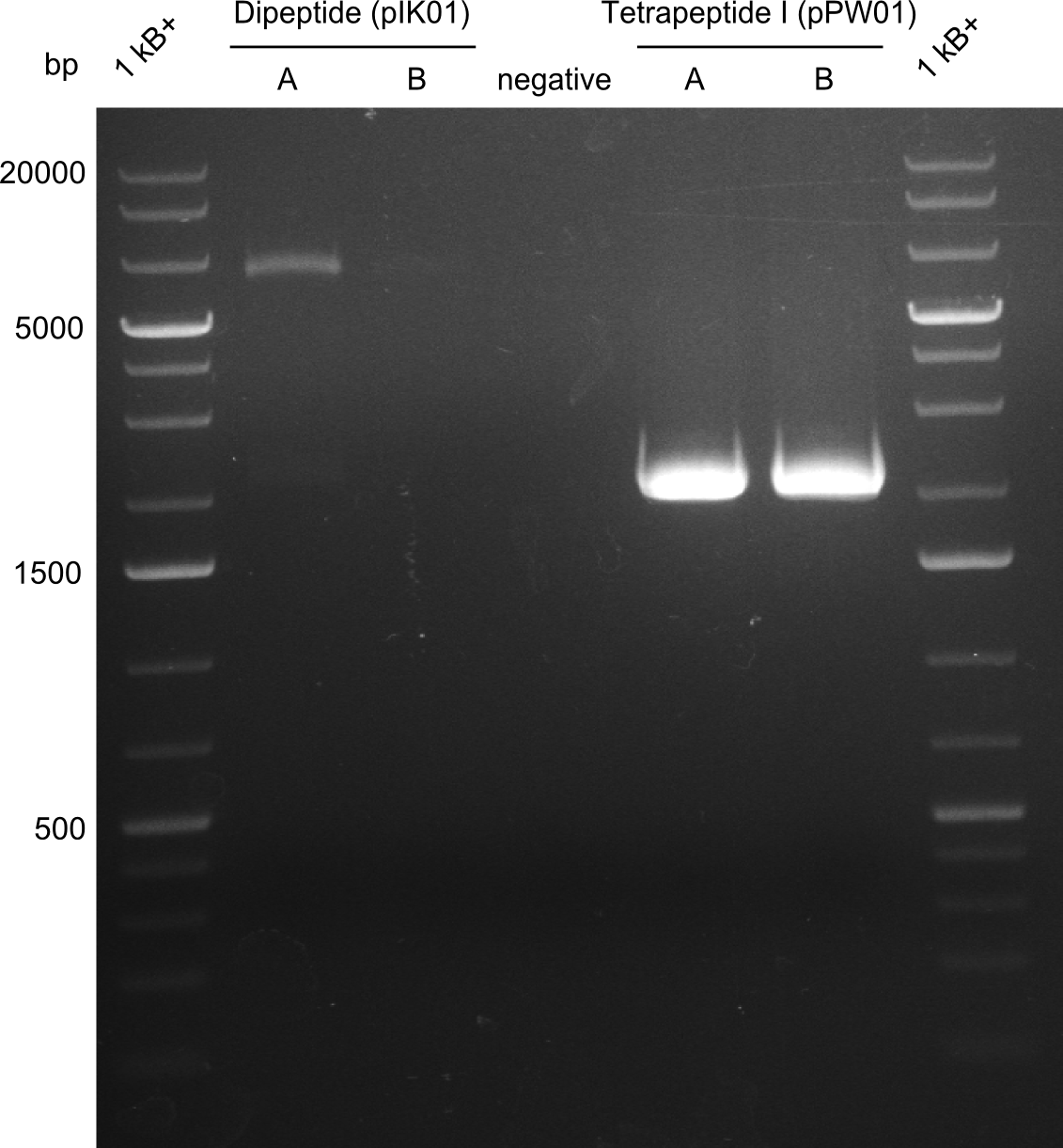
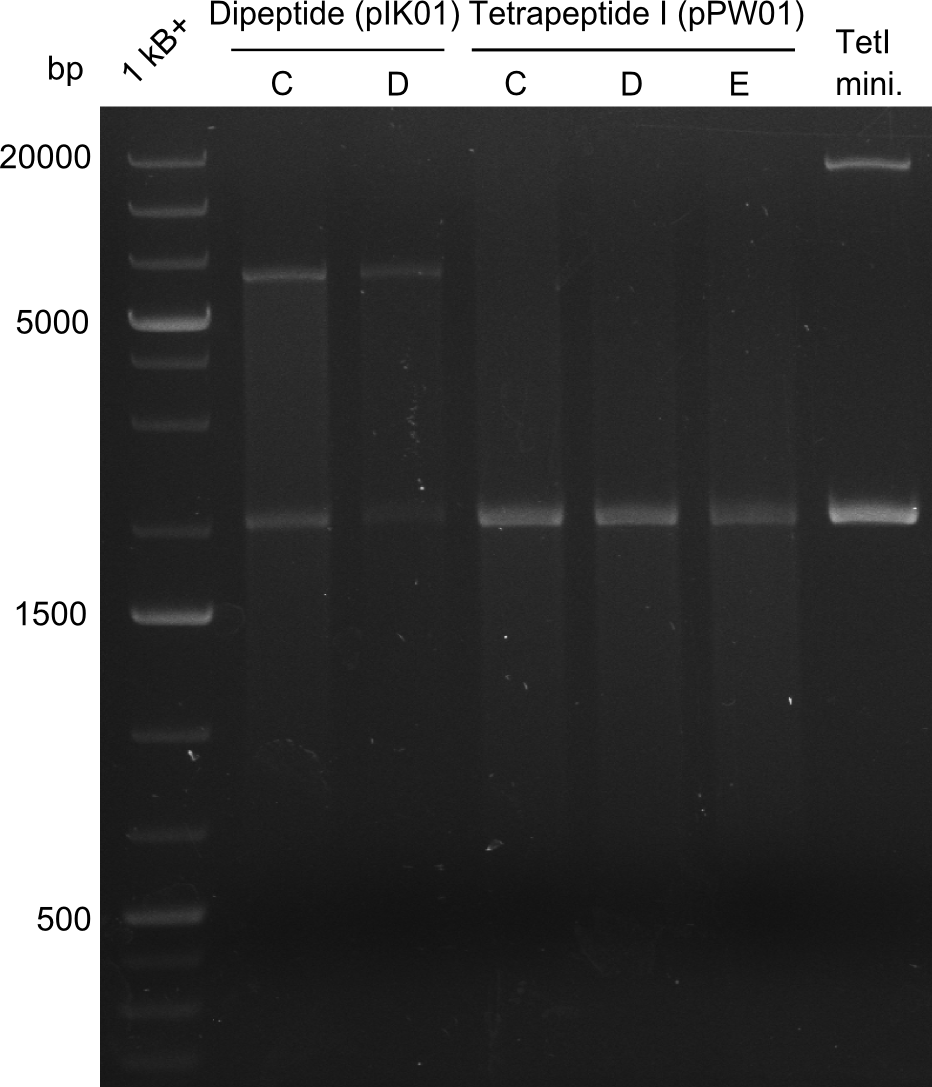
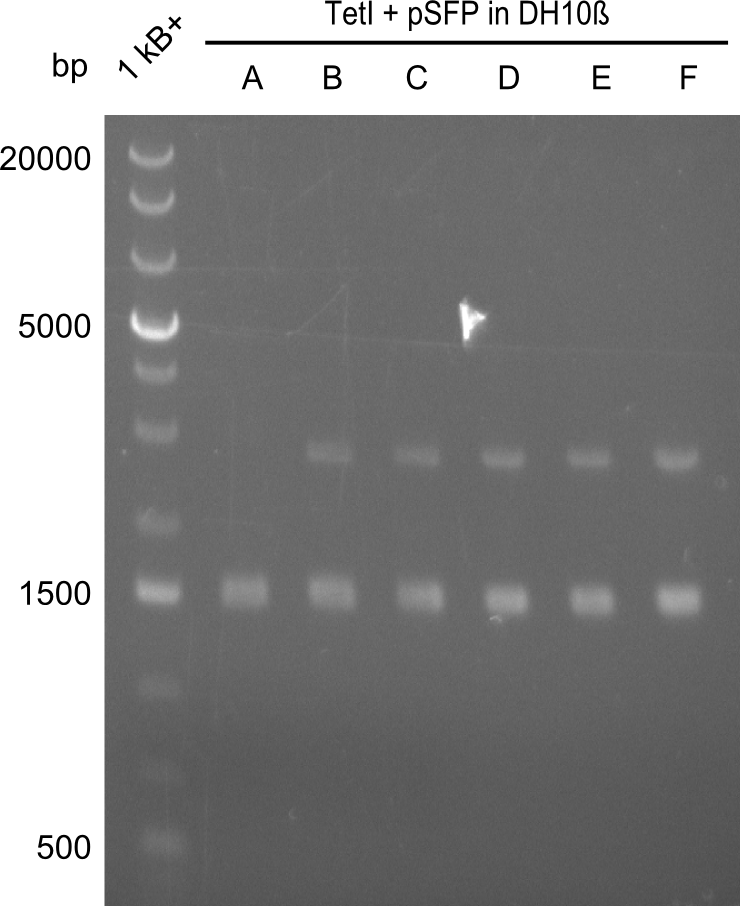
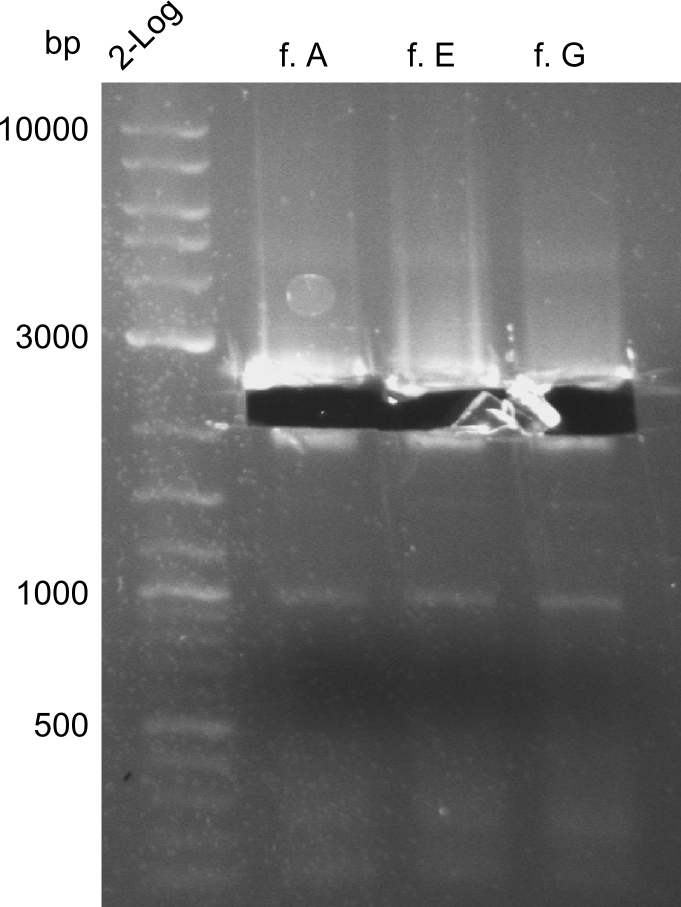
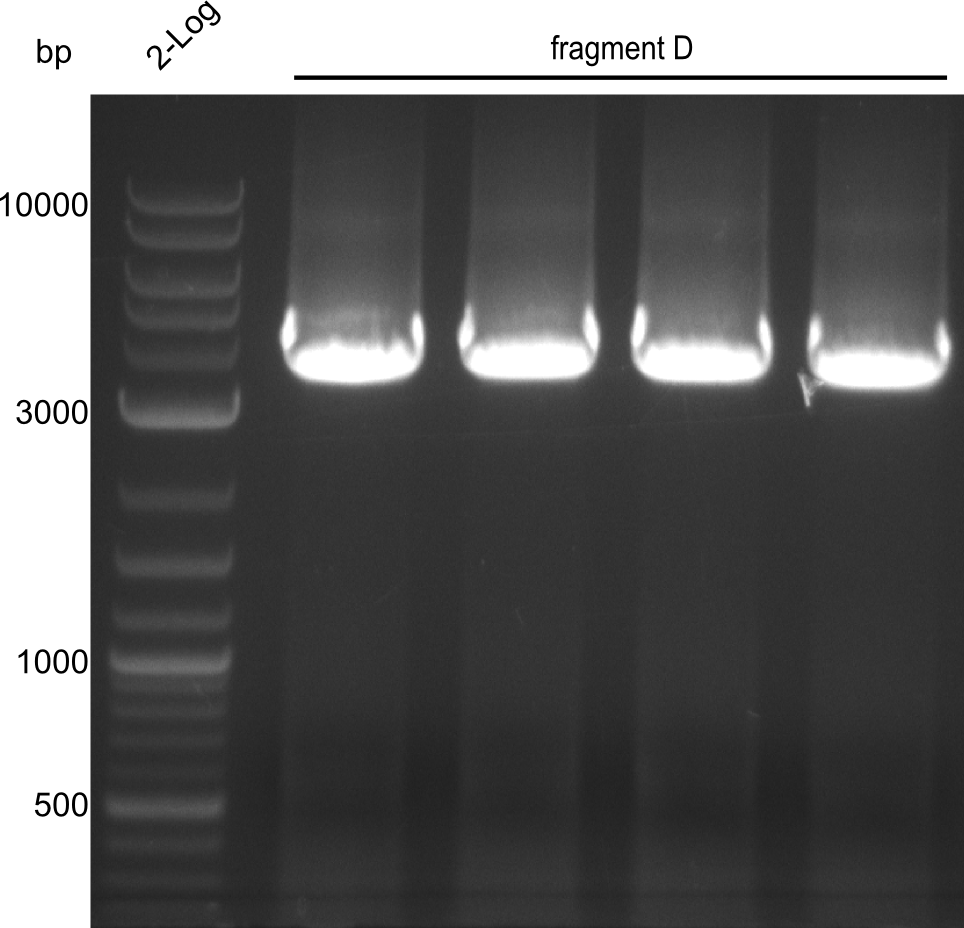
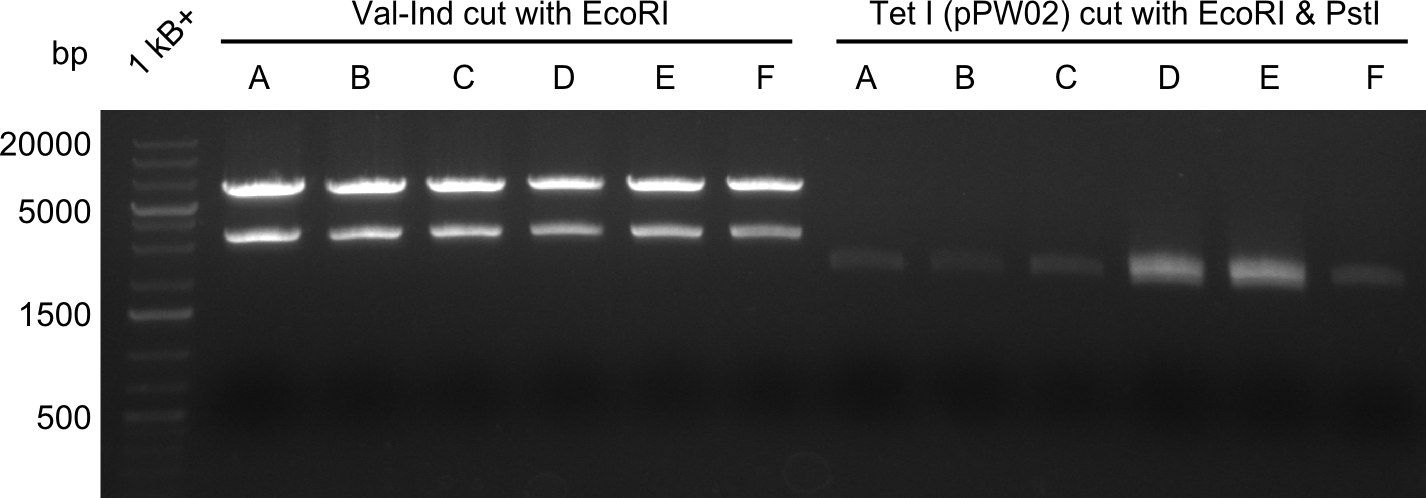
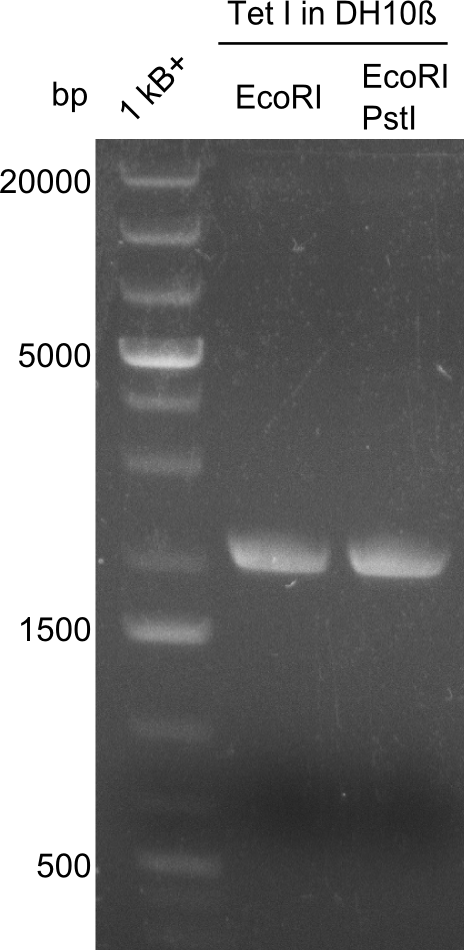
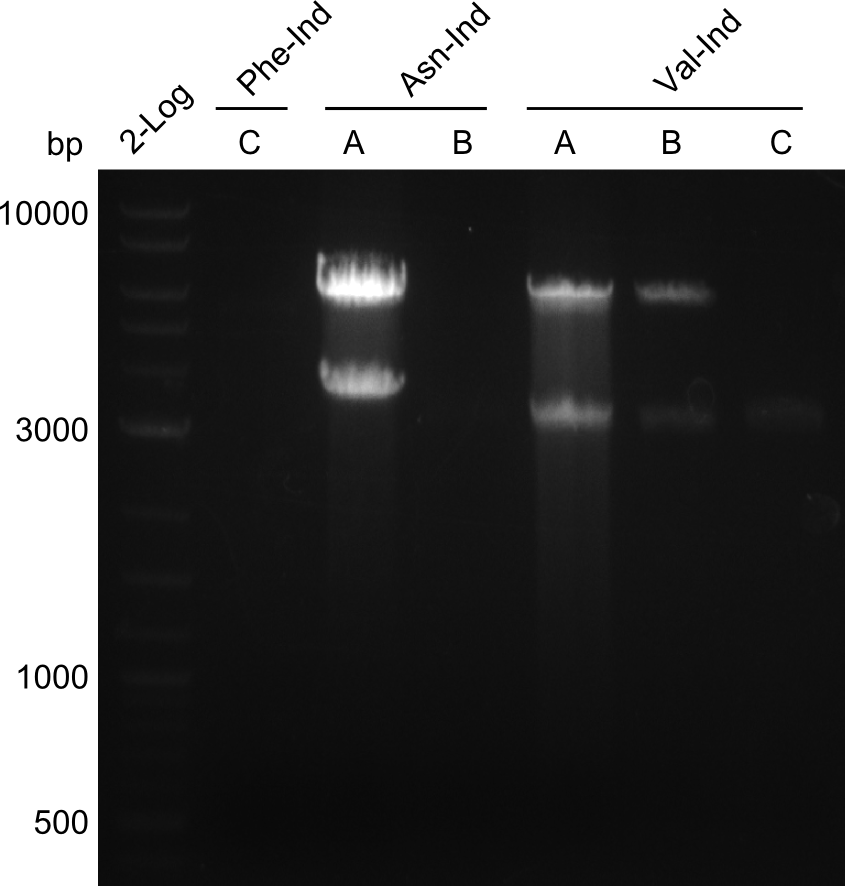
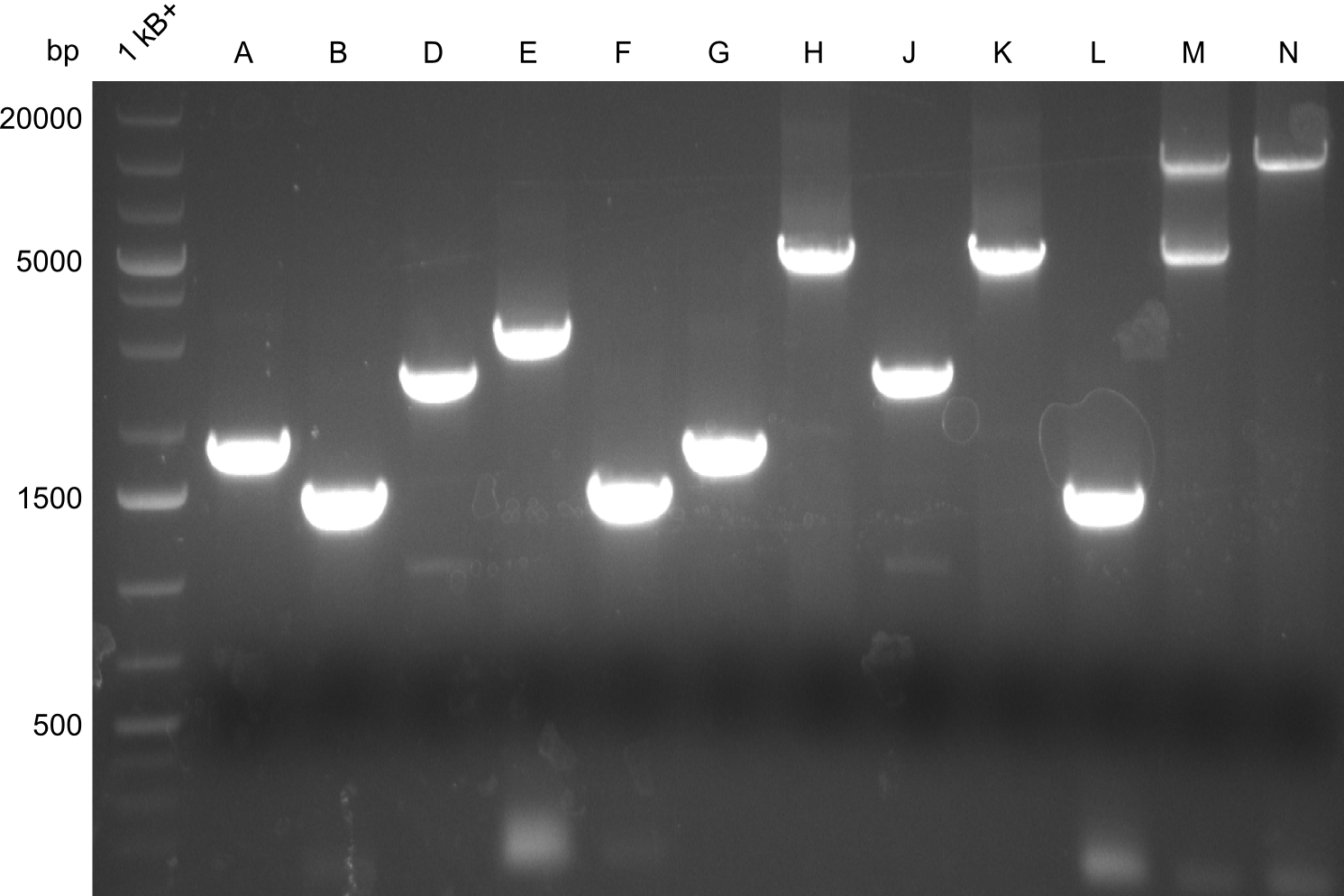
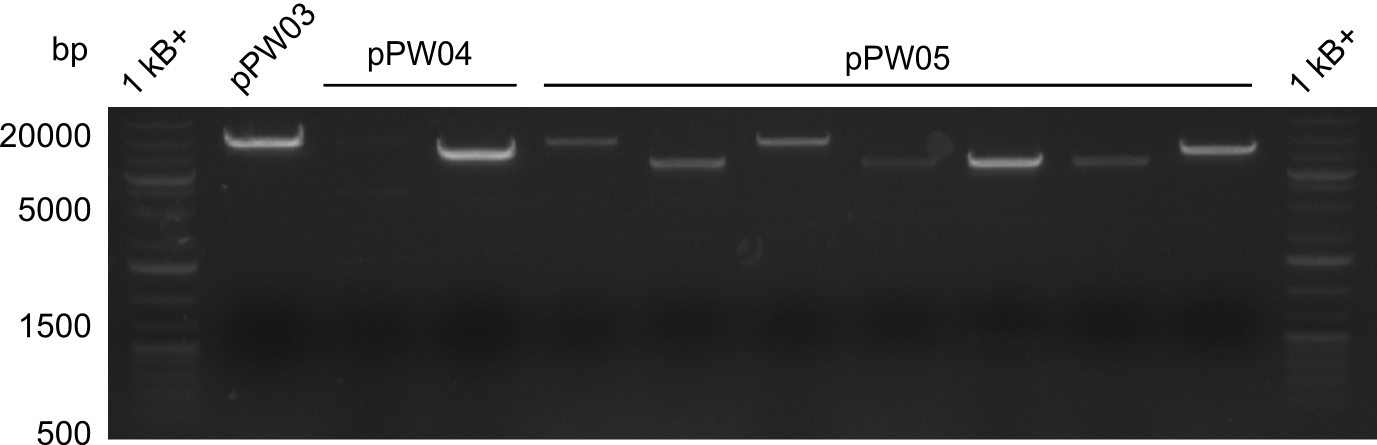
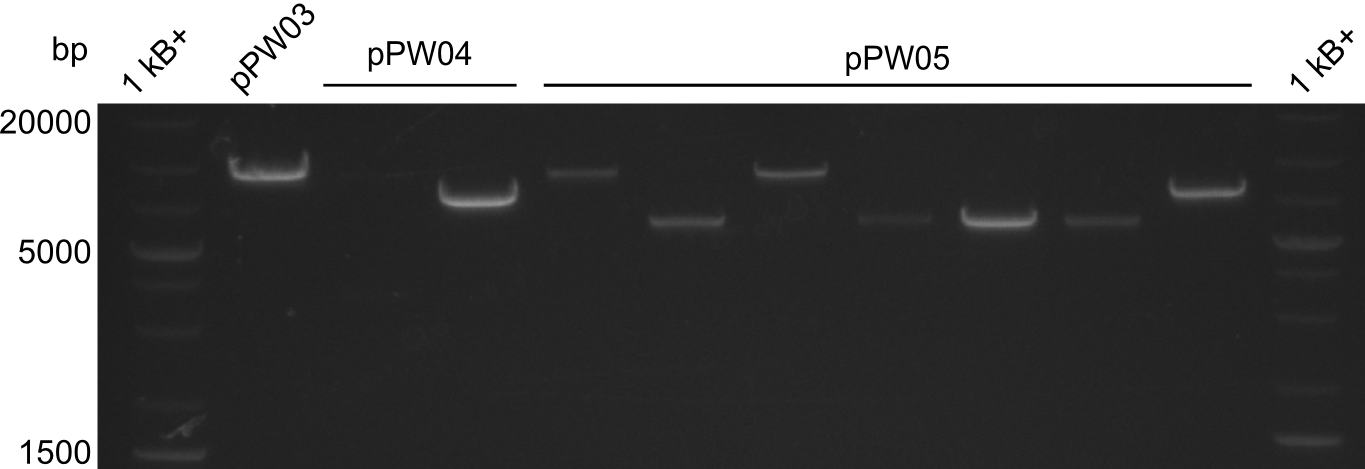
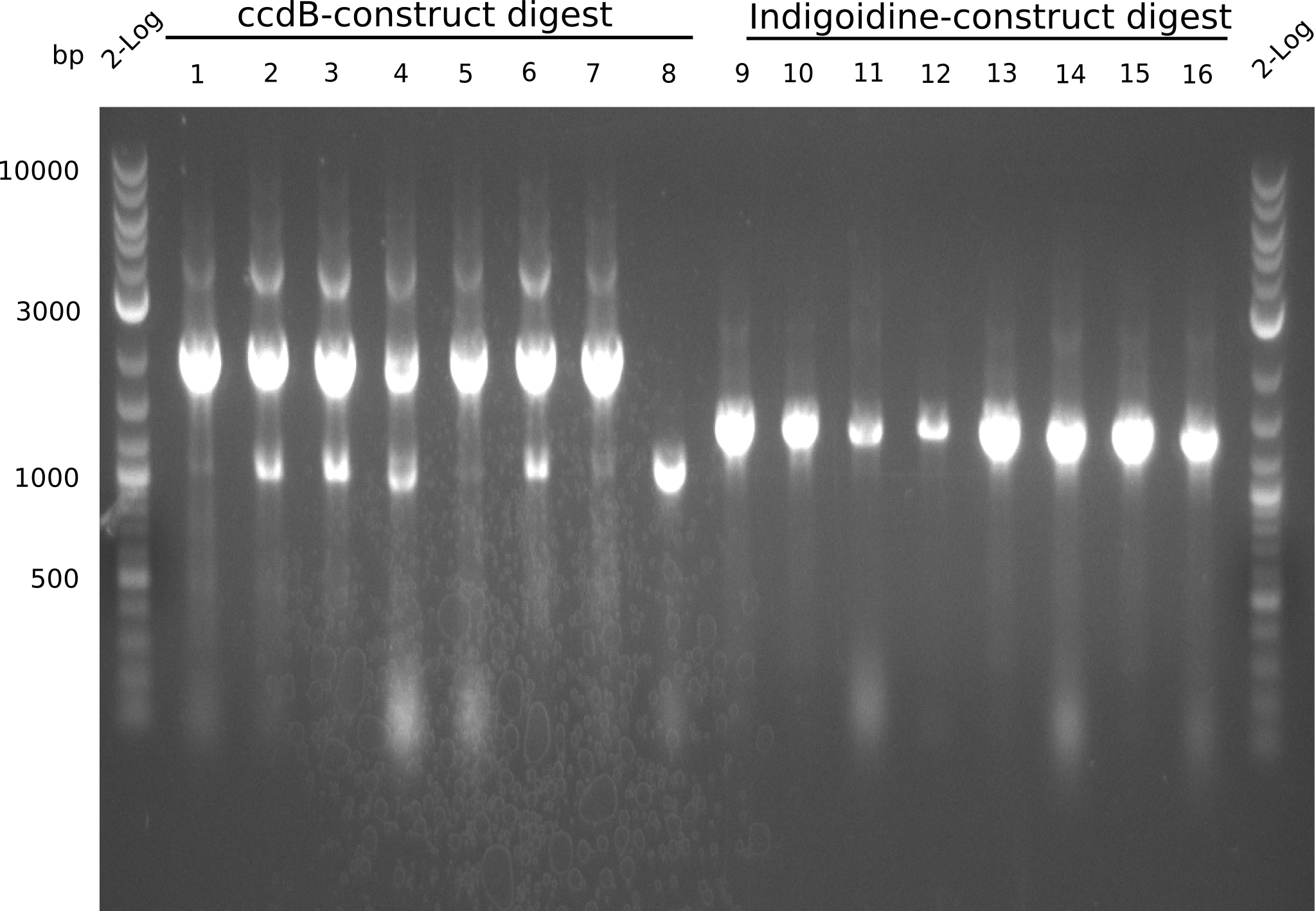
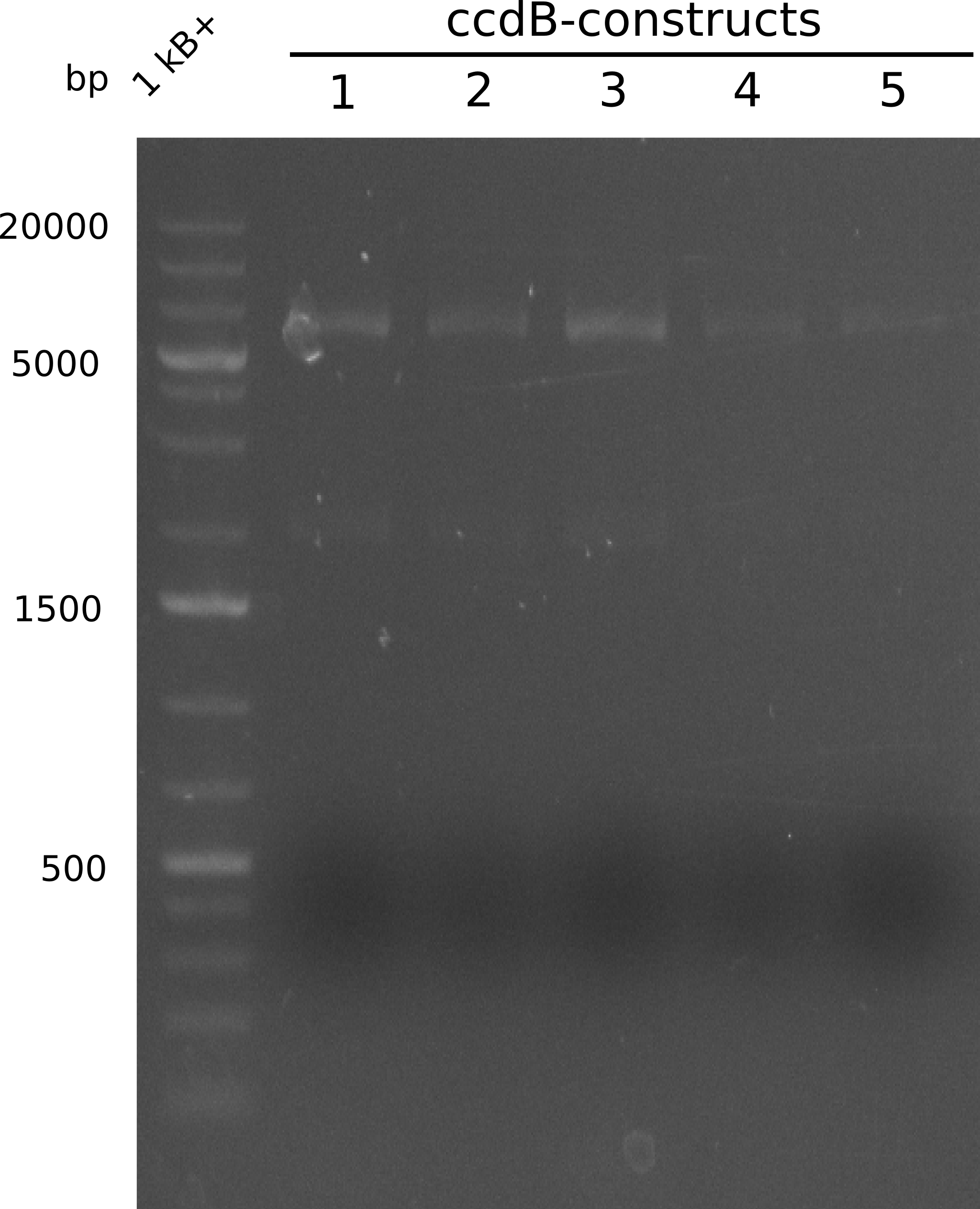
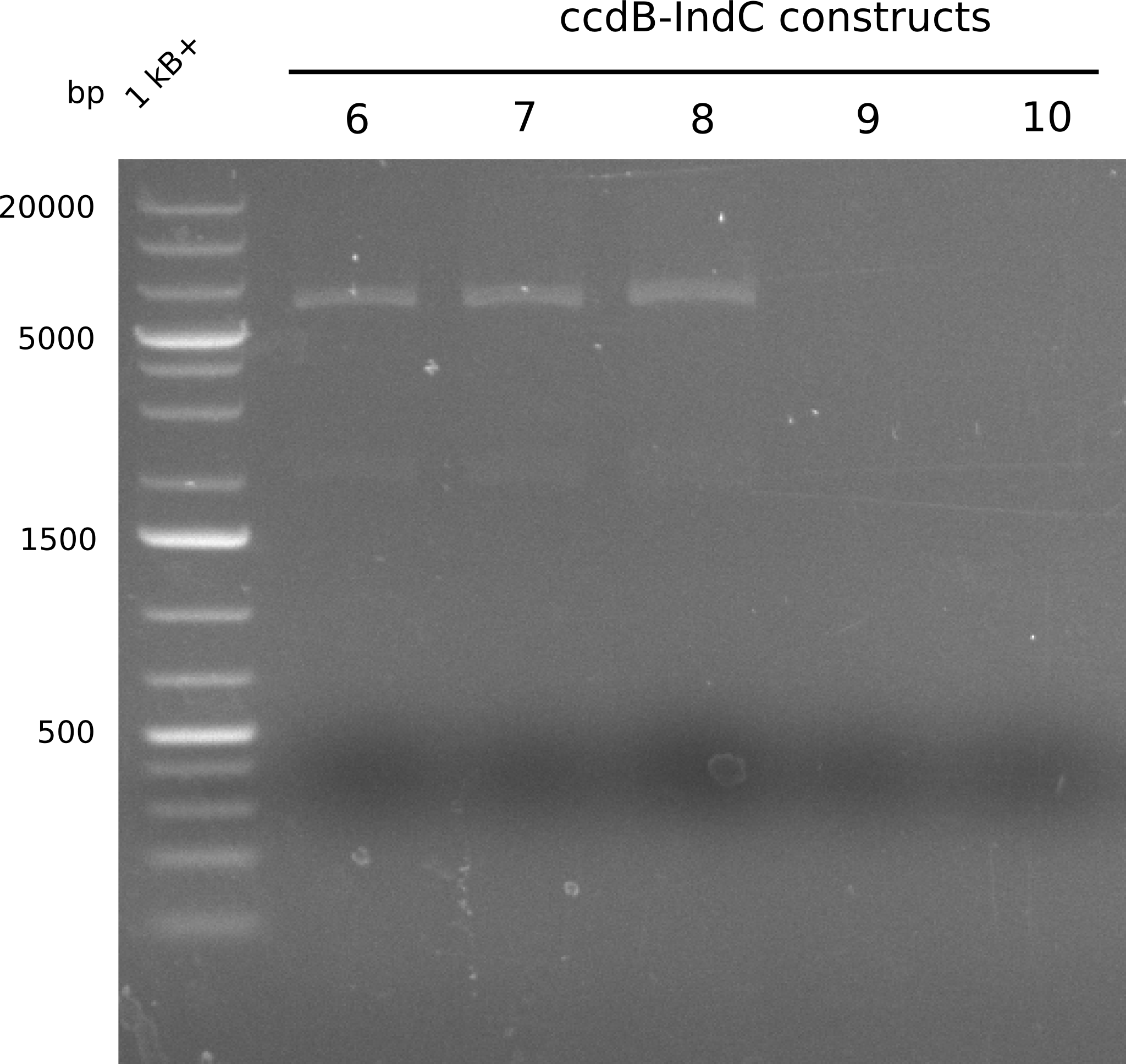
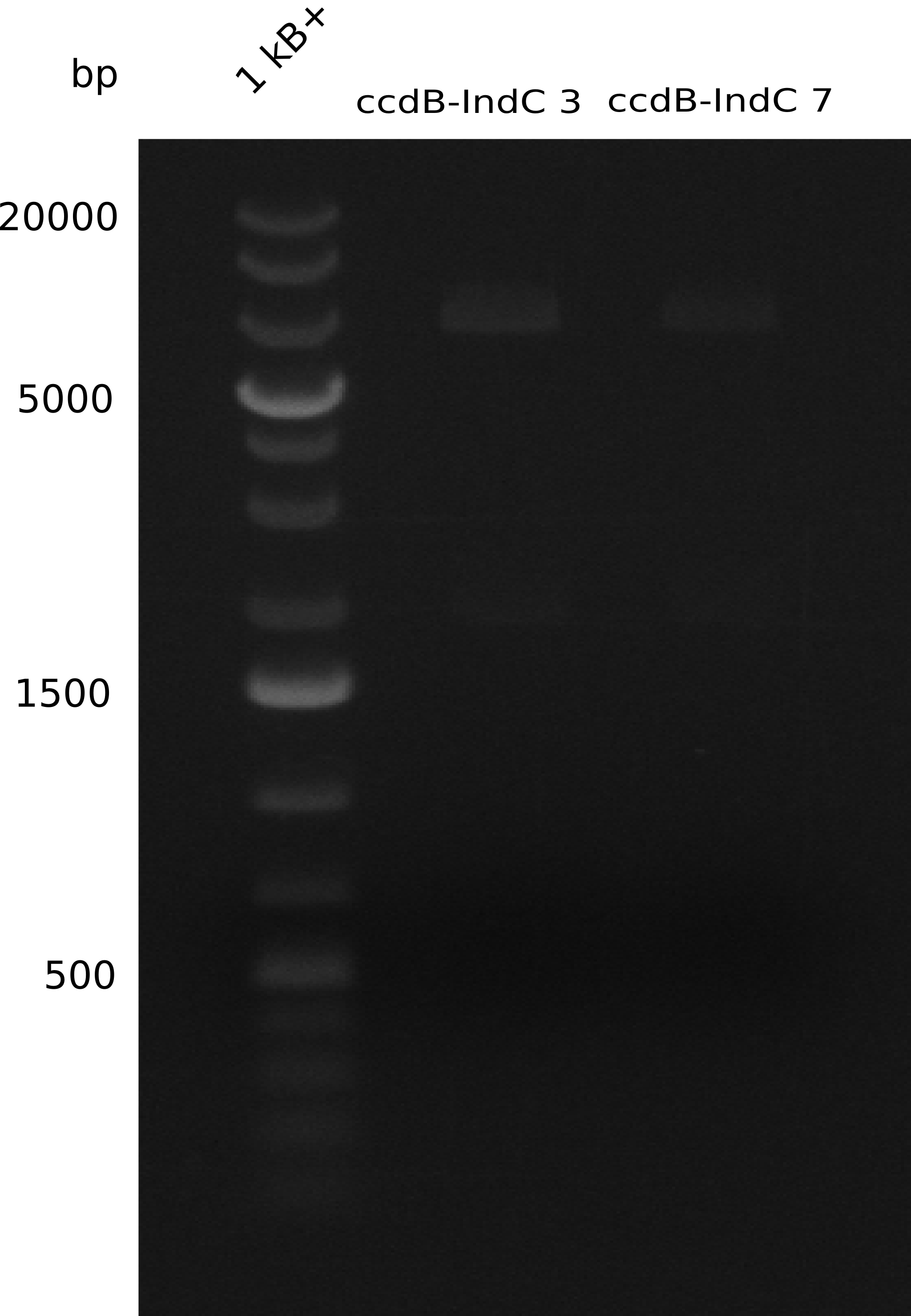
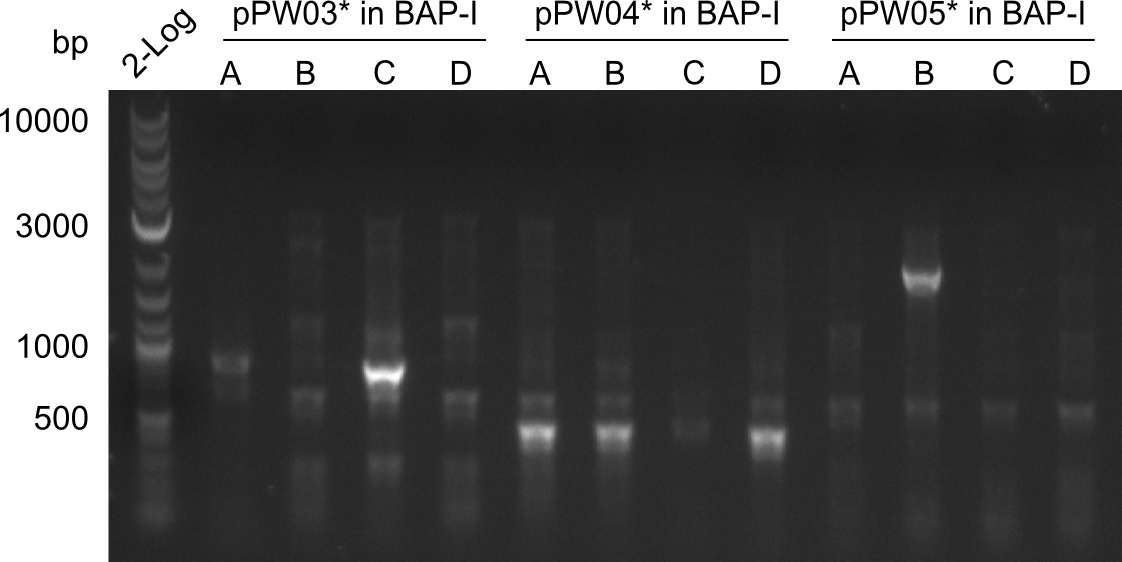
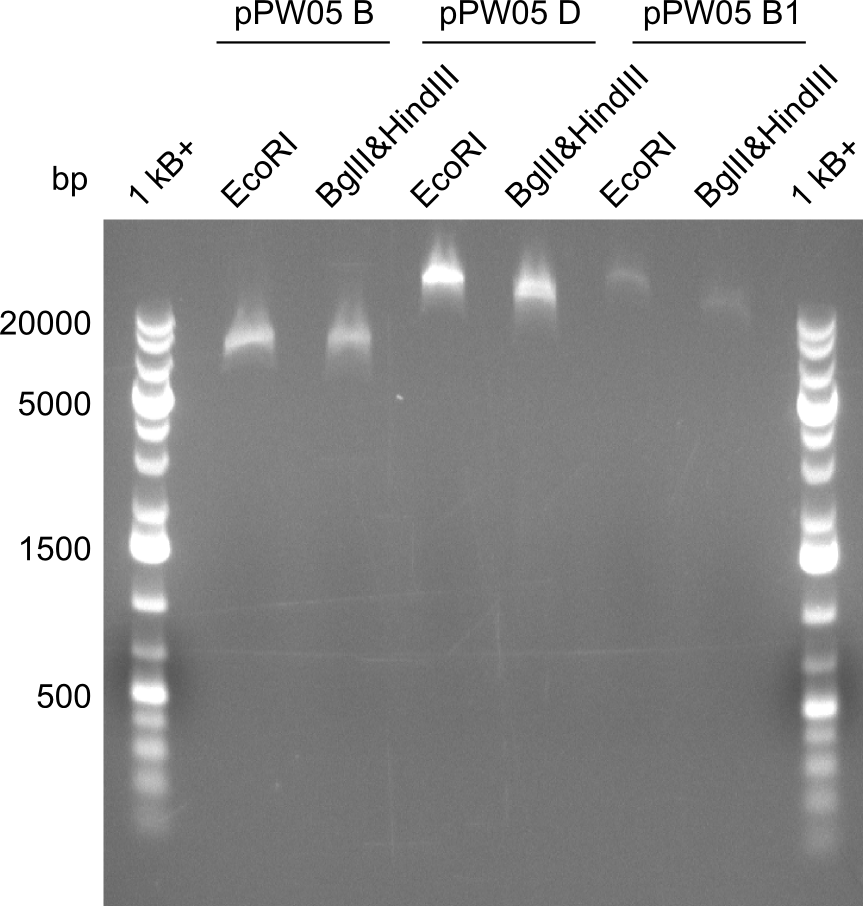
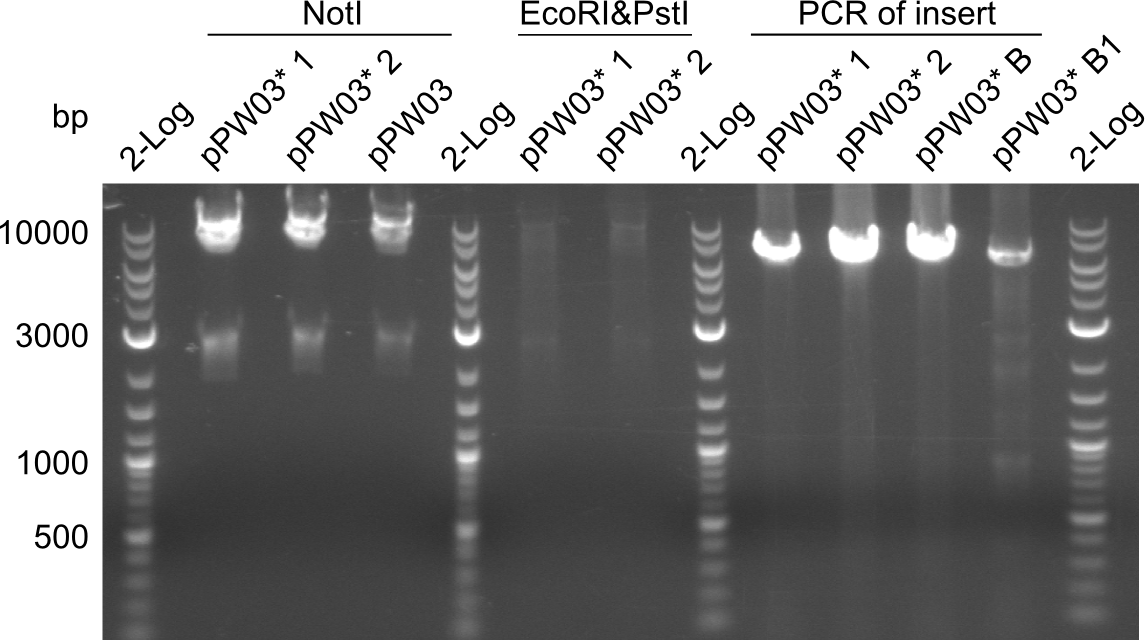
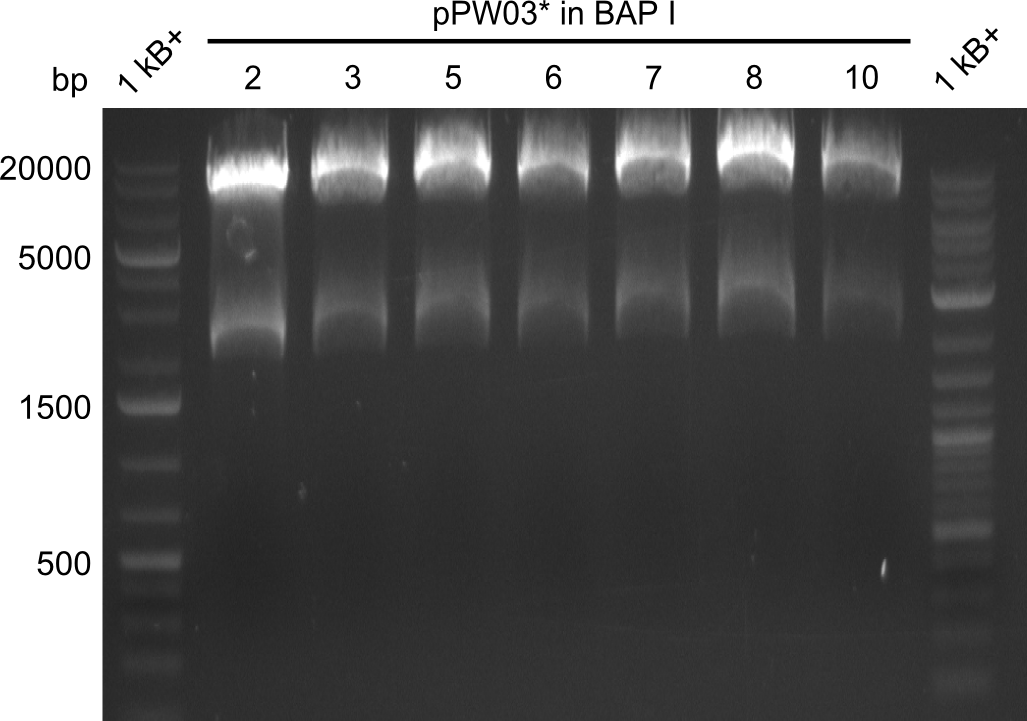
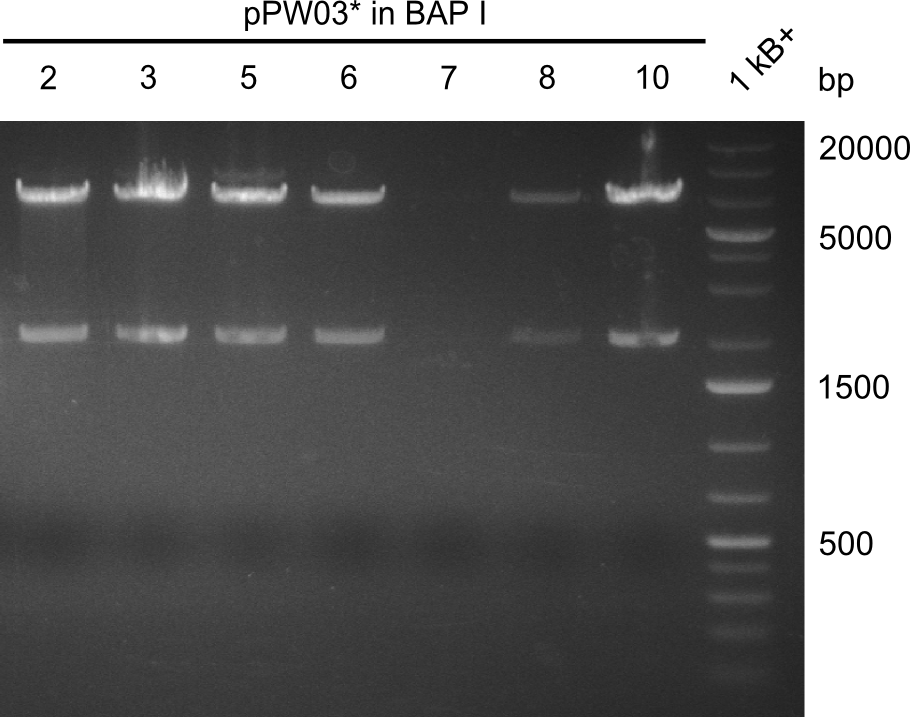
Our fusion experiment was continued for all three constructs with the Gibson Assembly into pSB1C3. The mix was electroporated into DH10β and plated onto Cm-LB. The analytical restriction digest of picked colonies was positive for all constructs. Selected samples of each were proven to be positive by sequencing. Thereafter, the constructs were chemically transformed into our expression strain BAPI and spread onto Cm-LB plates. The colonies turned blue after less than two days, indicating the expression of Indigoidine. | Our fusion experiment was continued for all three constructs with the Gibson Assembly into pSB1C3. The mix was electroporated into DH10β and plated onto Cm-LB. The analytical restriction digest of picked colonies was positive for all constructs. Selected samples of each were proven to be positive by sequencing. Thereafter, the constructs were chemically transformed into our expression strain BAPI and spread onto Cm-LB plates. The colonies turned blue after less than two days, indicating the expression of Indigoidine. | ||
<br/><br/> | <br/><br/> | ||
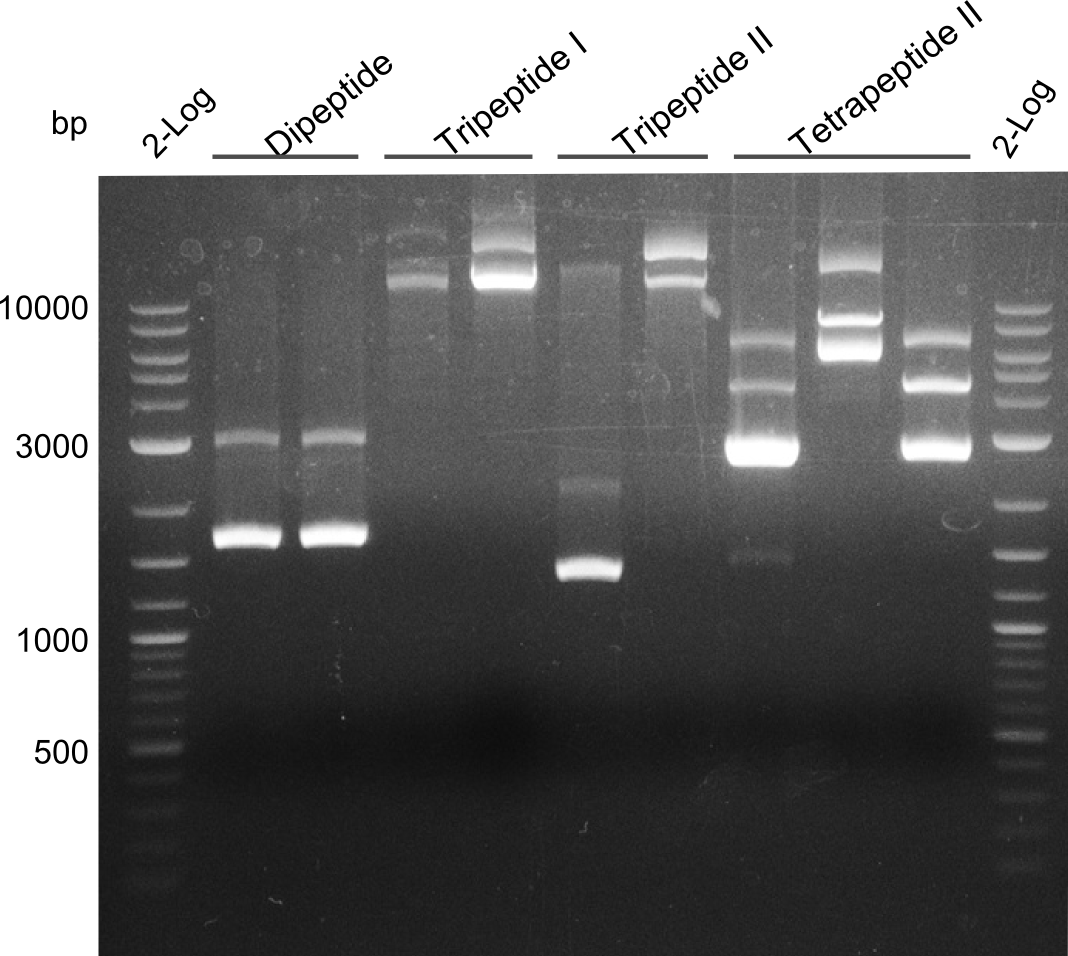
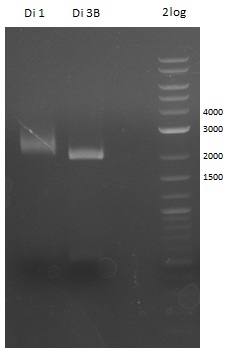
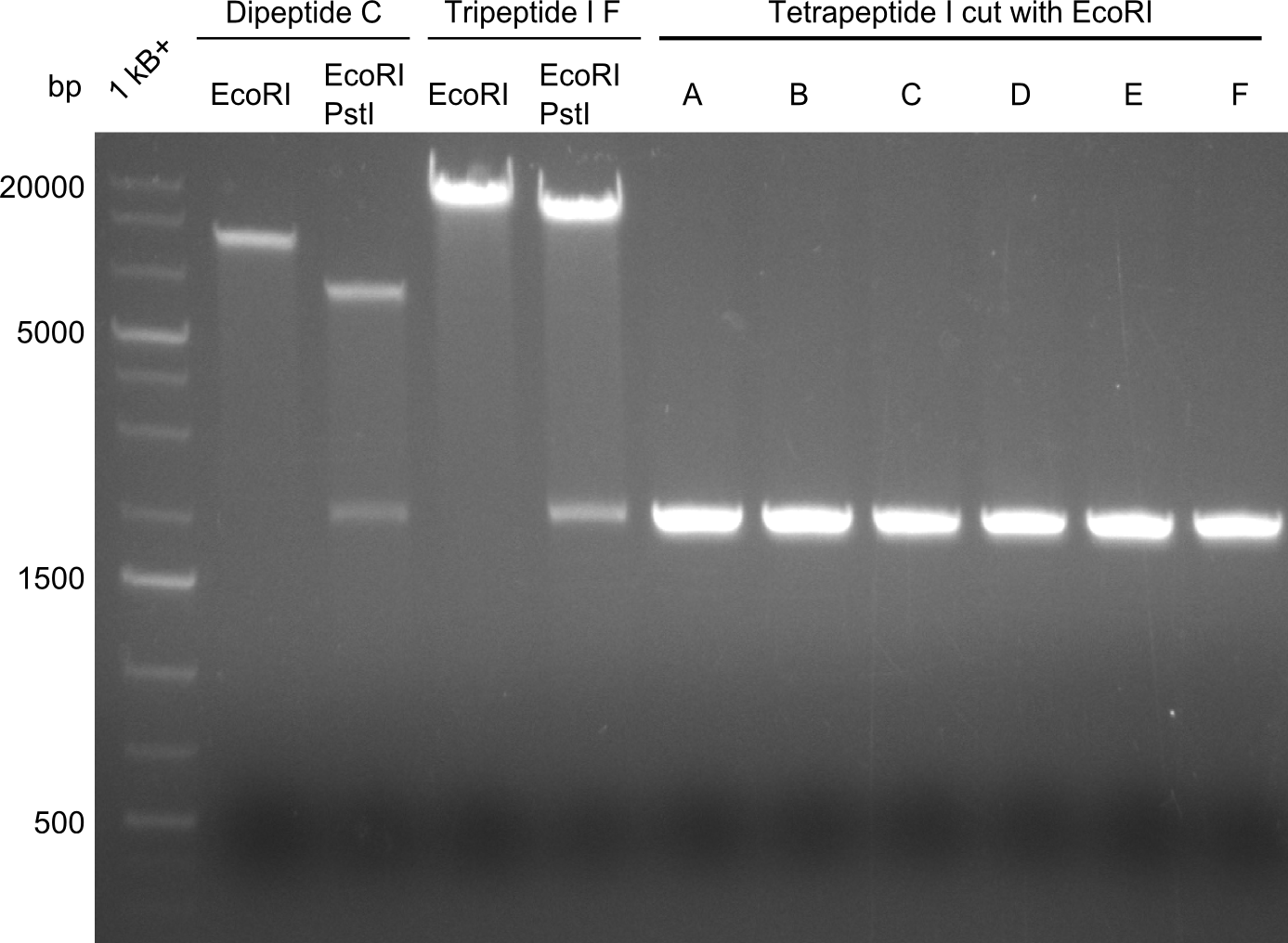
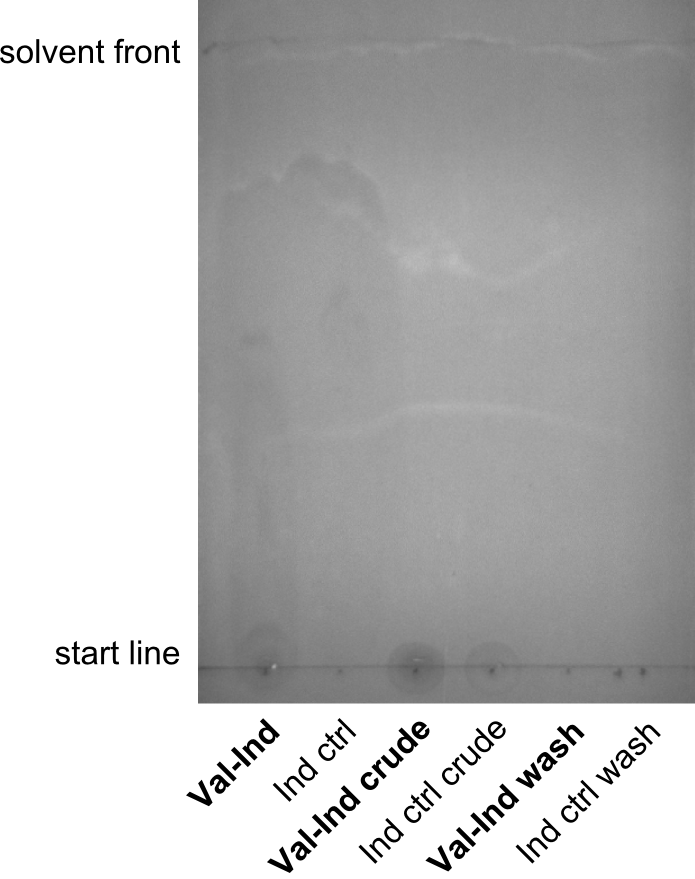
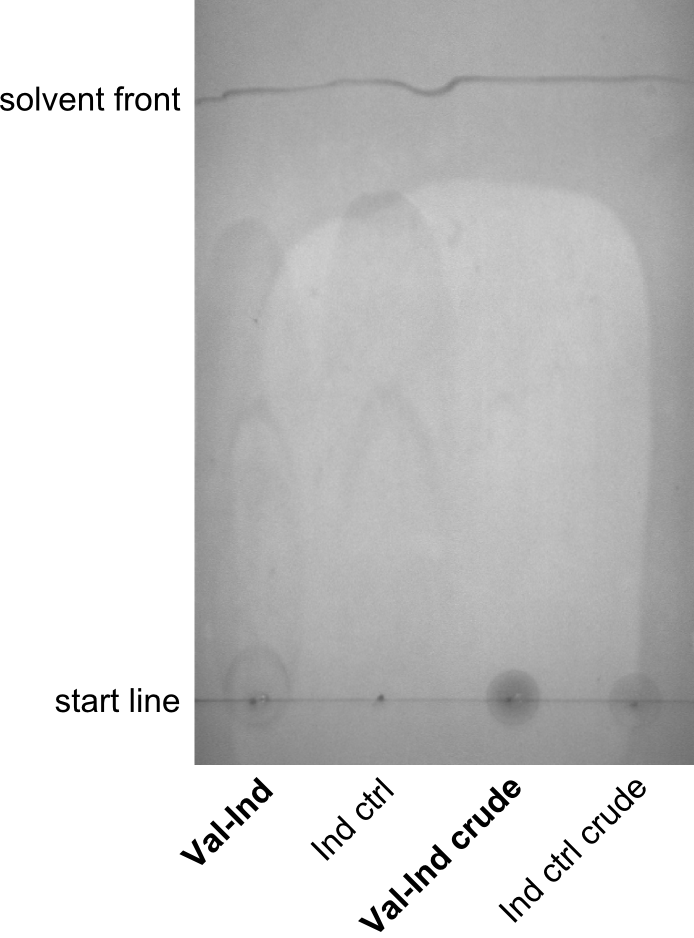
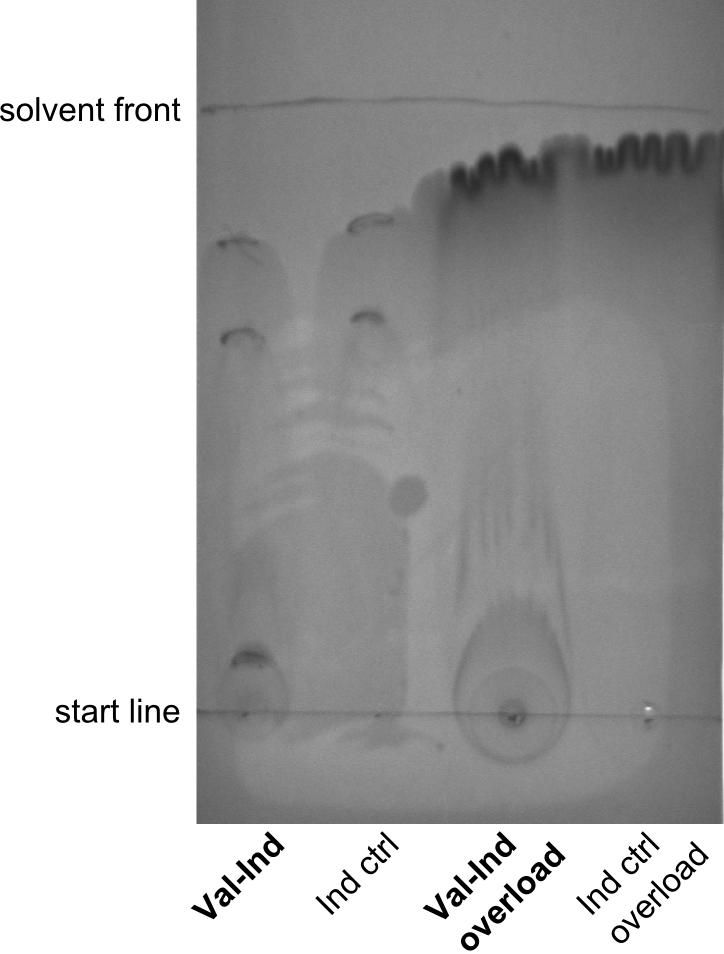
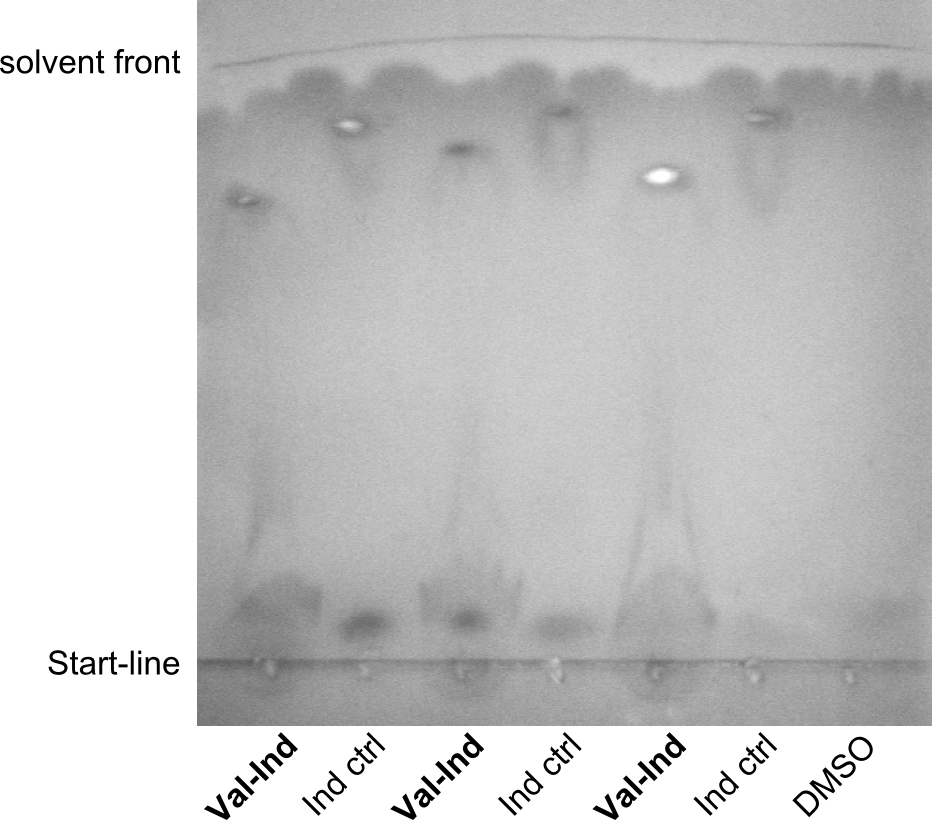
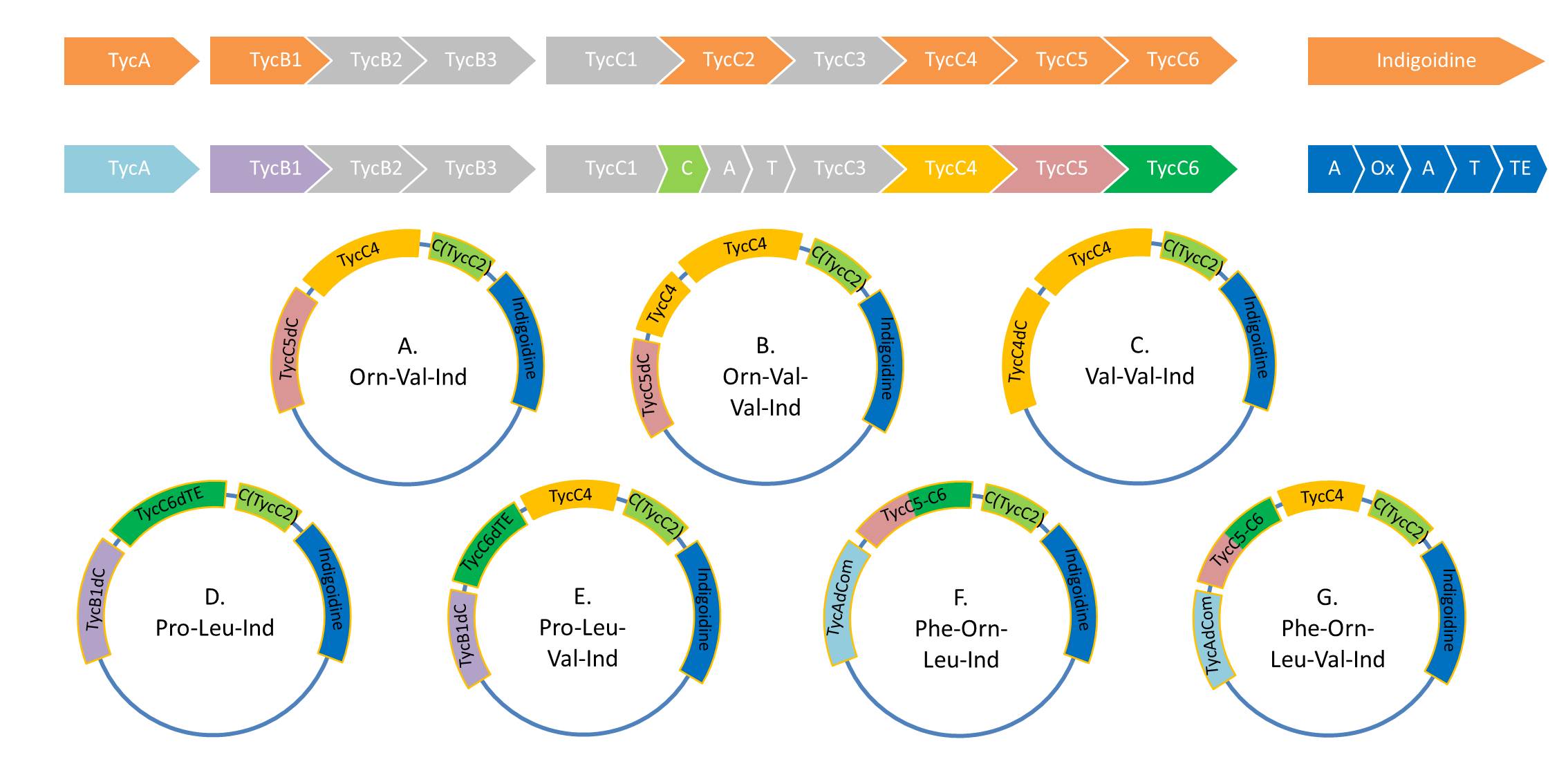
| - | First a SDS-PAGE was executed showing that only the band for the Fusion NRPS producing Val-Ind was positive. For this clone purified Valine-Indigoidine from liquid culture was run next to native Indigoidine in a comparative TLC. | + | First a SDS-PAGE was executed showing that only the band for the Fusion NRPS producing Val-Ind was positive. For this clone purified Valine-Indigoidine from liquid culture was run next to native Indigoidine in a comparative TLC. We were able to show that the product from the Val-Ind-Synthetase runs slower than native Indigoidine. As a result Indigoidine can be used to tag other NRP synthetase products by fusing it at the end of NRPS coding genes. To prove this assumption we designed a new strategy for the fusion of the indC to up to four Tyrocidine modules. For this experiment new Gibson Assembly compatible primers were designed and ordered. |
<br/><br/> | <br/><br/> | ||
<b>BioBrick Assembly</b><br/> | <b>BioBrick Assembly</b><br/> | ||
| Line 182: | Line 182: | ||
<b>Module Shuffling</b><br/> | <b>Module Shuffling</b><br/> | ||
| - | Two months after the start of the tyrocidine project and a lot of planning concerning the verifiability of the existence of small synthetic non-ribosomal peptides, the purification procedure for mass spectrometry was conducted. Since our backup strategies were rather complex, the simplest purification procedure was chosen to begin with. This procedure was performed in order to get a general idea of whether further desalting steps were needed. Purified samples were delievered to the neonate screening facility (tandem MS) at the university medical centre and the mass spectrometry facility at the Institute for Chemistry (HR-ESI MS). <br/><br/> | + | Two months after the start of the tyrocidine project and after a lot of planning concerning the verifiability of the existence of small synthetic non-ribosomal peptides, the purification procedure for mass spectrometry was conducted. Since our backup strategies were rather complex, the simplest purification procedure was chosen to begin with. This procedure was performed in order to get a general idea of whether further desalting steps were needed. Purified samples were delievered to the neonate screening facility (tandem MS) at the university medical centre and the mass spectrometry facility at the Institute for Chemistry (HR-ESI MS). <br/><br/> |
After a first measurement with a tandem mass spectrometer (neonate screening facility) reasonable amino acid levels were observed, accounting for a well suited sample work up process. Therefore, samples of different constructs were taken at several time points after induction and delievered as well to assess time dependet expression. <br/><br/> | After a first measurement with a tandem mass spectrometer (neonate screening facility) reasonable amino acid levels were observed, accounting for a well suited sample work up process. Therefore, samples of different constructs were taken at several time points after induction and delievered as well to assess time dependet expression. <br/><br/> | ||
| Line 202: | Line 202: | ||
<h1>Week 20</h1> | <h1>Week 20</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> | ||
| - | |||
<b> Module Shuffling</b> <br/> | <b> Module Shuffling</b> <br/> | ||
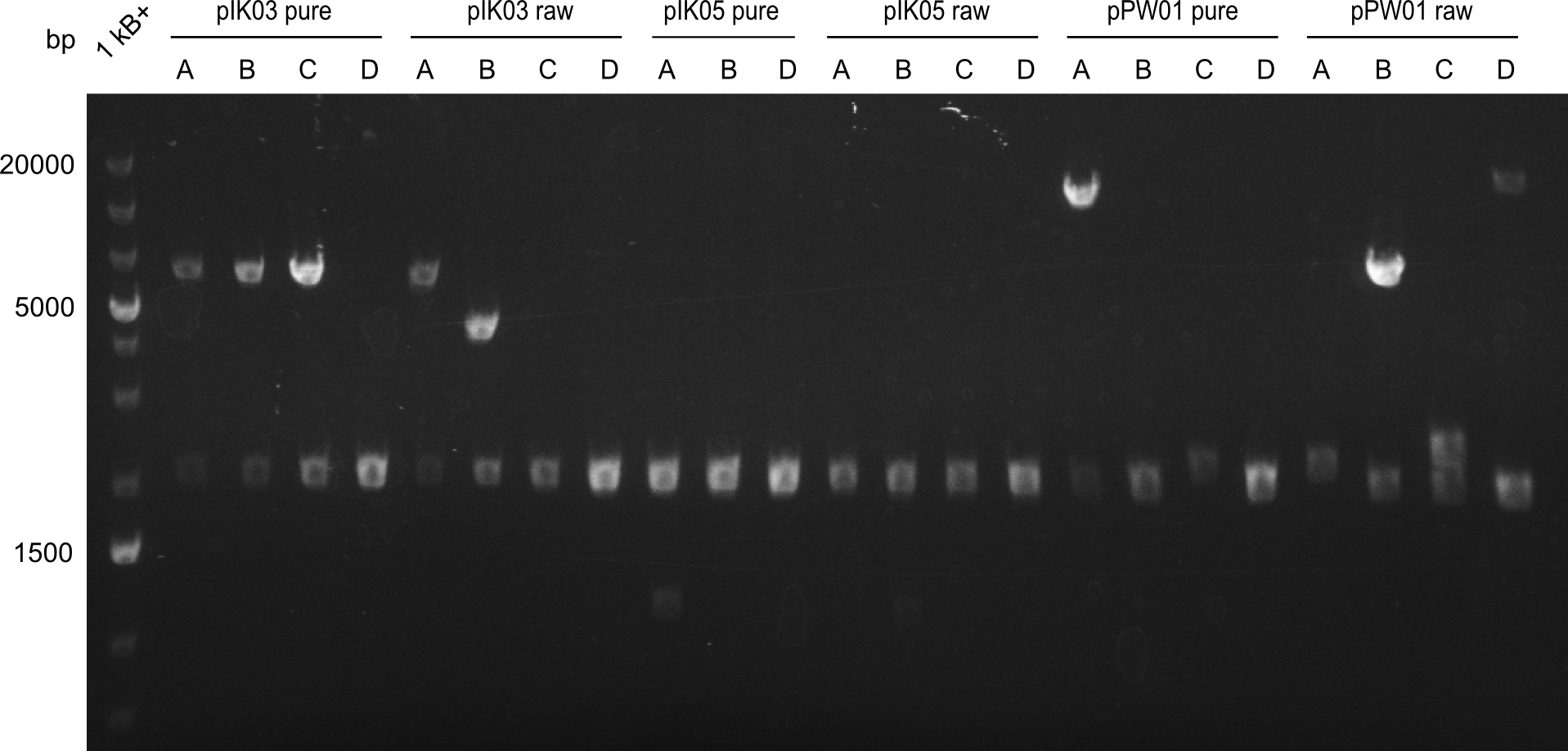
In this week we sent two samples to the mass spectrometry facility of the Institute for Chemistry on campus. We have been offered only a couple of free measurements, so we decided to send two pellet samples containing the Dipeptide (pIK03) and Tripeptide-I (pIK04). Even though we did not receive a conclusive spectrum we can be sure that the salt concentrations were low enough to not overlap the signals of our peptides. We assume that either the solvent or cell lysis debris could be the reason for the unspecific profile. We now want to test samples of the supernatant, as well. <br/><br/> | In this week we sent two samples to the mass spectrometry facility of the Institute for Chemistry on campus. We have been offered only a couple of free measurements, so we decided to send two pellet samples containing the Dipeptide (pIK03) and Tripeptide-I (pIK04). Even though we did not receive a conclusive spectrum we can be sure that the salt concentrations were low enough to not overlap the signals of our peptides. We assume that either the solvent or cell lysis debris could be the reason for the unspecific profile. We now want to test samples of the supernatant, as well. <br/><br/> | ||
| - | |||
<b>Interspecies Module Shuffling </b><br/> | <b>Interspecies Module Shuffling </b><br/> | ||
| Line 215: | Line 213: | ||
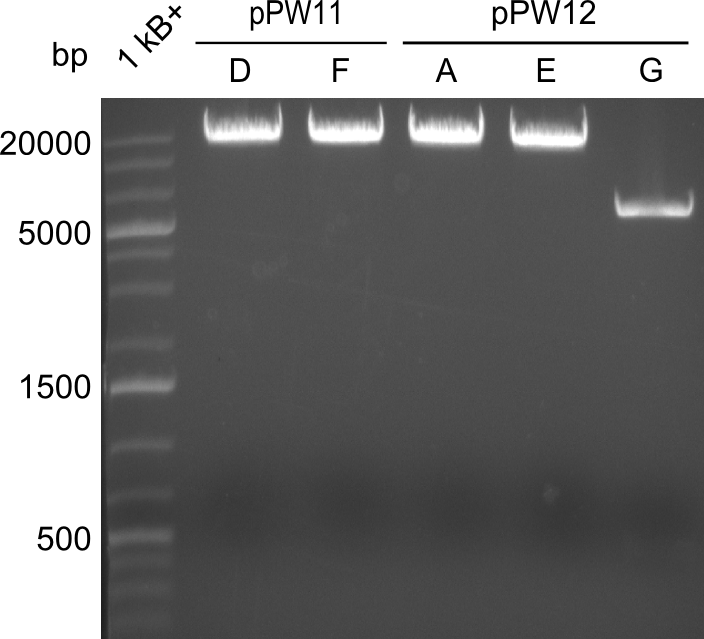
We started the preparation of the positive plasmids for shipping to the parts-registry.<br/><br/> | We started the preparation of the positive plasmids for shipping to the parts-registry.<br/><br/> | ||
<i>Interspecies Module Shuffling </i><br/> | <i>Interspecies Module Shuffling </i><br/> | ||
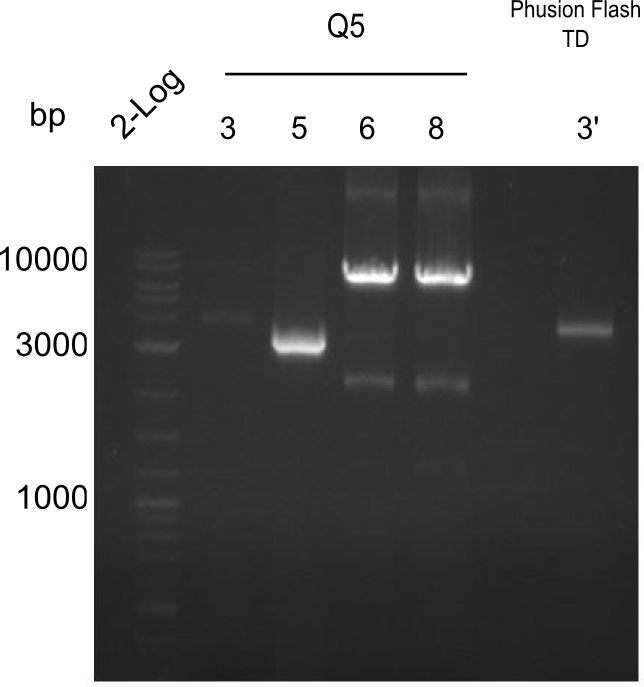
| - | With regard to the week before, the CPEC approach was resumed with a mutation at the desired position. In parallel as an alternative approach the indC fragment of the domain shuffling experiments was used. <br/><br/ | + | With regard to the week before, the CPEC approach was resumed with a mutation at the desired position. In parallel as an alternative approach the indC fragment of the domain shuffling experiments was used.<br/><br/></p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
<div class="item september"> | <div class="item september"> | ||
| - | |||
<div class="container"> | <div class="container"> | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| Line 245: | Line 236: | ||
</div> | </div> | ||
<div class="item september"> | <div class="item september"> | ||
| - | |||
<div class="container"> | <div class="container"> | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 22</h1> | <h1>Week 22</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Week of | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Week of SUBMISSION DEADLINE (2013-09-25) |
| - | + | Tyrocidine-Indigoidine-fusion extended<br/> | |
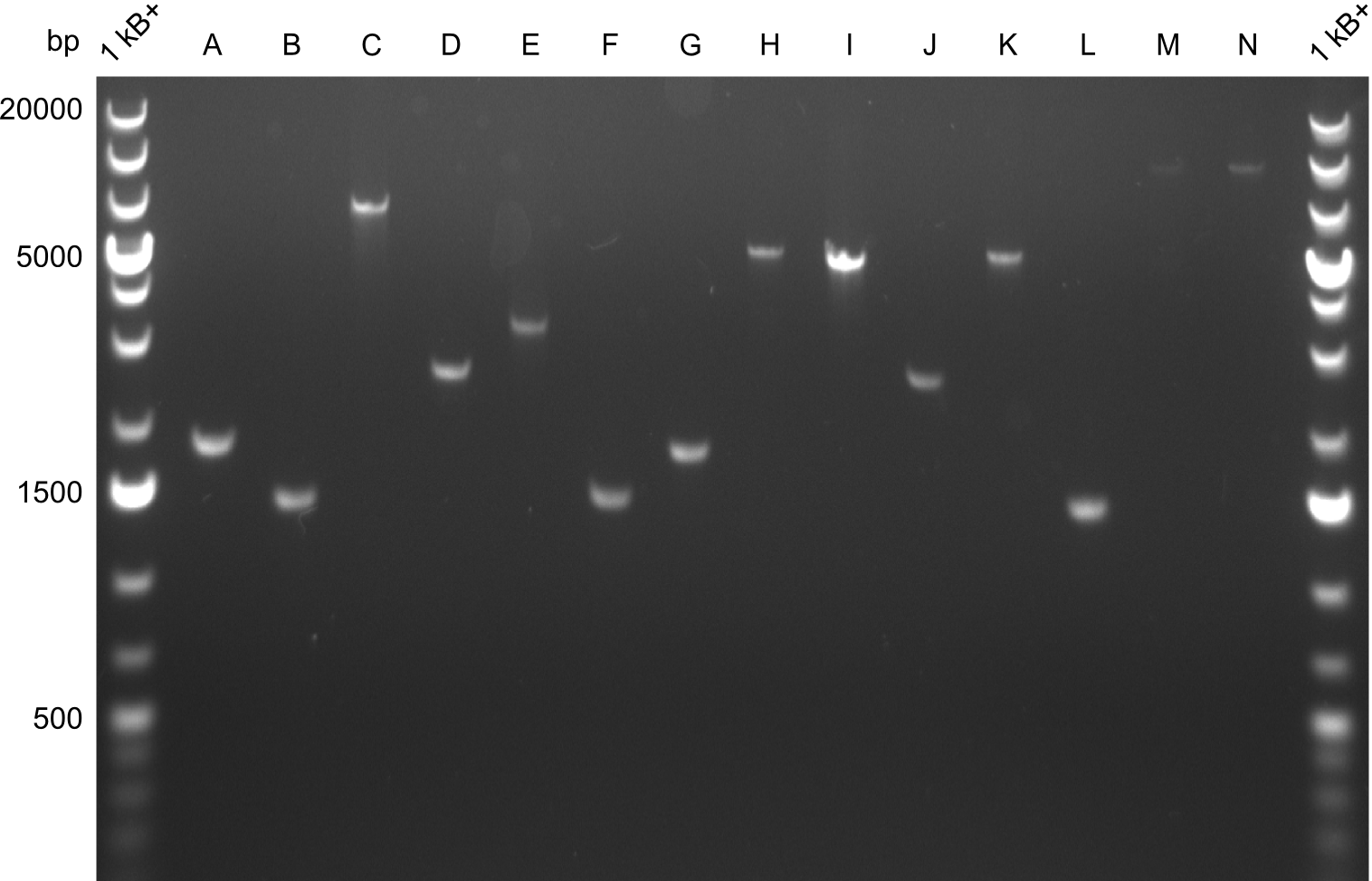
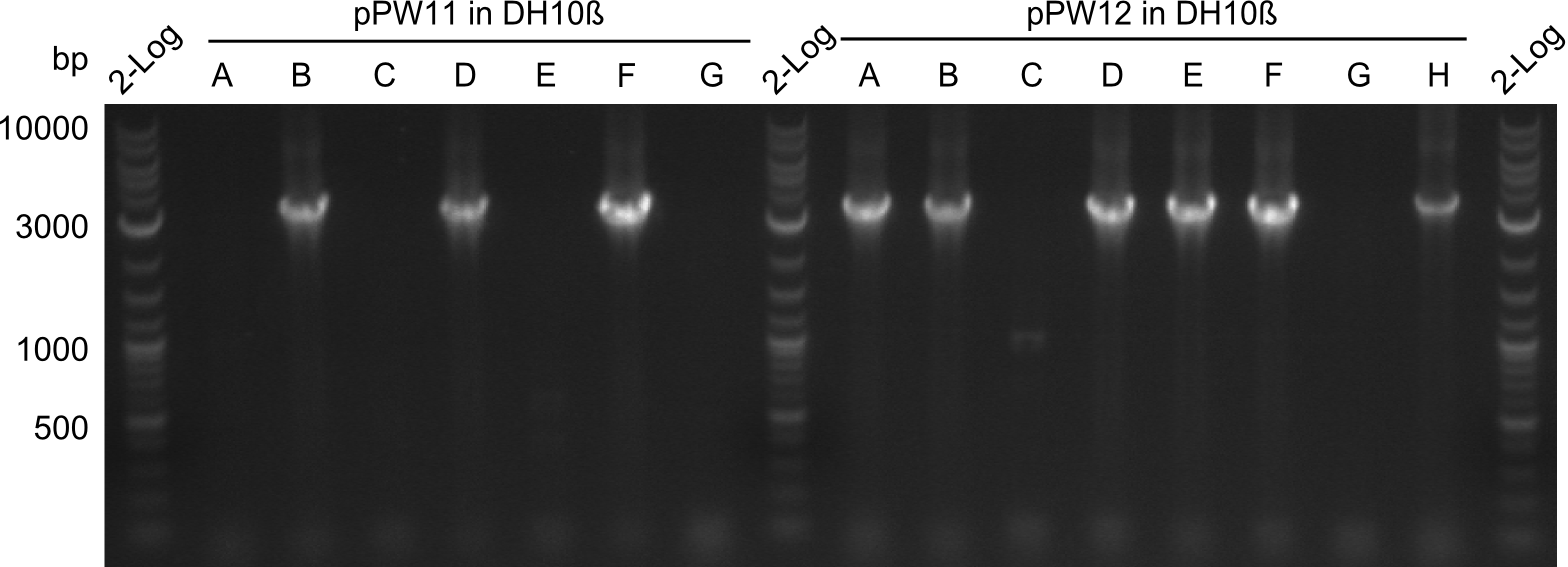
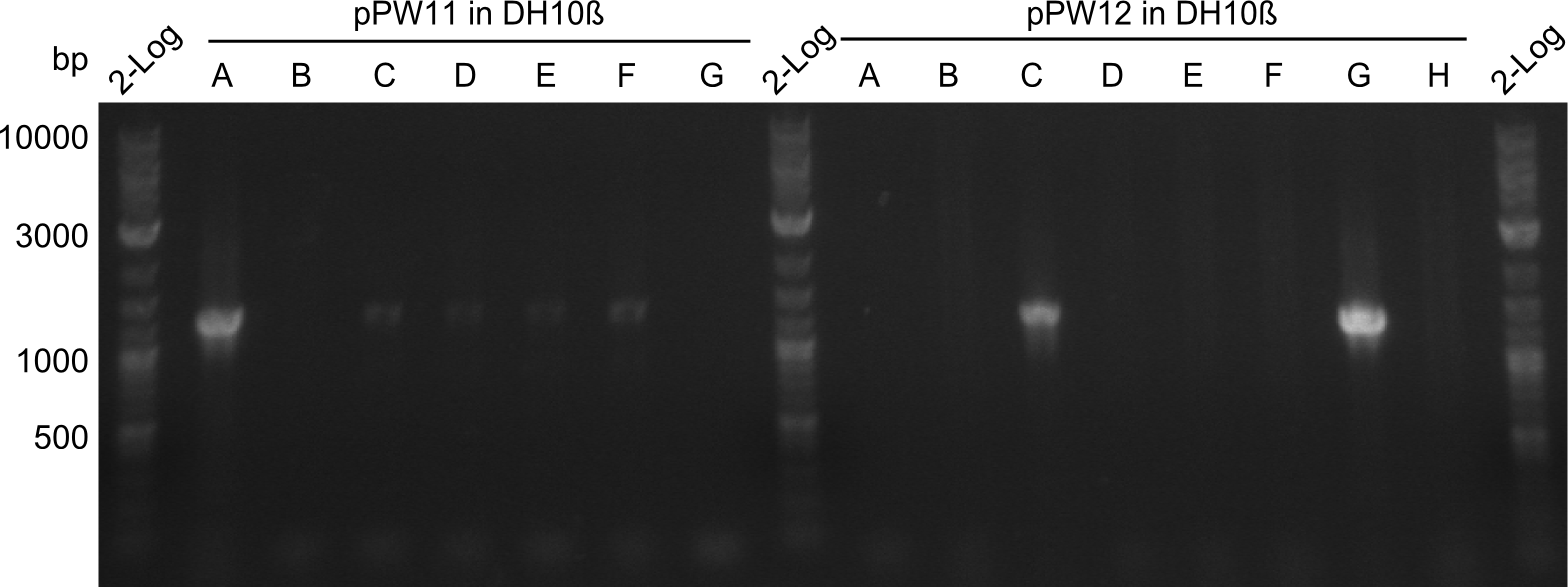
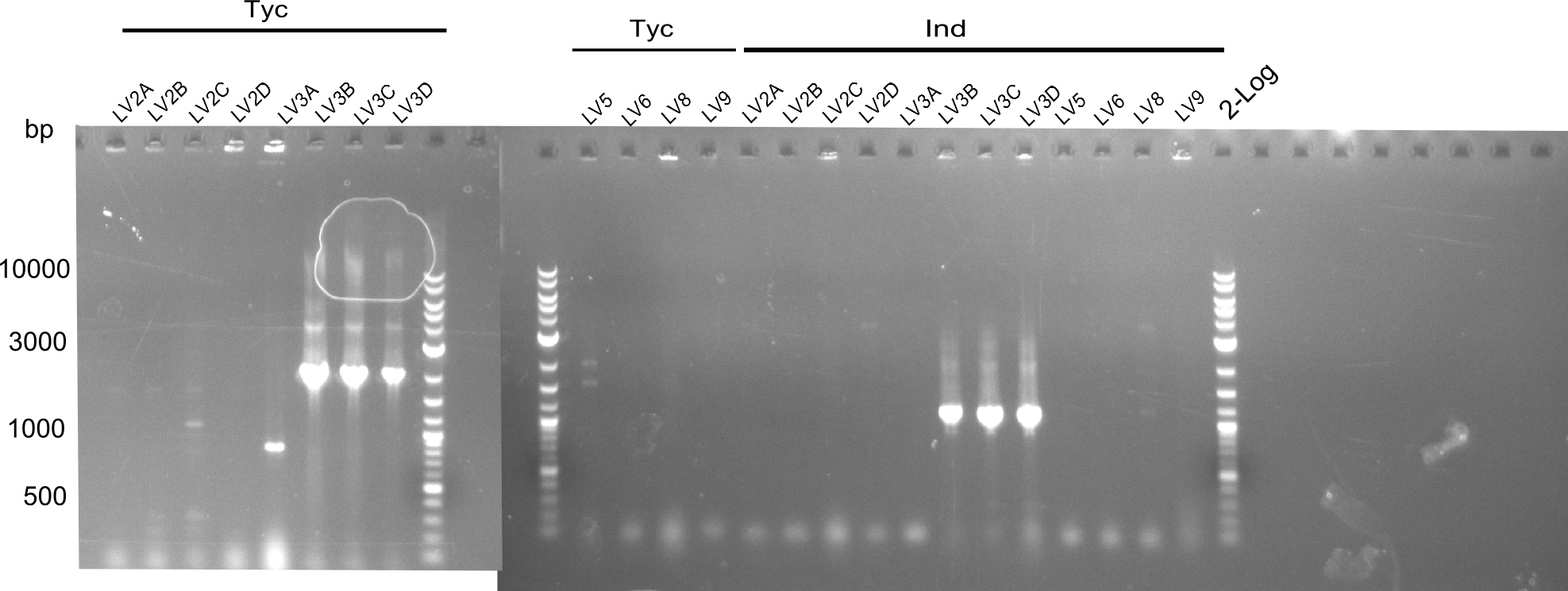
| - | As we focused mainly on the parts submission the week before, the Tyrocidine-Indigoidine fusion was picked up again this week. Four samples (pPW06, pPW09, pPW10 and pPW11) were transformed and colonies screened by colony PCRs. Except for pPW12G, all constructs showed expected cutting profiles after enzymatic digest. The sample pPW06 newC was transformed into BAP-I cells. The liquid culture was induced with IPTG and turned blue. The indigoidine-tagged peptide was run on a TLC to proof the basic working principle of this tagging method. | + | As we focused mainly on the parts submission the week before, the Tyrocidine-Indigoidine fusion was picked up again this week. Four samples (pPW06, pPW09, pPW10 and pPW11) were transformed and colonies screened by colony PCRs. Except for pPW12G, all constructs showed expected cutting profiles after enzymatic digest. The sample pPW06 newC was transformed into BAP-I cells. The liquid culture was induced with IPTG and turned blue. The indigoidine-tagged peptide was run on a TLC to proof the basic working principle of this tagging method.<br/> |
| - | + | Linker variation<br/> | |
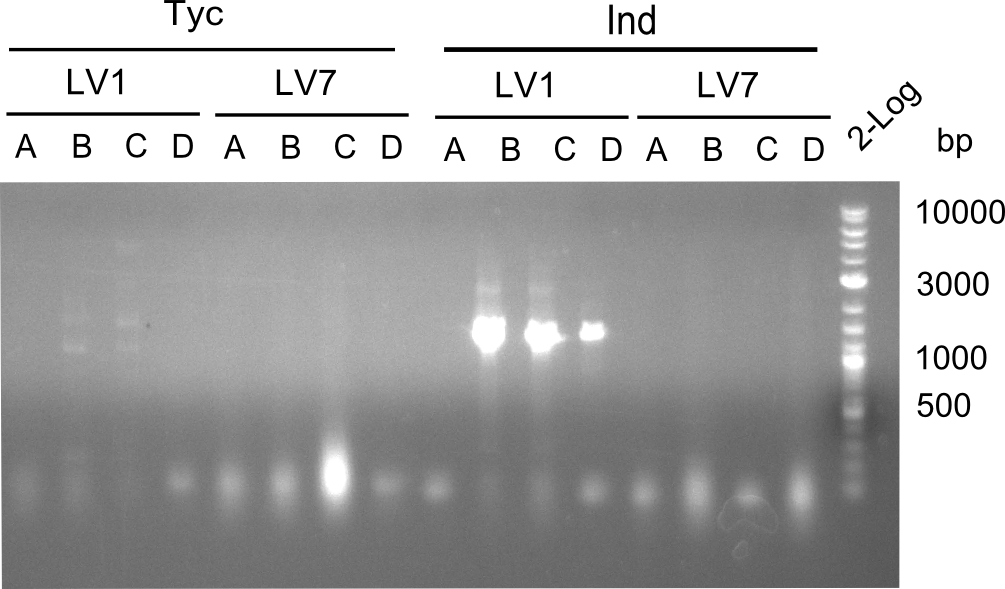
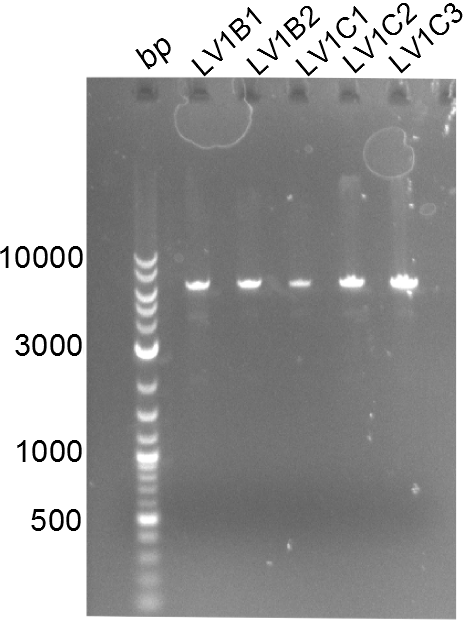
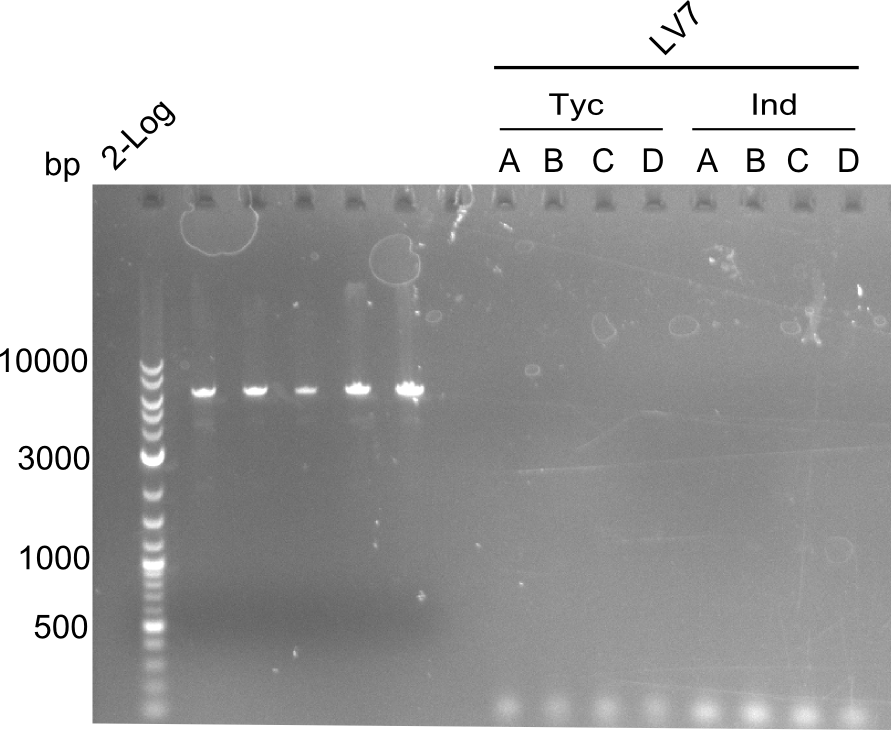
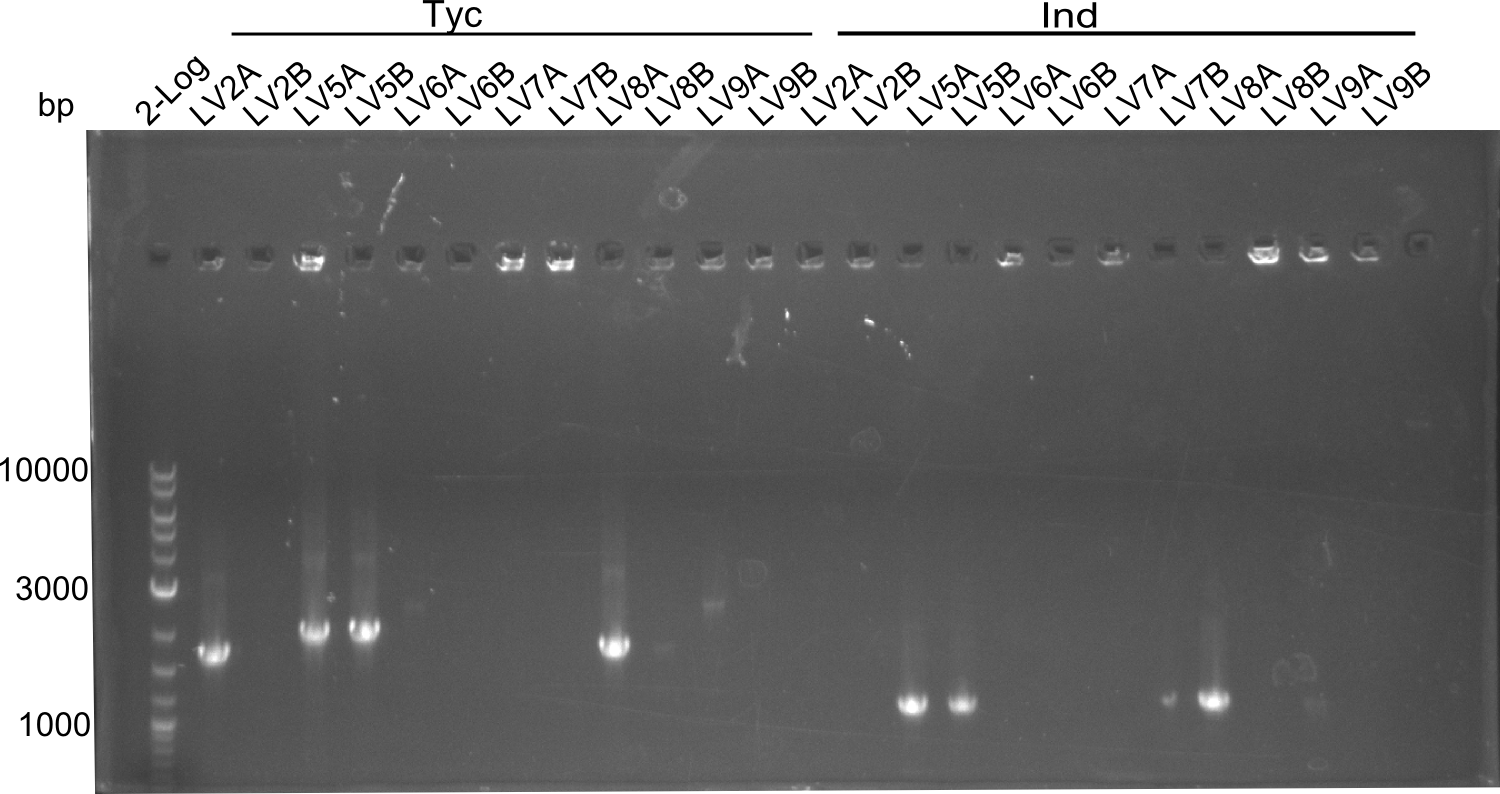
Following up the week before, two of the constructs, which comprised the most wide apart linker positions, were assembled. <br/> | Following up the week before, two of the constructs, which comprised the most wide apart linker positions, were assembled. <br/> | ||
Both plasmids were transformed via electroporation in competent DH10ß cells, spread on plates and picked for colony PCRs. As only LV1 showed appropriate bands, further colonies of LV7 were picked. In between, LV1 was chemically transformed into BAPI.<br/> | Both plasmids were transformed via electroporation in competent DH10ß cells, spread on plates and picked for colony PCRs. As only LV1 showed appropriate bands, further colonies of LV7 were picked. In between, LV1 was chemically transformed into BAPI.<br/> | ||
| Line 260: | Line 250: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
<div class="item september last"> | <div class="item september last"> | ||
<img src="data:image/png;base64,"/> | <img src="data:image/png;base64,"/> | ||
| Line 265: | Line 256: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 23</h1> | <h1>Week 23</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"><b>WIKIFREEZE (04.10.2013)</b> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"><b>WIKIFREEZE (04.10.2013)</b><br/> |
| - | <b>Linker variation</b> | + | <b>Linker variation</b><br/> |
| - | In this week we aim to produce results out of our –putative- positive constructs. Therefore the left presumably positive samples induced and turned blue. Our goal was to evaluate, whether even through the variation of our Linkers our desired Val-Ind was expressed. Our samples were compared to native Indigoidine by TLC. | + | In this week we aim to produce results out of our –putative- positive constructs. Therefore the left presumably positive samples induced and turned blue. Our goal was to evaluate, whether even through the variation of our Linkers our desired Val-Ind was expressed. Our samples were compared to native Indigoidine by TLC.</p> |
| - | </p> | + | |
</div> | </div> | ||
| Line 279: | Line 269: | ||
</div> | </div> | ||
</div><!-- /.carousel --> | </div><!-- /.carousel --> | ||
| + | </div> | ||
| + | <div class="col-sm-12 col-md-6"> | ||
| - | <div class="jumbotron methods" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | + | <div class="jumbotron methods" style="max-height:360px; margin-top:10%" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| - | < | + | <a class="fancybox fancyGraphical" rel="group" href="https://static.igem.org/mediawiki/2013/f/f6/Heidelberg_pepind_NB.png" style="height:260px; display:block;"> |
| - | + | <img style="margin-bottom:10px; height:100%; padding:4%; margin-left:5%;border-style:solid;border-width:1px;border-radius: 4px; border-color: grey;" src="https://static.igem.org/mediawiki/2013/f/f6/Heidelberg_pepind_NB.png"></img> | |
| - | + | </a> | |
</div> | </div> | ||
</div> | </div> | ||
| - | <div class="col-sm- | + | <div class="col-sm-12"> |
<!--Start Weekly Labjournal--> | <!--Start Weekly Labjournal--> | ||
| Line 301: | Line 293: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
| Line 336: | Line 328: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 368: | Line 360: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 403: | Line 395: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 436: | Line 428: | ||
<li class="active"><a href="#a16" data-toggle="tab">Overview</a></li> | <li class="active"><a href="#a16" data-toggle="tab">Overview</a></li> | ||
<li><a href="#b16" data-toggle="tab">Module Shuffling</a></li> | <li><a href="#b16" data-toggle="tab">Module Shuffling</a></li> | ||
| - | + | ||
<li><a href="#d16" data-toggle="tab">BioBrick Assembly</a></li> | <li><a href="#d16" data-toggle="tab">BioBrick Assembly</a></li> | ||
| Line 442: | Line 434: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 458: | Line 450: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 482: | Line 468: | ||
<div> | <div> | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | |||
<li><a href="#b17" data-toggle="tab">Module Shuffling</a></li> | <li><a href="#b17" data-toggle="tab">Module Shuffling</a></li> | ||
<li><a href="#c17" data-toggle="tab">Interspecies Module Shuffling</a></li> | <li><a href="#c17" data-toggle="tab">Interspecies Module Shuffling</a></li> | ||
| Line 490: | Line 475: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 527: | Line 512: | ||
<div> | <div> | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | + | ||
<li><a href="#b18" data-toggle="tab">Module Shuffling</a></li> | <li><a href="#b18" data-toggle="tab">Module Shuffling</a></li> | ||
<li><a href="#c18" data-toggle="tab">Interspecies Module Shuffling</a></li> | <li><a href="#c18" data-toggle="tab">Interspecies Module Shuffling</a></li> | ||
| Line 534: | Line 519: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 569: | Line 554: | ||
<div> | <div> | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | + | ||
<li><a href="#b19" data-toggle="tab">Module Shuffling</a></li> | <li><a href="#b19" data-toggle="tab">Module Shuffling</a></li> | ||
| Line 576: | Line 561: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 616: | Line 601: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 652: | Line 637: | ||
<div> | <div> | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | + | ||
| - | + | ||
<li><a href="#c21" data-toggle="tab">Interspecies Module Shuffling</a></li> | <li><a href="#c21" data-toggle="tab">Interspecies Module Shuffling</a></li> | ||
<li><a href="#d21" data-toggle="tab">BioBrick Assembly</a></li> | <li><a href="#d21" data-toggle="tab">BioBrick Assembly</a></li> | ||
| Line 660: | Line 644: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 709: | Line 693: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
| Line 758: | Line 742: | ||
</div> | </div> | ||
<div class="jumbotron weekly"> | <div class="jumbotron weekly"> | ||
| - | <div class=" | + | <div class="nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> |
<div class="tab-content"> | <div class="tab-content"> | ||
Latest revision as of 03:25, 29 October 2013


 "
"