Team:Heidelberg/NRPSDesigner
From 2013.igem.org
(Difference between revisions)
JuliaS1992 (Talk | contribs) |
JuliaS1992 (Talk | contribs) |
||
| (7 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
<h1><span style="font-size:180%;color:#074E0B;">NRPS-Designer.</span><span class="text-muted" style="font-family:Arial, sans-serif; font-size:100%"> Designing custom peptides.</span></h1> | <h1><span style="font-size:180%;color:#074E0B;">NRPS-Designer.</span><span class="text-muted" style="font-family:Arial, sans-serif; font-size:100%"> Designing custom peptides.</span></h1> | ||
<p>This tool allows you to design any customized peptide you can build using the most common NRPS domains. This gives you the oppurtunity to include non-proteinogenic amino acids and secondary modifications without going through chemical synthesis or tRNA reprogramming. On top of providing you with a target pathway sequence the tool is also integrated with the parts registry to include any further BioBricks and with Gibthon to directly go from NRP design to Gibson cloning strategy within 10 minutes.</p> | <p>This tool allows you to design any customized peptide you can build using the most common NRPS domains. This gives you the oppurtunity to include non-proteinogenic amino acids and secondary modifications without going through chemical synthesis or tRNA reprogramming. On top of providing you with a target pathway sequence the tool is also integrated with the parts registry to include any further BioBricks and with Gibthon to directly go from NRP design to Gibson cloning strategy within 10 minutes.</p> | ||
| - | <p>You should also read our <a href="https://2013.igem.org/Team:Heidelberg/Project_Software">documentation</a> and our <a href=" | + | <p>You should also read our <a href="https://2013.igem.org/Team:Heidelberg/Project_Software">documentation</a> and our <a href="https://2013.igem.org/Team:Heidelberg/RFCs"><b>RFC100</b></a>! |
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| Line 90: | Line 90: | ||
<h1>Week 15</h1> | <h1>Week 15</h1> | ||
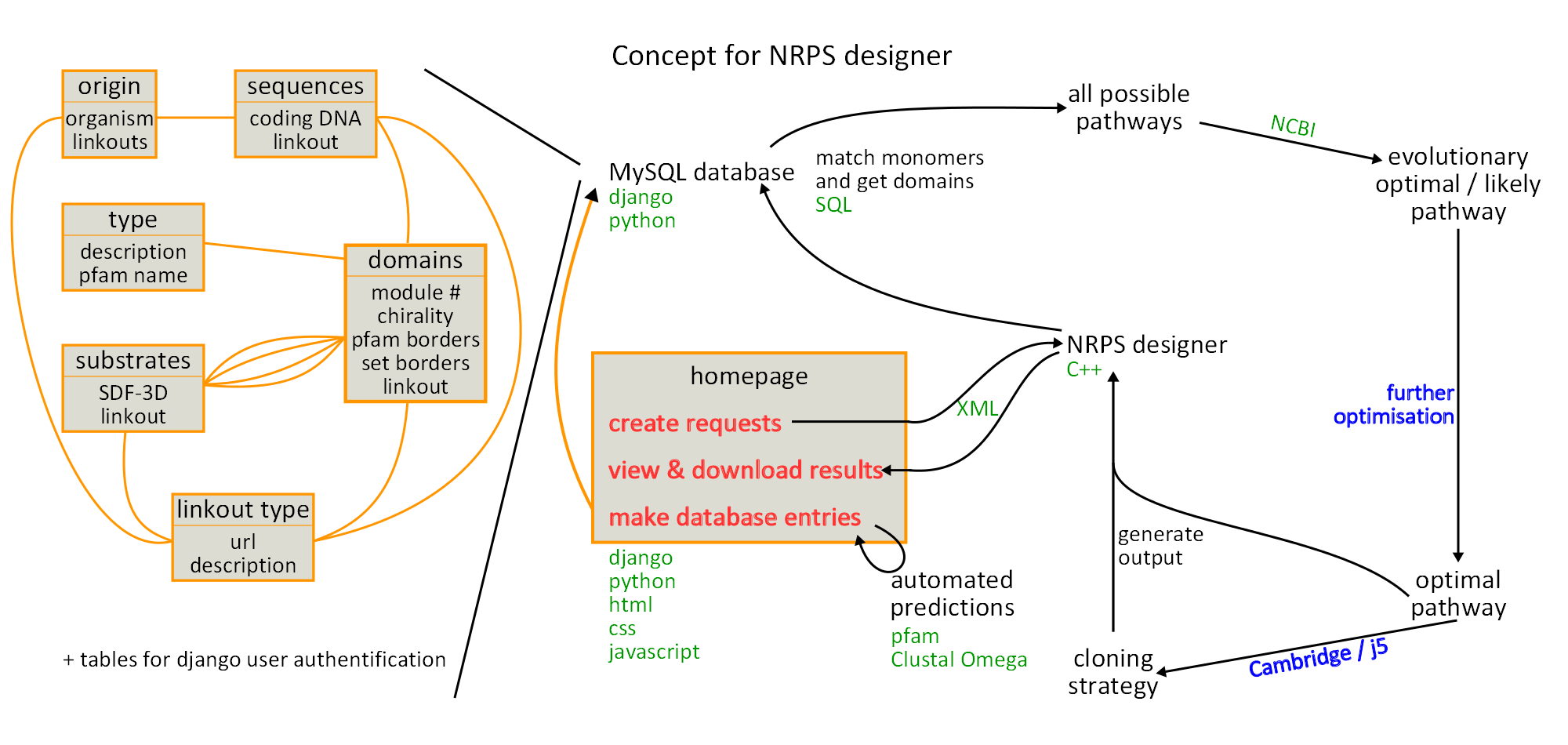
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Besides the regular changes to the database design, we integrated the graphical user interface in Django and also included OpenBable for displaying chemical structures. Furthermore we visualised our concept this week.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Besides the regular changes to the database design, we integrated the graphical user interface in Django and also included OpenBable for displaying chemical structures. Furthermore we visualised our concept this week.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 99: | Line 99: | ||
<h1>Week 16</h1> | <h1>Week 16</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we mainly made changes to the NRP-Design interface and integrated ajax.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we mainly made changes to the NRP-Design interface and integrated ajax.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 109: | Line 109: | ||
<h1>Week 17</h1> | <h1>Week 17</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Besides fixing the relations in our database and improving th interface, we integrated Gibthon - the tool for Gibson primer design developed by the 2011 Cambridge team.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Besides fixing the relations in our database and improving th interface, we integrated Gibthon - the tool for Gibson primer design developed by the 2011 Cambridge team.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 119: | Line 119: | ||
<h1>Week 18</h1> | <h1>Week 18</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we started filling our database, using the NRPS-PKS database. Besides that we introduced session data and updated the designer algorithm.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we started filling our database, using the NRPS-PKS database. Besides that we introduced session data and updated the designer algorithm.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 129: | Line 129: | ||
<h1>Week 19</h1> | <h1>Week 19</h1> | ||
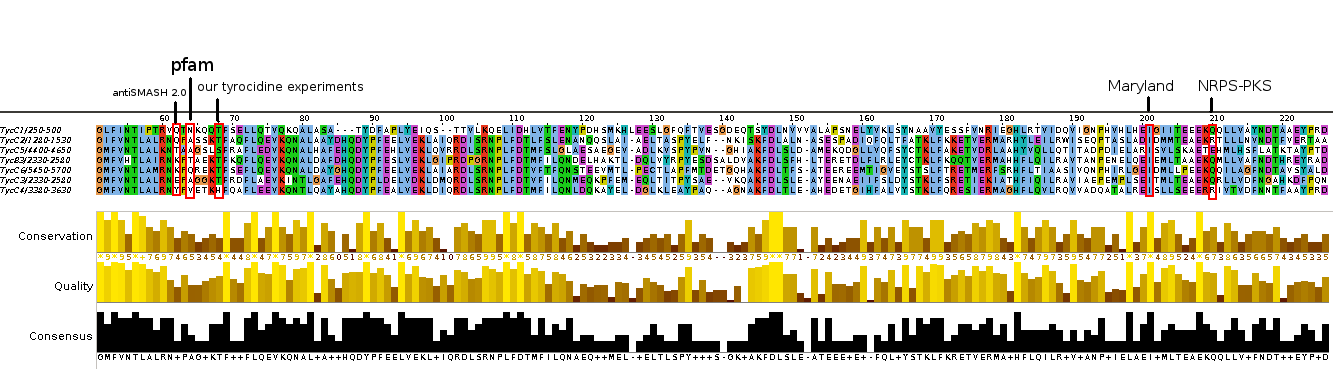
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">As we noticed, that different prediction tools give different domain borders, we further evaluated them and finally integrated antiSMASH</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">As we noticed, that different prediction tools give different domain borders, we further evaluated them and finally integrated antiSMASH</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 139: | Line 139: | ||
<h1>Week 20</h1> | <h1>Week 20</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we started the manual sequence annotation and database curation and integrated the multiple sequence alignment in the database input interface.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week we started the manual sequence annotation and database curation and integrated the multiple sequence alignment in the database input interface.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 149: | Line 149: | ||
<h1>Week 21</h1> | <h1>Week 21</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">During this week we integrated our tool with the parts regisrty, did more manual database curation, added more hidden Markov-Models to antiSMASH and improved Gibthon.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">During this week we integrated our tool with the parts regisrty, did more manual database curation, added more hidden Markov-Models to antiSMASH and improved Gibthon.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 159: | Line 159: | ||
<h1>Week 22</h1> | <h1>Week 22</h1> | ||
<p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to do request management we introduced celery to our system. Furthermore we optimised the multiple sequence alignment and did lots of manual database curation.</p> | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to do request management we introduced celery to our system. Furthermore we optimised the multiple sequence alignment and did lots of manual database curation.</p> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 168: | Line 168: | ||
<div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
<h1>Week 23</h1> | <h1>Week 23</h1> | ||
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In the final week we added several linkouts, including SBOL and GenBank files and fixed lots of bugs.</p> | + | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In the final week we added several linkouts, including SBOL and GenBank files and fixed lots of bugs. After the Regional Jamboree in Lyon, we fixed several bugs and most important we introduced a library function. This allows for construction of many similar NRPs at a time, which could for example be useful for medical compund screening.</p> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 350: | Line 350: | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
<li class="active"><a href="#a18" data-toggle="tab">Interface</a></li> | <li class="active"><a href="#a18" data-toggle="tab">Interface</a></li> | ||
| - | <li><a href="#b18" data-toggle="tab">Database</a></li> | + | <!--<li><a href="#b18" data-toggle="tab">Database</a></li>--> |
<li><a href="#c18" data-toggle="tab">Gibthon</a></li> | <li><a href="#c18" data-toggle="tab">Gibthon</a></li> | ||
</ul> | </ul> | ||
| Line 362: | Line 362: | ||
</html>{{:Team:Heidelberg/Templates/NRPS-W-18a}}<html> | </html>{{:Team:Heidelberg/Templates/NRPS-W-18a}}<html> | ||
</p> | </p> | ||
| - | </div> | + | </div><!-- |
<div class="tab-pane" id="b18"> | <div class="tab-pane" id="b18"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/NRPS-W-18b}}<html> | </html>{{:Team:Heidelberg/Templates/NRPS-W-18b}}<html> | ||
</p> | </p> | ||
| - | </div> | + | </div>--> |
<div class="tab-pane" id="c18"> | <div class="tab-pane" id="c18"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| Line 382: | Line 382: | ||
<div> | <div> | ||
<ul class="nav nav-tabs"> | <ul class="nav nav-tabs"> | ||
| - | <li class="active"><a href="#a19" data-toggle="tab">Algorithm</a></li> | + | <!--<li class="active"><a href="#a19" data-toggle="tab">Algorithm</a></li>--> |
| - | <li><a href="#b19" data-toggle="tab">Database</a></li> | + | <li class="active"><a href="#b19" data-toggle="tab">Database</a></li> |
<li><a href="#c19" data-toggle="tab">Frontend</a></li> | <li><a href="#c19" data-toggle="tab">Frontend</a></li> | ||
</ul> | </ul> | ||
| Line 390: | Line 390: | ||
<div class="scrollContent nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | <div class="scrollContent nav navbar" data-spy="scroll" data-target="#navbarExample" data-offset="0"> | ||
| - | <div class="tab-content"> | + | <div class="tab-content"><!-- |
<div class="tab-pane active" id="a19"> | <div class="tab-pane active" id="a19"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/NRPS-W-19a}}<html> | </html>{{:Team:Heidelberg/Templates/NRPS-W-19a}}<html> | ||
</p> | </p> | ||
| - | </div> | + | </div>--> |
<div class="tab-pane" id="b19"> | <div class="tab-pane" id="b19"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| Line 401: | Line 401: | ||
</p> | </p> | ||
</div> | </div> | ||
| - | <div class="tab-pane" id="c19"> | + | <div class="tab-pane active" id="c19"> |
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
</html>{{:Team:Heidelberg/Templates/NRPS-W-19c}}<html> | </html>{{:Team:Heidelberg/Templates/NRPS-W-19c}}<html> | ||
| Line 453: | Line 453: | ||
<div class="tab-pane active" id="a21"> | <div class="tab-pane active" id="a21"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/NRPS-W-21a}}<html> | |
</p> | </p> | ||
</div> | </div> | ||
| Line 497: | Line 497: | ||
<div class="tab-pane active" id="a23"> | <div class="tab-pane active" id="a23"> | ||
<p style="font-size:12pt; text-align:justify;"> | <p style="font-size:12pt; text-align:justify;"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/NRPS-W-23}}<html> | |
| + | After the Regional Jamboree in Lyon, we fixed several bugs and most important we introduced a library function. This allows for construction of many similar NRPs at a time, which could for example be useful for medical compund screening. | ||
</p> | </p> | ||
</div> | </div> | ||
Latest revision as of 03:57, 29 October 2013


NRPS-Designer. Designing custom peptides.
This tool allows you to design any customized peptide you can build using the most common NRPS domains. This gives you the oppurtunity to include non-proteinogenic amino acids and secondary modifications without going through chemical synthesis or tRNA reprogramming. On top of providing you with a target pathway sequence the tool is also integrated with the parts registry to include any further BioBricks and with Gibthon to directly go from NRP design to Gibson cloning strategy within 10 minutes.
You should also read our documentation and our RFC100!
 "
"