Team:Heidelberg/Delftibactin/MMCoA
From 2013.igem.org
Nils.kurzawa (Talk | contribs) m |
Nils.kurzawa (Talk | contribs) m |
||
| Line 211: | Line 211: | ||
</div> | </div> | ||
</div><!-- /.carousel --> | </div><!-- /.carousel --> | ||
| + | </div> | ||
<div class="col-xs-12 col-sm-6"> | <div class="col-xs-12 col-sm-6"> | ||
| - | <div class="jumbotron methods" data-spy="scroll" data-target="#navbarExample" data-offset="0" style="margin-top: | + | <div class="jumbotron methods" data-spy="scroll" data-target="#navbarExample" data-offset="0" style="margin-top:10%"> |
<h3>Graphical Abstract</h3> | <h3>Graphical Abstract</h3> | ||
<div style="width:100%;"> | <div style="width:100%;"> | ||
| Line 221: | Line 222: | ||
</a> | </a> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
Revision as of 09:17, 18 October 2013


Methylmalonyl-CoA Pathway. Making the whole thing work.
Week 6
In this week the first experiments for the genomic integration of the methyl-malonyl-CoA synthetase were set up. First of all the plasmids were transformed into TOP10 grown and purified. The first attempts on the amplification of the fragment didn't work at all and we ended up with a specific product of the wrong size.
Week 7
After a validation of the template plasmid via restriction digest the PCR reaction was optimised. Unfortunately none of the cells transformed with the PCR product using electroporation could be positively screened, no matter how the PCR fragment was amplified.
Week 8
As the repetition of the colony PCR under different conditions was still negative, new PCR products were prepared and pooled. To eliminate possible errors fresh competent cells were prepared.
Week 9
After doing colony PCR again and getting inconclusive results, it was decided that the genomic integration failed using this experimental setup. Thus we started from scratch using a new primer pair and finally it turned out that the polymerase was causing the problems. By the end of the week BAP1-pKD46 could be transformed.
Week 10
Even after running several colony PCRs there were no positive clones to be found. Thus the pKD46 was digested for validation. On the gel a miniprep from the PCR-transformed cells was included. The identity of the pKD46 was fine and the miniprep didn't contain any DNA, as expected.
Week 11
In this week the whole procedure was repeated again from fragment amplification to extensive colony PCR. This week several different colony types were encountered and one that grew only after 3 days actually gave a band at 1.2 kb, but it turned out to be unspecific.
Week 12
Several possible error sources such as unselective plates were investigated, but all turned out to be fine. The colony PCR was then repeated using yet another primer pair, but it still only gave unspecific or no bands.
Week 13
In order to check whether the cell line used was actually the right one primers against sfp were used and cell from other subgroups were included. It turned out that something really weird was going onwith the cells, since sfp could not be detected. Thus the entirely new strategy of generating a plasmid instead of doing genomic integration was initialised.
Week 14
To be really sure about the sfp a new plasmid preparation was made and cells were tranformed again. In parallel the pLF03 was also prepared and sent for sequencing. It turned out, that there is a deletion in the sequence and thus the primers can't bind properly.
Week 15
A final try on integrating the methyl-malonyl-CoA pathway was made. At the same time new plasmids carrying the pathway arrived and had to be validated first via PCR and restriction digestion. Several PCR reaction were run and also the first Gibson assembly and transformation was done for both plasmids. The positive colonies were sent for sequencing by the end of the week.
Week 16
After finally giving up on the genomic integration we focused on the plasmid strategy, where the sequencing results of the two plasmids arrived. As they were already positive glycerol stocks were prepared.
Week 17
In order to check for sfp expression pMM64, carrying an indigoidine synthetase was cotransformed, but didn't give any blue colonies. Thus the plasmid backbone had to be changed to allow for cotransformation with our own indigoidine construct. One of the new plasmids was sent for sequencing.
Week 18
The sequencing results of pIK1.3 revealed a point mutation in the permeability device. All the other plasmids sent for sequencing also carried some kind of mutation that introduced a stop codon within the permeability device, which is thus non-functional. Moreover there were still no blue colonies even though we now used our own construct.
Week 19
After doing some research in 2007's Cambridge team wiki it was found, that the permeability device is probably toxic to the cells and can thus only be succesfully cloned behind a very weak promotor.
Week 20
This week we grew the first blue colonies with an updated version of the indigoidine construct. We also put the permeability device behind a very weak promotor and prepared new plasmids for sequencing.
Week 21
When the sequencing results were analysed there were no mutations in the permeability device. Now the cloning of the plasmids for the parts submission started and by the end of the week we sent them for sequening.
Week 22
Unfortunately we noticed that we accidentally introduced an additional restriction site and could thus not submit the part to the registry. We already removed the restriction site and validated it by restriction digestion. Anyway we are going to try different restriction enzymes and send it for sequencing after wiki freeze.
Goal
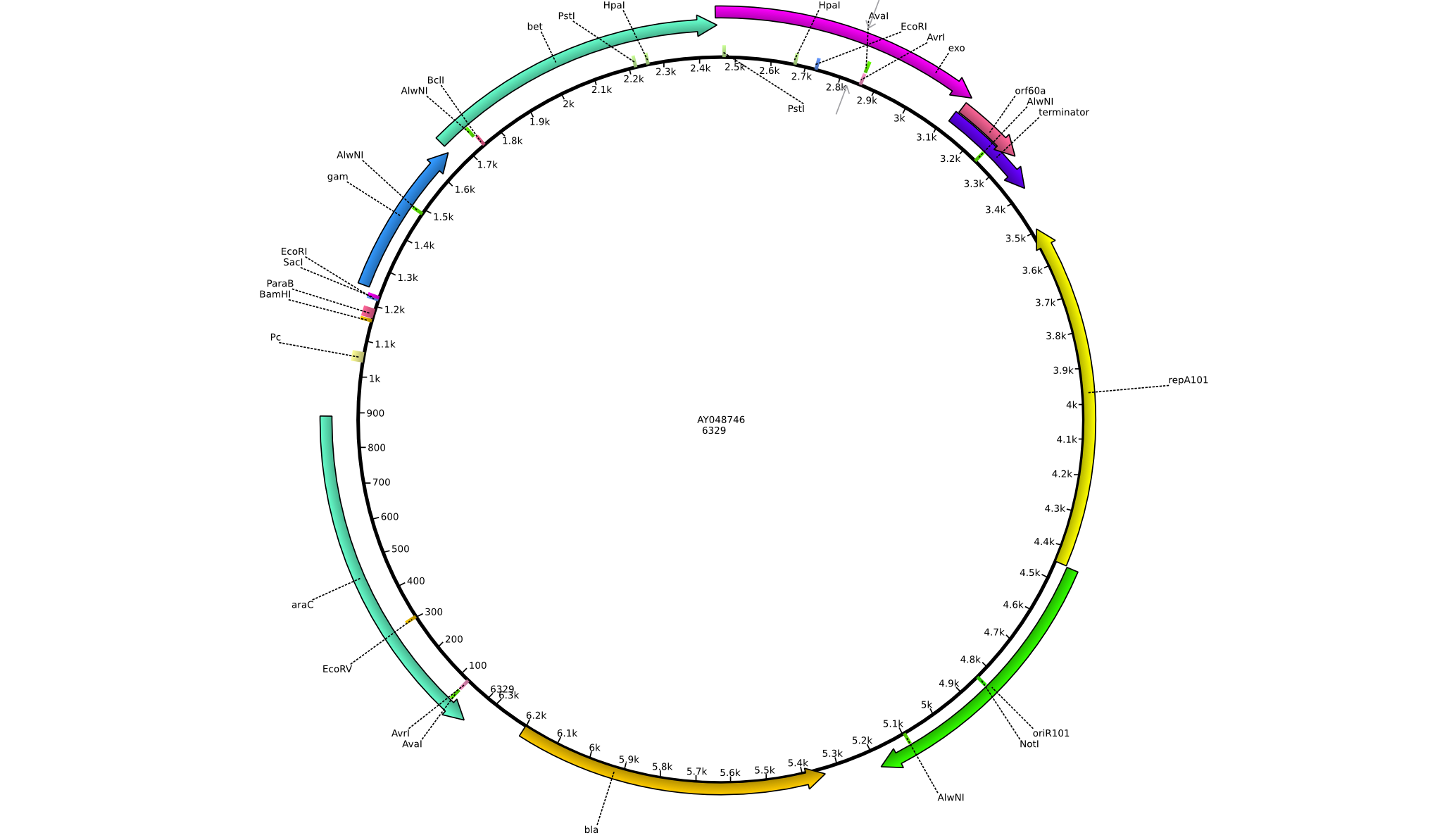
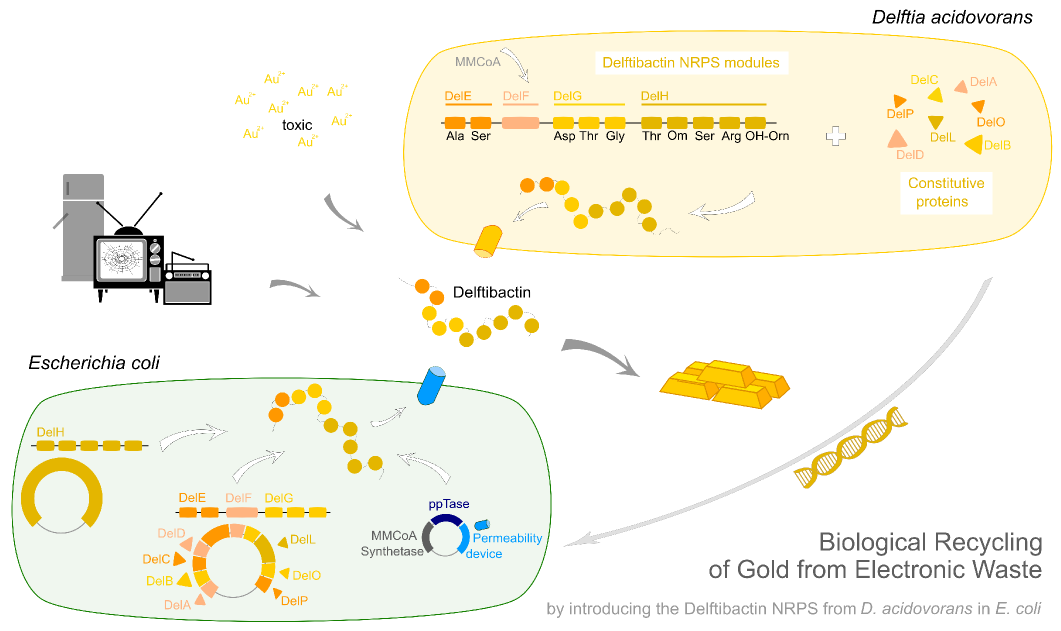
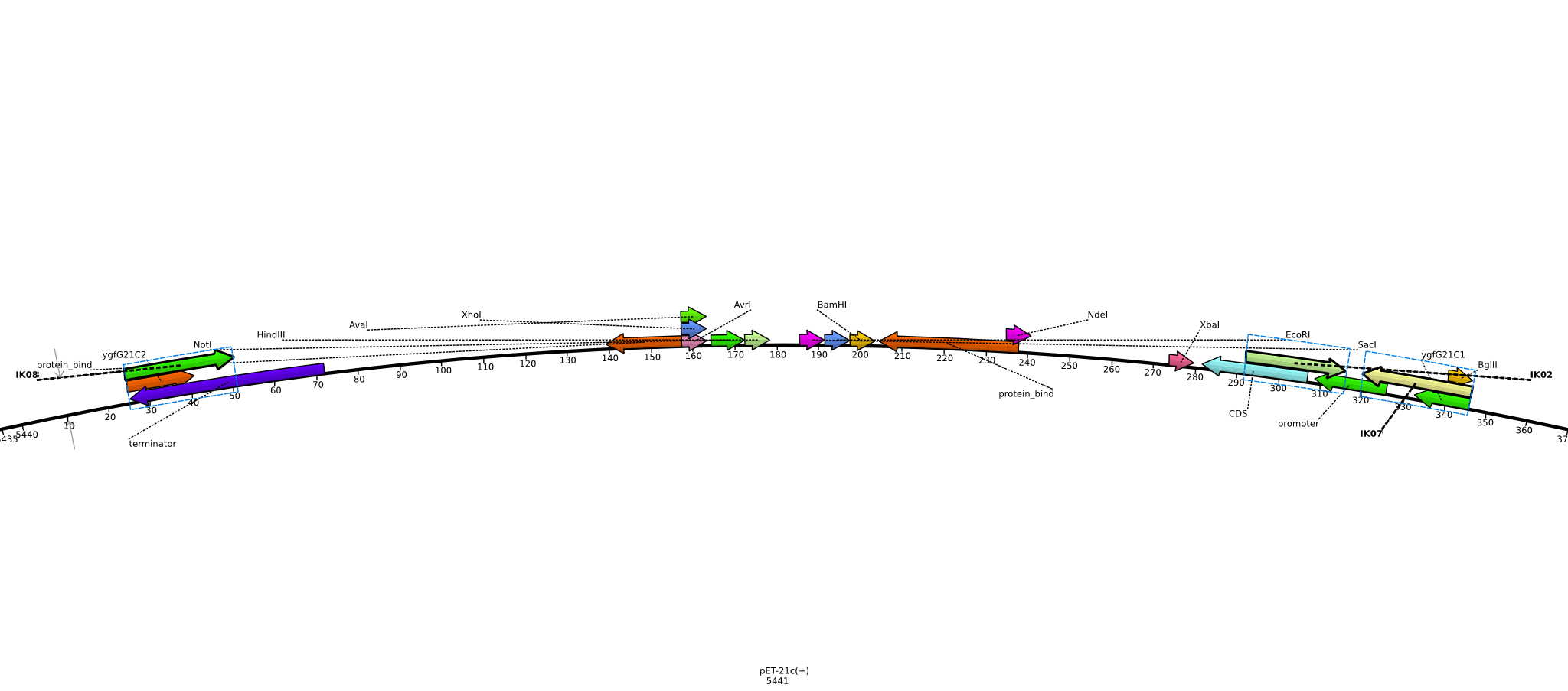
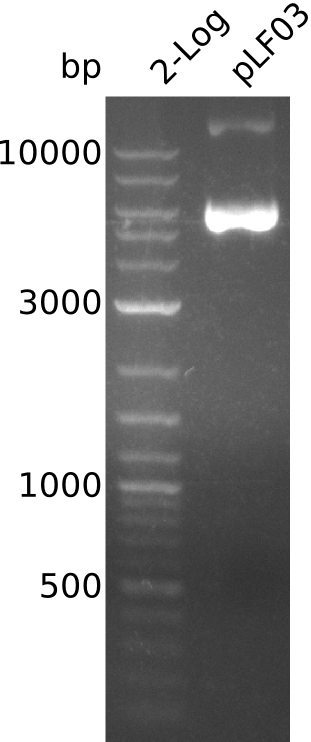
Integration of the methylmalonyl-CoA sythesis pathway into the BAP1 genome. This is a reproduction of a part of <bib id="pmid17959404"/>. For the integration, the λ-red protocol established by Datsenko and Wanner<bib id="pmid10829079"/> is used. pKD20/pKD46 plasmids contain the λ-red genes begind an arabinose inducible promoter, a temperature-sensitive origin, and an ampicillin selection marker. The part to be integrated is on the pLF03 plasmid, which is identical to pYW201<bib id="pmid17959404"/> in every way except for the selection marker in the insert (pYW201 has kanamycin resistance, pLF03 chloramphenicol). pLF03 contains an ampicillin marker that is expressed constitutively, chloramphenicol is only used to test for successful integration, as it is behind an IPTG-inducible T7 promoter (as are the other two genes, pccB and accA1). pLF03 is derived from pET-21c. The expected amplificate size for PCR of pLF03 is 4kb. pccB-accA will replace the ygfG gene in the BAP1 genome.
2013-06-03
- inoculate 50 ml LB with BAP1, grow at 37°C until OD=0.57
- prepare BAP1 glycerol stocks
- prepare competent BAP1
- recover pKD46, pCP20, pLF03
- transform TOP10 with pKD46, plate on Amp, grow at 30°C
- transform TOP10 with pLF03, plate on Amp, grow at 37°C
- transform TOP10 with pCP20, plate on Amp, grow at 30°C
2013-06-04
- inoculate 4 ml LB+Amp with TOP10-pLF03, grow at 37°C
- make miniPrep of pLF03 => 97 ng/µl in 27 µl
- transform BAP1 with pKD46, plate on Amp, grow at 30°C
2013-06-05
- prepare 50 ml LB + Amp + Arabinose(0.1%), inoculate with BAP1-pKD46
- prepare 4 ml LB + Amp, inoculate with TOP10-pKD46 (for miniPrep)
- no colonies with pCP20 => repeat transformation with three times the amount of plasmid (30 µl)
- BAP1-pKD46 reached OD=0.68 at 22:30 => put in fridge over night
2013-06-06
- inoculate 50 ml LB + Amp + Arabinose(0.1%) with 500 µl BAP1-pKD46 from ON culture (fridge)
- at OD=0.49: make glycerol stocks, prepare electrocompetent BAP1-pKD46
- make miniPrep of TOP10-pKD46 ON culture -> 31 ng/µl
- low yield of TOP10 miniPrep: make 2 miniPreps of BAP1-pKD46 ON culture -> 45.5 ng/µl and 46.0 ng/µl
- evening: no colonies with pCP20 -> write Lei Fang (Pfeifer lab) -> their stock is bad, we should get pCP20 somewhere else
2013-06-07
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) of with primers ygfG21C1 and ygfG21C2 using Phusion polymerase:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 10 |
| 66 | 30 | |
| 72 | 90 | |
| 1 | 72 | 450 |
| 1 | 4 | inf |
- using hot start at 98°C
- two PCR samples were run, in one the annealing step at 66°C was left out
- => no amplificate
2013-06-08
- Looking in more detail at the primers, the parts binding to the template have melting temperatures of 40 and 45°C (the rest is homologous to E. coli genome for recombination)
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion polymerase:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 5 | 98 | 10 |
| 39 | 45 | |
| 72 | 90 | |
| 5 | 98 | 10 |
| 45 | 45 | |
| 72 | 90 | |
| 20 | 98 | 10 |
| 58 | 45 | |
| 72 | 90 | |
| 1 | 4 | inf |
- using hot start at 98°C
- two PCR samples were run, in one the annealing step at 58°C was left out
- => no amplificate
2013-06-09
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion polymerase:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 35 | 98 | 10 |
| 40 | 60 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- using hot start at 98°C
- amplificate has 1.3kb -> too short, but specific (one distinct band)
2013-06-10
- need to verify plasmid identity: digest pLF03 with EcoRI (for linearization) and XhoI+NdeI (verify insert size); 388 ng (4 µl of miniPrep from 2013-06-04) each
- insert only 3.3 kb in length, expected >4 kb ([http://www.uniprot.org/uniprot/Q9RGQ6 AccA1]: 590AS * 3 = 1770 bp, [http://www.uniprot.org/uniprot/Q9X4K7 PccB]: 530AS * 3 = 1590 bp, [http://www.uniprot.org/uniprot/P62577 CatR]: 219 AS = 657 bp; adds up to 4017 bp, not counting RBS and spacers between the individual genes)
- contacted Lei Fang (Pfeifer Lab), there are NdeI and EcoRI sites in catR (they were not indicated in his plasmid map)
- to verify presence of chloramphenicol acetyl transferase in insert: transform BAP1 with 97 ng pLF03 (1 µl of miniPrep from 2013-06-04), plate on Amp + IPTG
- evening: pick BAP1-pLF03 colonies, transfer to Cm + IPTG
- inoculate 4 and 5 ml of liquid culture (Amp) with BAP1-pLF03
2013-06-11
- BAP1-pLF03 grew on Cm + IPTG
- try to optimize PCR:
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion Flash F548 polymerase (performed by Dominik):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 45 | 5 | |
| 72 | 120 | |
| 25 | 98 | 1 |
| 55 | 5 | |
| 72 | 120 | |
| 1 | 72 | 120 |
| 1 | 4 | inf |
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Taq polymerase (cheaper):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 94 | 180 |
| 35 | 94 | 20 |
| 41.5/43 | 60 | |
| 72 | 480 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
- prepare glycerol stock of BAP1-pLF03
- make miniPreps of BAP1-pLF03 -> 21 ng / µl and 27 ng / µl in 27.5 µl
- received just plated DH5α-pCP20 from Sourjik Lab, grow at 30°C
2013-06-12
- purify Phusion Flash PCR product with Qiagen PCR purification kit -> 120 ng / µl in 27.5 µl
- inoculate 2 x 5 ml LB + Amp with DH5α-pCP20, grow at 30°C
2013-06-13
- miniPreps of DH5α-pCP20 -> 7.8 ng / µl and 14.6 ng / µl in 27.5 µl, respectively
- low yield of miniPreps: inoculate 2 x 5 ml LB + Amp with DH5α-pCP20, grow at 30°C
2013-06-14
- miniPreps of DH5α-pCP20 -> 25 ng / µl and 29 ng / µl in 27.5 µl, respectively
- prepare glycerol stocks of DH5α-pCP20
- to avoid electroporation with intact pLF03: add 10 µl of PCR amplificate (1.2 mg) on gel, perform gel extraction (QiaGen gel extraction kit) -> 3.2 ng / µl
- add 240 ng (2 µl) raw PCR product to 100 µl electrocompetent BAP1-pKD46
- add 16 ng (5 µl) gel-extracted PCR product to 100 µl electrocompetent BAP1-pKD46
- electroporate 50 µl each, add to 1 ml SOC with 1 mM IPTG
- shake at 300 rpm, 37°C for 1 h
- spin cells down (3 min 10 000 rpm = 8000 g), decant supernatant (leave about 200 µl supernatant in eppendorf tube)
- resuspend pellet, plate 100 µl on Cm-IPTG, leave rest at RT
- grow at 37°C
2013-06-15
- one colony on plate with gel-extracted PCR product
- many colonies on plate with PCR-purified PCR product
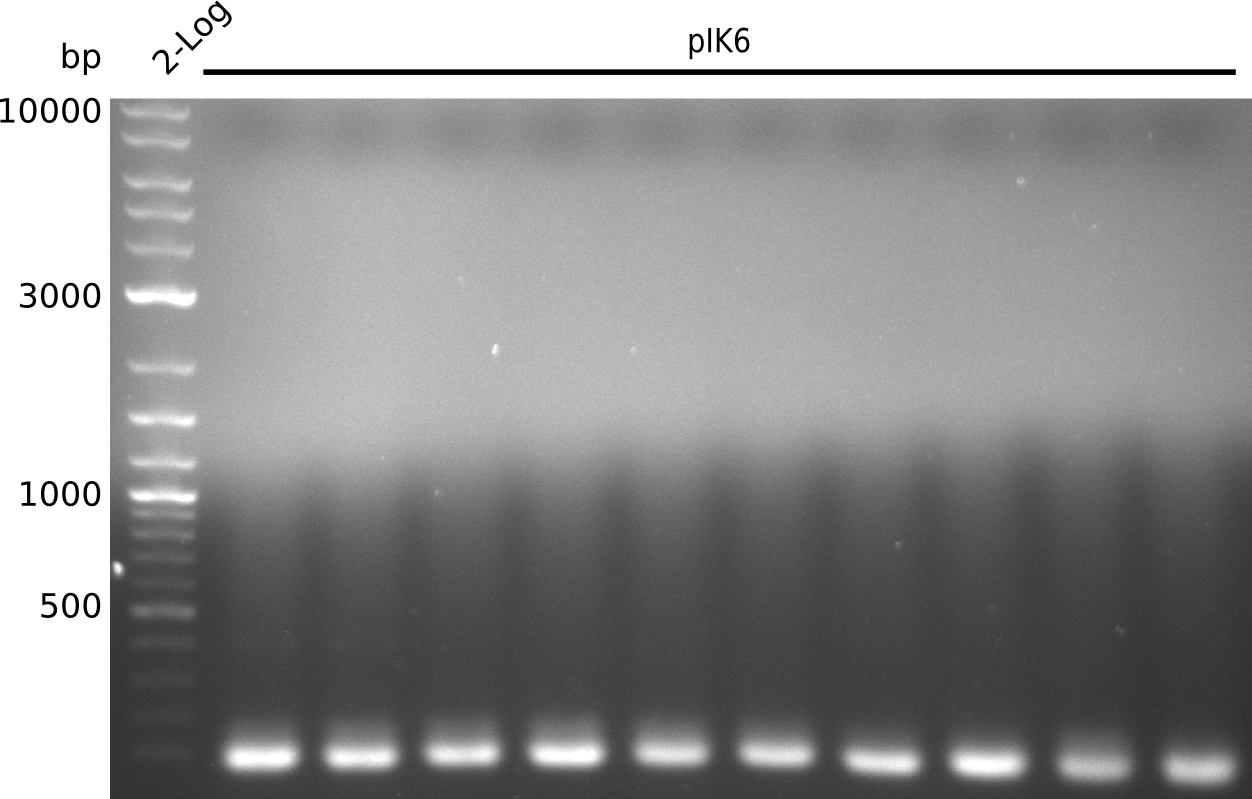
- pick colonies from plate with PCR-purified PCR product, run colony-PCR (Taq, 25 µl total volume) with primers IK01 and IK02 (expected product: 465 bp):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 180 |
| 30 | 95 | 30 |
| 61 | 30 | |
| 72 | 30 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product -> pick colony from gel-extracted plate, pick colonies from PCR-purified plate, run colony-PCR (Taq, 25 µl total volume) with primers IK01 and IK02:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 240 |
| 30 | 95 | 30 |
| 61 | 30 | |
| 72 | 30 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product -> 2 possibilities:
- all colonies bear complete pLF03 plasmid (PCR was run with 5 ng template, DNA concentration in PCR product: 120 ng / µl, electroporated with 2 µl -> 1/6 ng pL03 vs. 240 ng amplificate)
- PCR did not work
- for colony from gel-extracted plate + 3 colonies from PCR-purified plate: add up to 3 ml medium, add IPTG (1 mM) + Cm, grow at 37°C
- plate rest of bacteria electroporated with gel-extracted PCR product on Cm + IPTG
2013-06-16
- no colonies on gel-extracted plate
- make miniPreps of ON cultures, run gel
- pLF03: 9.2 kb, bands above 10 kb, bands smear, low intensity (applied all 30 µl of miniPrep to gel) -> fragments of genomic DNA?
- => PCR went wrong?
2013-06-17

- repeat colony PCR, use 1 µl of liquid culture from gel-extracted plate, pick colonies from PCR-purified plate (Taq, 25 µl total volume)
- 2 conditions:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 600 |
| 30 | 95 | 240 |
| 64 | 30 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 600 |
| 12 | 95 | 240 |
| 66°C ↓0.5°C | 30 | |
| 72 | 60 | |
| 18 | 95 | 240 |
| 60°C | 30 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no positives
- plate samples of liquid cultures from 2013-06-15 on Amp, grow at 37°C
2013-06-18
- colonies grew on Amp
2013-06-19
- prepare for repetition of experiment: run 4 PCRs of 1 ng pLF03 (miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion Flash (performed by Dominik), one reaction = 50 µl total volume
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 45 | 5 | |
| 72 | 130 | |
| 25 | 98 | 1 |
| 55 | 5 | |
| 72 | 120 | |
| 1 | 72 | 120 |
| 1 | 4 | inf |
- pool products, purify (digestion only 3 h)
- nanoDrop: 2088 ng/µl -> remeasure: 532 ng/µl
- load 1 µl purified PCR product on gel -> very weak band -> nanoDrop wrong
- to eliminate error: make fresh electrocompetent cells:
- pick colony from BAP1-pKD46 plate from 2013-06-04
- inoculate 1 ml LB + Amp
- grow at 30°C
2013-06-20
- too little cells: use aliquot from 2013-06-06
- use all available DNA (14 µl) and 100 µl cells (complete aliquot)
- grow in SOC + IPTG(1mM) at 300 rpm for 3h
- spin cells down, decant supernatant, resuspend in remainder of medium
- plate 10 µl, rest on Cm + IPTG
2013-06-24

- no colonies on 10 µl plate, 2 colonies on rest plate
- colony-PCR using primer pairs IK01+IK02, IK01+IK03 (positive control), IK05+IK06 (negative control) (Taq, 20 µl total volume)
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 30 | 95 | 60 |
| 62 | 30 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- inconclusive results: no band for IK05+IK06 in BAP1-pLF03 (negative control)
- grow liquid cultures at 37°C
2013-06-25
- repeat PCR with 1 µl of ON cultures:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 (IK01+IK02; IK01+IK03;IK05+IK06) / 240 (IK01+IK06) | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 (IK01+IK02; IK01+IK03;IK05+IK06) / 240 (IK01+IK06) | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- genomic integration failed
- pick colony from BAP1-pKD46 plate from 2013-06-04, grow in 3 ml LB+Amp at 30°C
2013-06-26
- inoculate 50 ml LB + Amp + Ara(0.5%) with 1.1 ml of ON culture
- grow at 30°C to OD=0.73, prepare electrocompetent BAP1-pKD46
2013-06-27
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers IK07 and IK08 using Phusion polymerase (20 µl total volume) on cycler 1 (does not cool down to 4°C) (using hot start):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product -> repeat PCR with cycler 2 (fully functional)
- no product -> run 2-step PCR with 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Phusion polymerase (20 µl total volume, using hot start) on cycler 2:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 72 | 150 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product
2013-06-28
- run PCR of 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Q5 polymerase (20 µl total volume, using hot start) on cycler 2:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 1800 (30 min) |
| 1 | 4 | inf |
- perfect PCR => Phusion is crappy
- run 4 PCRs of 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Q5 polymerase (50 µl total volume, using hot start) on cycler 2:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 45 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 1800 (30 min) |
| 1 | 4 | inf |
2013-06-29
- load 1 µl of PCR product on gel -> specific amplificate
- pool products (also include amplificate from 2013-06-28), purify (only first step, as no DpnI available)
2013-06-30
- inoculate 1 ml LB+Amp with 10 µl of ON culture from 2013-06-25, grow at 30°C
2013-07-01
- finish purification (digest for 5h, dissolve in 50 µl H2O) -> 1162 ng/µl (performed by Fanny)
- prepare electrocompetent cells from ON culture (add 0.5% arabinose at dilution) (performed by Fanny)
- electroporate fresh electrocompetent cells, 1 aliquot from 2013-06-26 (23 µl DNA each) (performed by Fanny)
- grow in SOC at 30°C for 30 min, transfer to falcons, add 1 ml LB, add IPTG (1 mM), grow at 37°C for 4h
- spin cell down (8000 rcf for 3 min), decant supernatant, resuspend pellet in remaining medium
- plate on Cm+IPTG (2 plates for each sample: 10µl + rest, plates marked F: from -80°C, marked H: newly prepared cells), grow at 37°C
2013-07-02

- bacteria lawn on rest-plate from -80°C, possible colonies on other plates (barely visible, may be bubbles)
- pick 2 colonies from F-rest, sample of lawn, 2 colonies from H-10µl, colony-PCR with primers IK01+IK02, IK01+IK03 (positive control) (20µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- very evening: colonies on F-10µl, H-rest, H-10µl plates appear -> leave at 37°C ON
2013-07-03

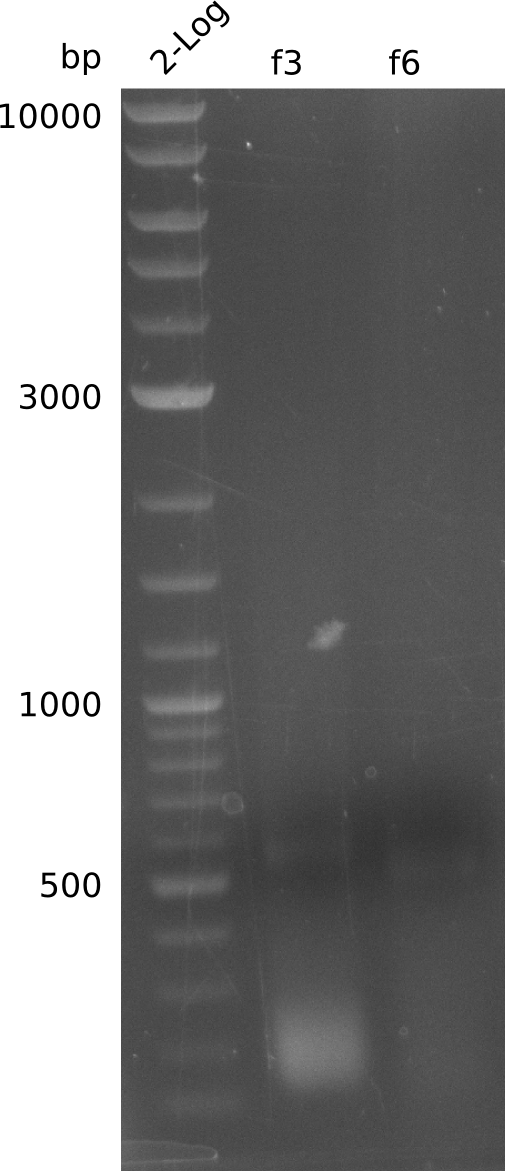
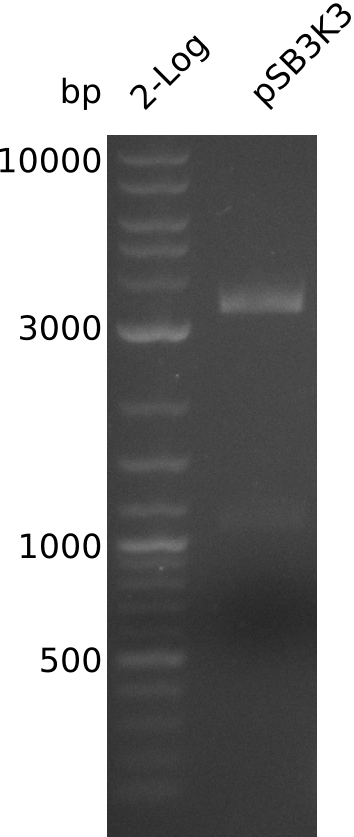
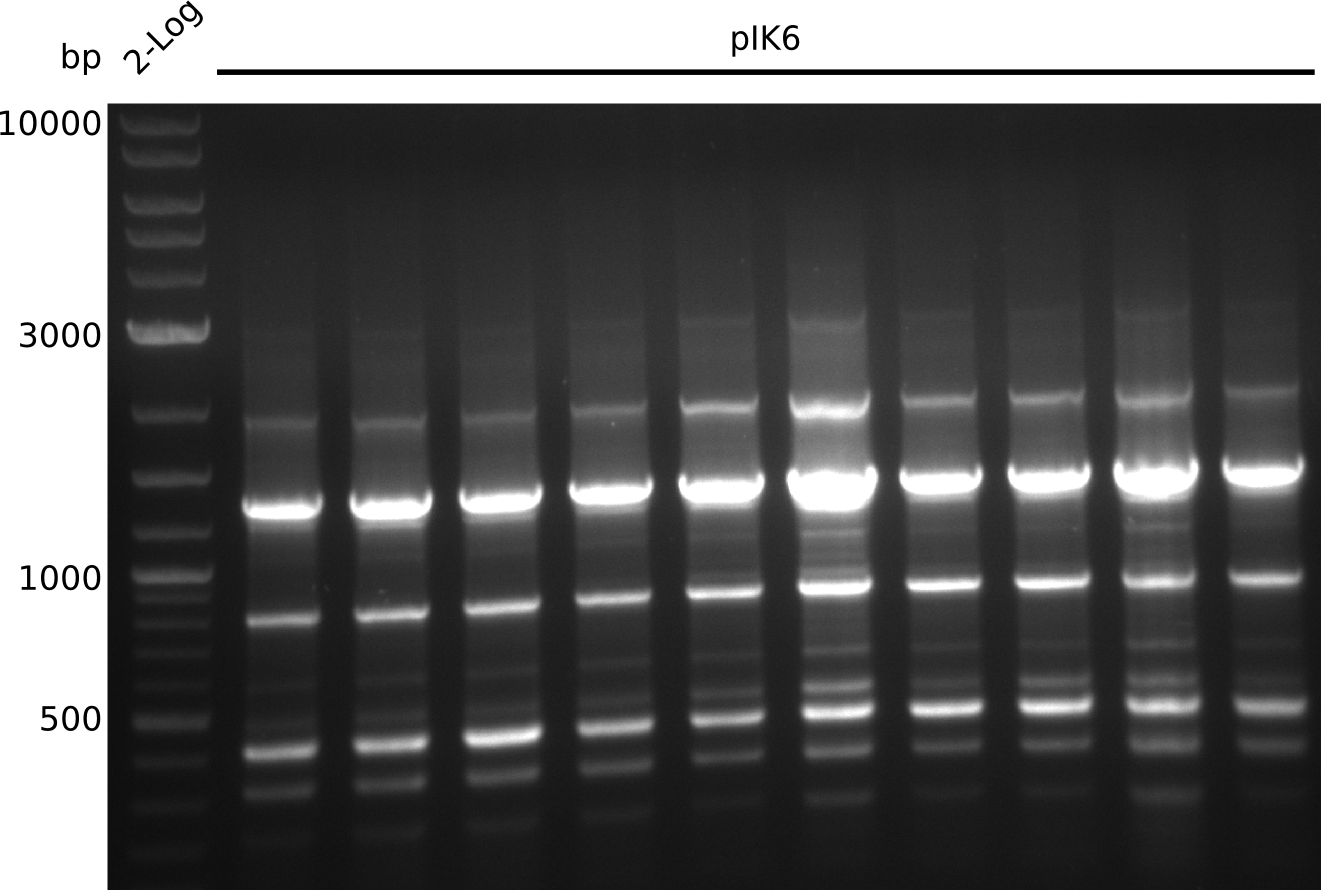
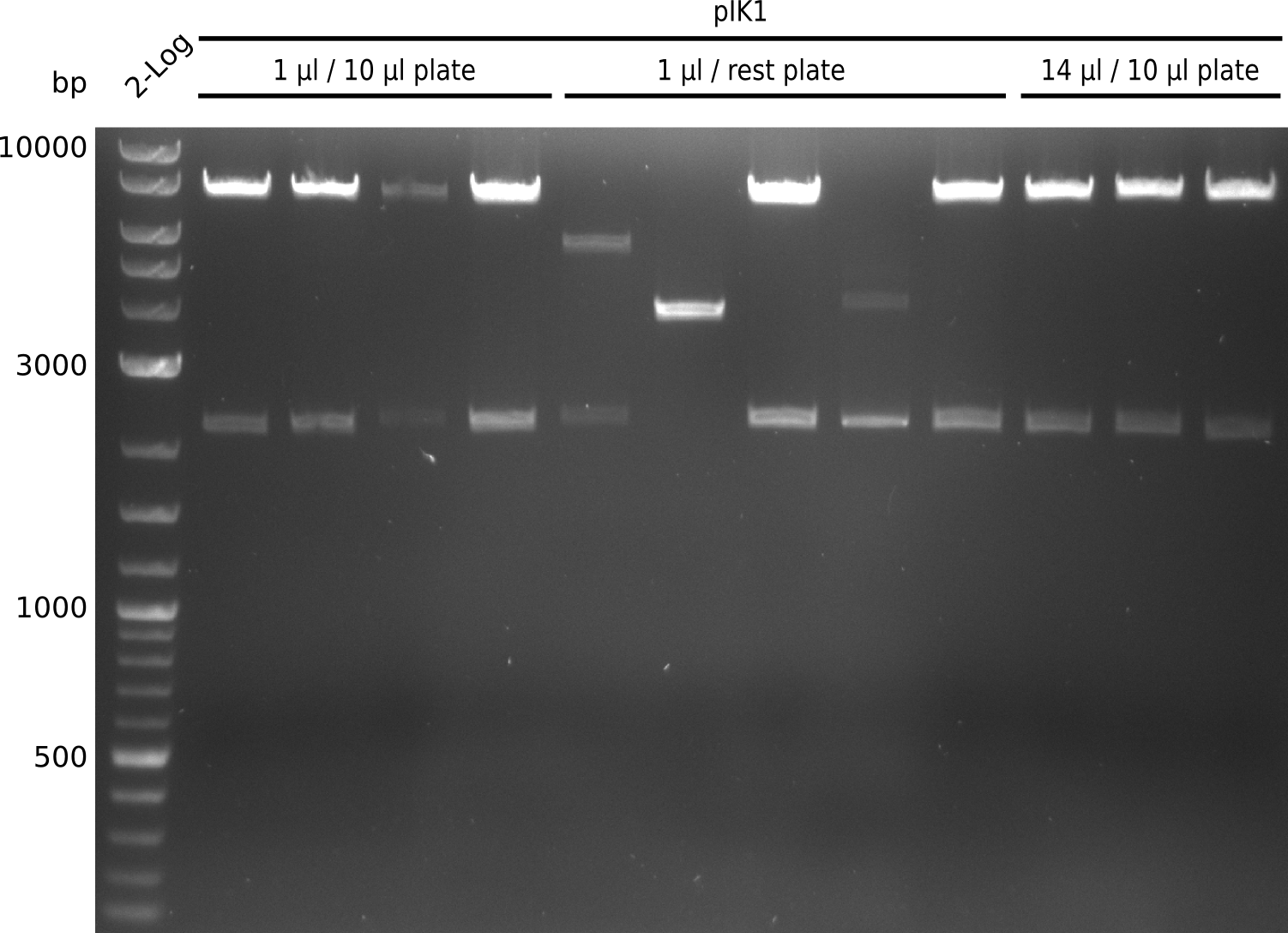
Lane 1: NEB 2-log
Top: lanes 2-4: negative control (BAP1-pLF03); lanes 5-13: colony-PCR F-10µl
bottom:lanes 2-4: colony-PCR of F-10µl; lanes 5-13: colony-PCR of H-10µl; lanes 2,5,11: primers IK01+IK02; lanes 3,6,12: primers IK01+IK03 (positive control); lanes 4,7,13: primers IK05+IK06 (negative control)
- colonies on F-rest plate appeared (plate left ON at RT)
- pick 4 colonies from F-10µl, 3 colonies from H-10µl, colony-PCR with primers IK01+IK02, IK01+IK03 (positive control), IK05+IK06 (negative control):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- F-10µl 1,2,4 show no negative control -> grow at 37°C
- continue growing plates at 37°C until evening, leave at RT ON
2013-07-04

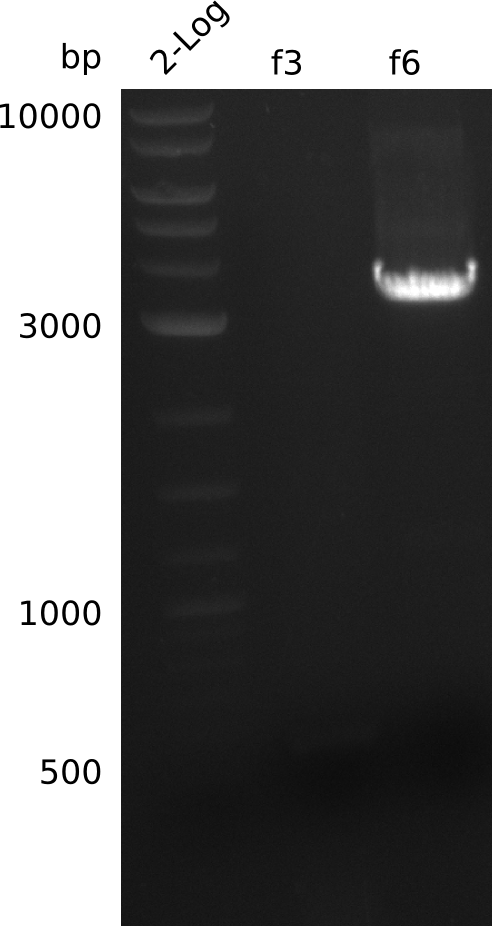
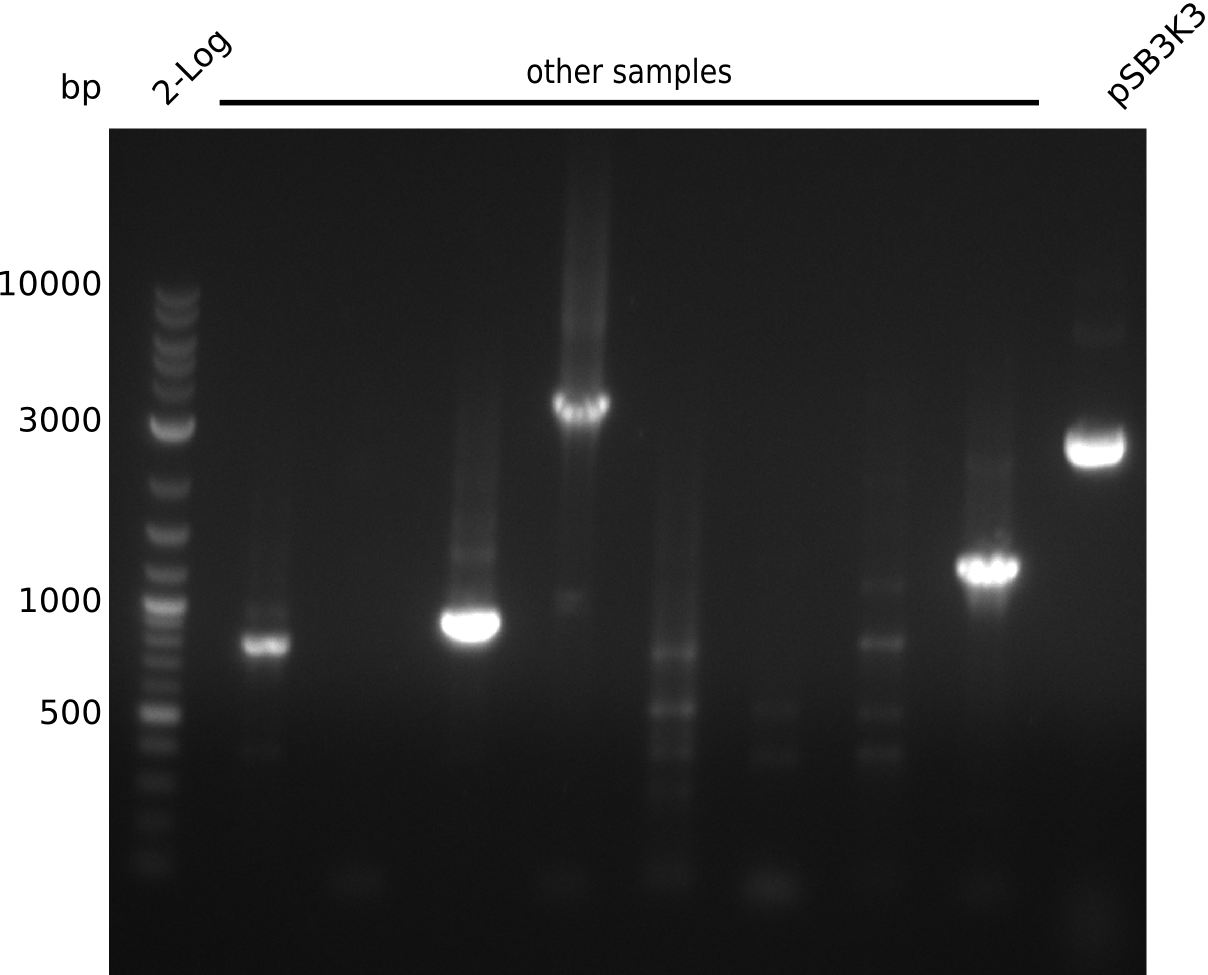
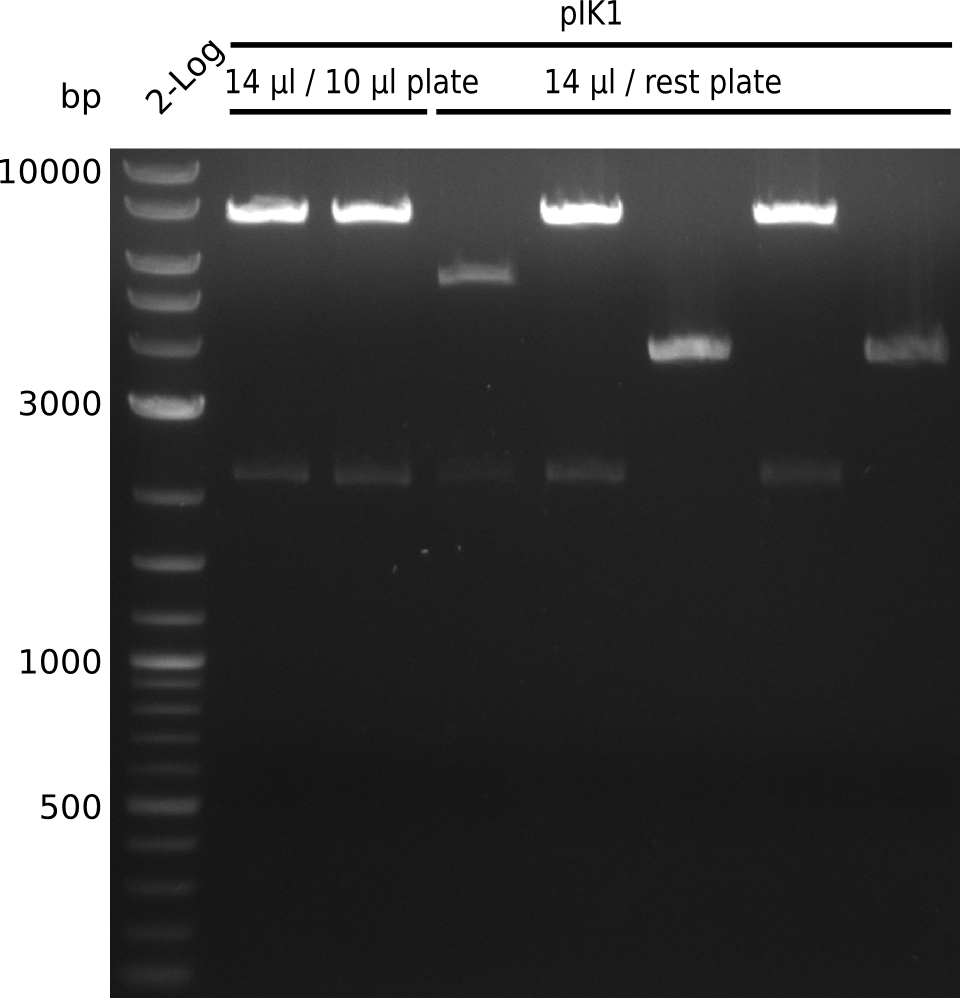
Lane 1: NEB 2-log
Top: lanes 2-10: cultures from 2013-07-03; lanes 2,5,8: primers IK01+IK02; lanes 3,6,9: primers IK01+IK03; lanes 4,7,10: primers IK05+IK06; lane 11: BAP1-pLF03 (negative control); lanes 12-13: colonies from F-10µl plate; lanes 11-13: primers IK05+IK06
bottom: lanes 2-13: primers IK05+IK06; lanes 2,3: colonies from F-10µl plate; lanes 4-6: colonies from F-rest plate; lanes 7-10: colonies from H-10µl plate; lanes 11-13: colonies from H-rest plate
- repeat colony-PCR for 3 ambiguous cultures form 2013-07-03 (use 1 µl of culture), pick 14 new colonies (only primers IK05+IK06); 20 µl total volume:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no positives
2013-07-05
- digest 450 ng pKD46 (10 µl of 45.5 and 46 ng/µl miniPreps from 2013-06-06) with EcoRI (expected: 4816+1505 bp), BamHI+NotI (expected: 3675+2646 bp), PstI+NotI (expected: 3711+2363+243 bp); 30 µl total volume
- right plasmid
- inoculate 2 x 7 ml LB+Cm+IPTG(1mM) with colonies from F-10µl plate, grow at 30°C
2013-07-06
- add Cm (1:3000), IPTG (1:1000) to liquid cultures, grow for 8h at 30°C
- make miniPreps -> 25 ng/µl and 14.4 ng/µl in 27.5 µl
- digest 25 ng/µl miniPrep with EcoRI (use all of miniPrep)
- load digest completely, 10 µl of 14.4 ng/µl miniPrep on gel
- no bands -> nanoDrop crappy, no plasmid
- wrong strain? Streak BAP1 on Cm, grow at 37°C
2013-07-08
- no colonies of BAP1 on Cm
- repeat PCR of pLF03:
- run 4 PCRs of 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Q5 polymerase (50 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 45 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 1800 (30 min) |
| 1 | 4 | inf |
- extract amplificate from gel (4 lanes pooled, extracted by Dominik -> 119 ng/µl; 4 lanes pooled, extracted by Ilia -> 111.5 ng/µl)
- load 1 µl of each extract on gel
2013-07-09
- electroporate electrocompetent BAP1-pKD46 with 1µl, 5µl, 25µl DNA
- resuspend in 1 ml SOC + IPTG (1mM) + Ara (0.1%)
- grow for 90 min at 37°C, 400 rpm
- spin cells down, plate 10µl, rest of each sample on Cm+IPTG
2013-07-10
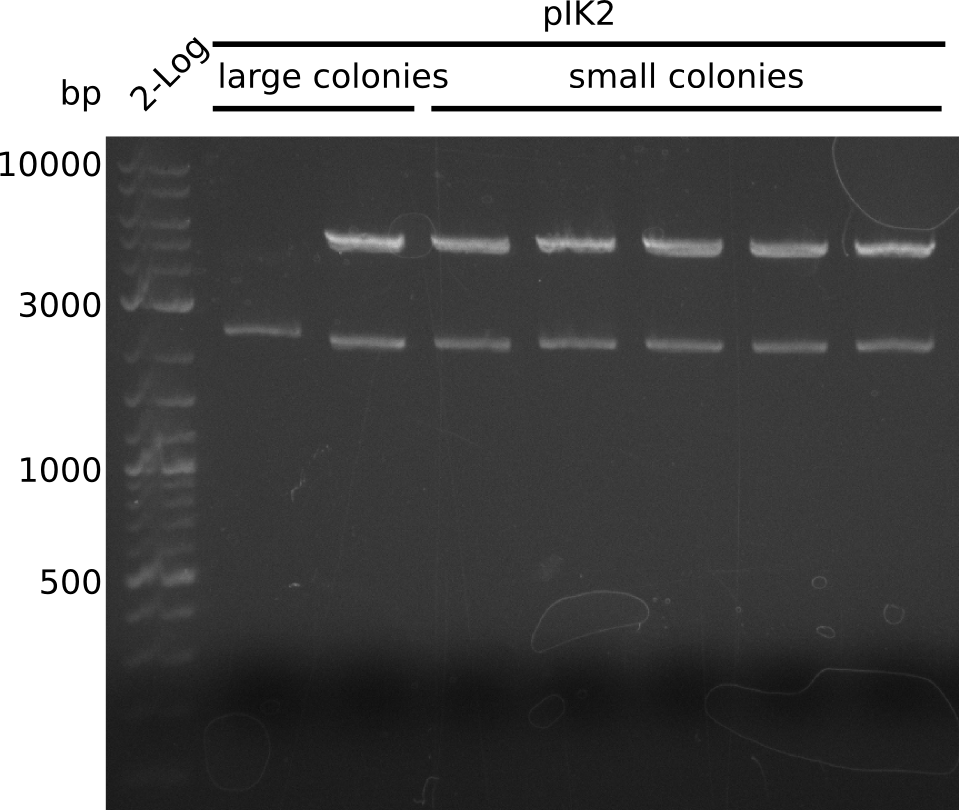
- on all plates: large + small colonies
- small colonies too small to pick => pick 10 large colonies from 25µl DNA / 10 µl bacteria plate, run colony-PCR with primers IK05+IK06 (Taq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no integration
- continue growing plates at 37°C

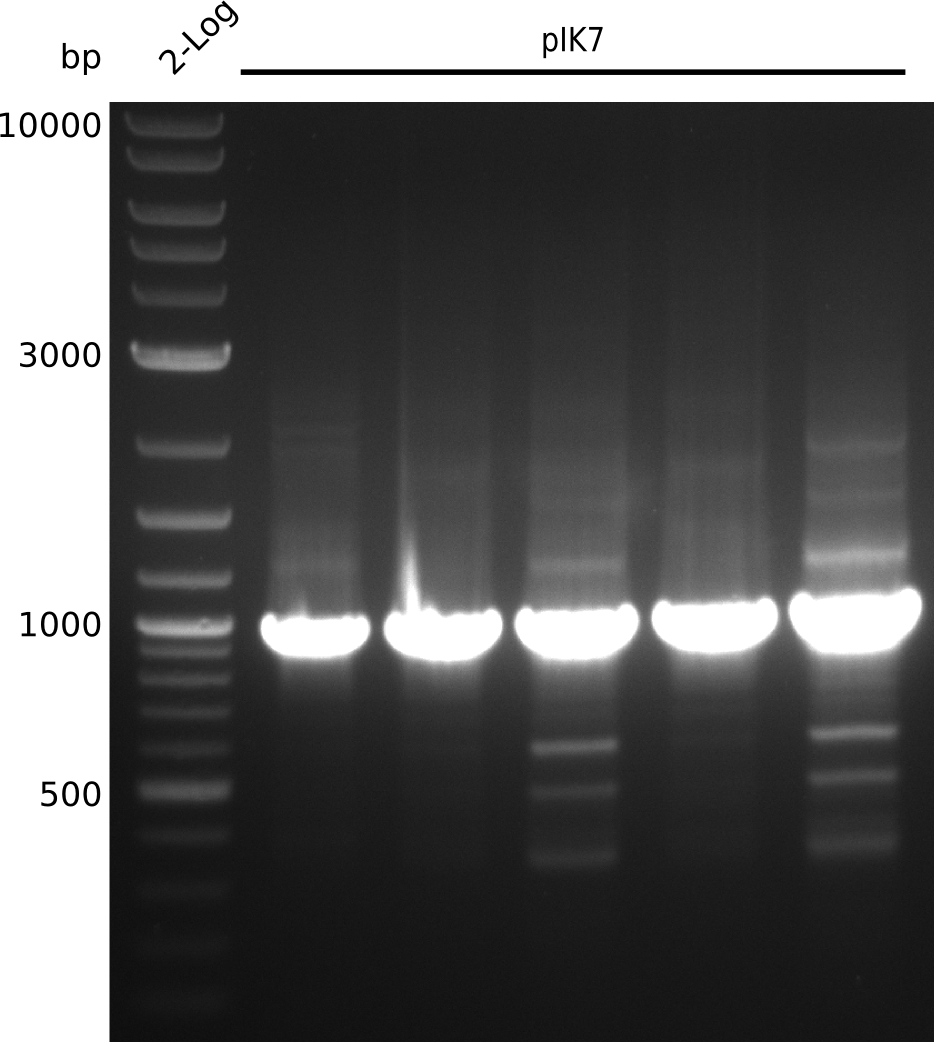
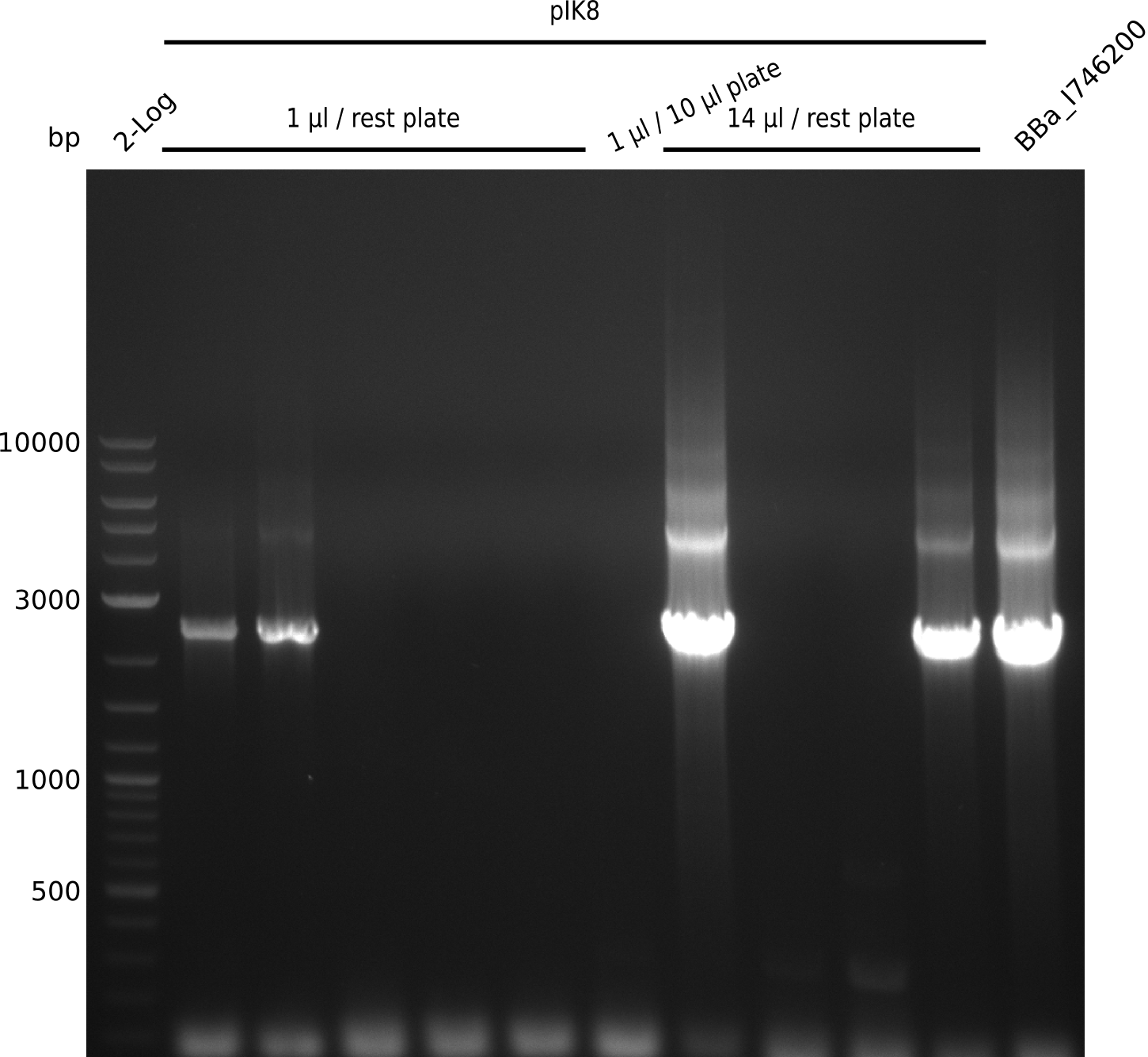
Top: lane 1: NEB 2-log; lanes 2,4,6,8,10,12: primers IK05+IK06; lanes 3,5,7,9,11,13: primers IK01+IK03 (positive control); lanes 2,3: BAP1-pLF03 (control); lanes 4-7: colonies from 1µl/10µl plate; lanes 8-11: 1µl/rest; lanes 12-13: 5µl/10µl
Bottom: lane 11: NEB 2-log; lanes 1,3,5,7,9: primers IK05+IK06; lanes 2,4,6,8,10: primers IK01+IK03 (positive control); lanes 1-2: 5µl/10µl; lanes 3-6: 5µl/rest; lanes 7-8: 25µl/10µl; lanes 9-10: 25µl/rest
- when small colonies large enough: pick, run colony-PCR (OneTaq, 20µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- 1st colony from 5µl/rest plate (marked 7) seems promising => grow at 37°C in 1 ml LB
2013-07-11
- run full colony-PCR of liquid culture (OneTaq, 20 µl total volume, use 1 µl of culture):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no integration
- 3rd population of colonies appeared on plates (stored at RT): very small, probably satellites

- run colony-PCR of liquid with primers IK07+IK08 (test for integration in wrong place) (OneTaq, 20µl total volume, use 1 µl liquid culture):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 30 | 95 | 60 |
| 66 | 30 | |
| 72 | 600 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- no integration at all

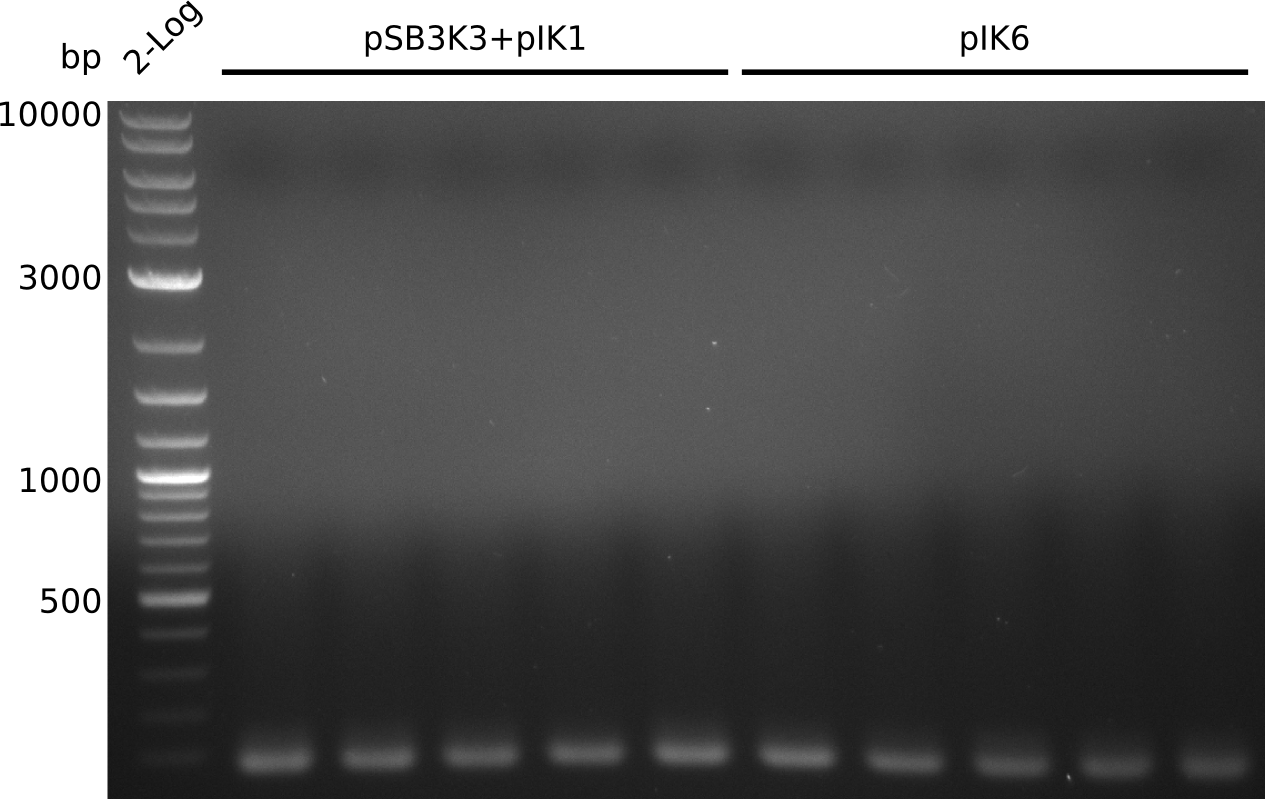
Top: lane 1: NEB 2-log; lane 2: BAP1-pLF03(control); lanes 3-9: colonies from 1µl/10µl plate; lanes 10-13: colonies from 1µl/rest plate
Bottom: lane 1: NEB 2-log; lanes 2-10: colonies from 1µl/rest plate (lane 5: swiped 3rd population of extremely small colonies)
- pick 20 new colonies, run colony-PCR with primers IK05+IK06 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- unspecific bands (also present in colony-PCR from 2013-07-10)
- unspecific bands not present in BAP1-pLF03 PCR (control) and bottom lane 5 (satellite colonies) => not due to iTaq
- assume: some amount of satellites was picked together with colonies from plates -> give band at 500 bp; partially amplified ygfG fragments prime integrated insert
- do multiple sequence alignment of ygfG, [http://www.ncbi.nlm.nih.gov/nuccore/AF113605 pccB], [http://www.ncbi.nlm.nih.gov/nuccore/AF113603 accA1], and [http://www.ncbi.nlm.nih.gov/nuccore/AY048742 catR] using ClustalO => long enough stretches of partially identical sequences present
- add 1mM IPTG to cultures with colonies from 1µl/10µl plate picked today (grew in 500 µl LB at 37°C for several hours), after 2h at 37°C: streak on Cm+IPTG
- pick 10 colonies each from 1µl/10µl and 1µl/rest plates, transfer to new Cm+IPTG plate, grow at 37°C
- pick 1 colony from 1µl/10µl plate, inoculate 1 ml LB + Cm + IPTG (1mM)
2013-07-12

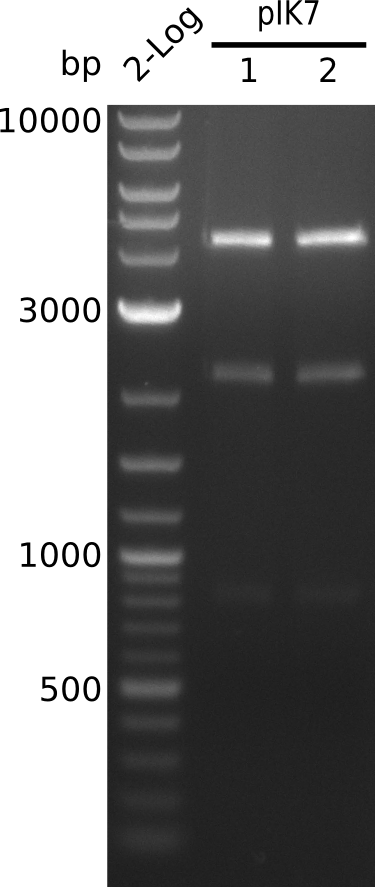
Top: lane 1: NEB 2-log; lane 2: BAP1-pLF03(control); lanes 3-12: colonies liquid culture that were plated on Cm+IPTG; lane 13: colony from 1µl/10µl plate that was transferred to new Cm+IPTG plate
Bottom: lane 1: NEB 2-log; lanes 2-6: colonies from 1µl/10µl plate that were transferred to new Cm+IPTG plate; lanes 7-12: colonies from 1µl/rest plate that were transferred to new Cm+IPTG plate; lane 13: liquid culture (Cm+IPTG(1mM)) from 1µl/10µl plate
- colonies grew slowly
- run colony-PCR with primers IK01+IK02 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no bands
- run colony-PCR with primers IK05+IK06 of 2 colonies from liquid culture that were transferred to Cm + IPTG plate (iTaq, 20µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- band at 1.2 kb is now specific -> WTF?!?
2013-07-13

- need to determine whether 1.2 kb band is specific: run gradient PCR of colony 2 (iTaq, 20 µl total volume) with primers IK05+IK06:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 63-68 (ΔT = 1) | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- intensity of 1.2kb band decreases, intensity of 0.5 kb band (control) does not -> 1.2 kb band is unspecific product
- check for presence of genomically integrated insert: run colony-PCR of colony 2 with primers IK07+IK08 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 66 | 30 | |
| 72 | 600 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- transfer some of colony 2 on Amp (check for presence of pLF03 as a whole), grow at 37°C
- transfer some of colony 2 to liquid culture: LB + Cm + IPTG (1mM), grow at 37°C
2013-07-14
- bacteria grew on Amp => 3 possibilities:
- Amp plates not selective
- bacteria have complete pLF03 plasmid
- bacteria still have pKD46, although grown at 37°C
- plate BAP1, BAP1-pKD46 on Amp, grow at 37°C
- colony-PCR did not work for the control -> repeat with OneTaq (use 1 µl of liquid culture)
- no amplificate for colony: possibly, Taq has problems amplifying 4.8 kb from genomic DNA

- run colony-PCR with primers IK01+IK06 (OneTaq, 20 µl total volume, use 1 µl of liquid culture):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 240 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 240 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- control shows bright band where expected, colony does not => something is integrated, unknown what
2013-07-15

- BAP1 did not grow on Amp, BAP1-pKD46 did grow on Amp at 37°C => no discrimination between pLF03 and pKD46 possible
- Taq might have problems amplifying 4.8 kb from genomic DNA => use Phusion Flash (primers IK07+IK08, 20 µl total volume, use 1 µl of liquid culture, old BioRad cycler):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 100 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- control did not work -> repeat in new BioRad cycler (pick from plate)
- same result
2013-07-18


- run colony-PCR with primers IK24, IK25 (OneTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 180 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- 1.2 kb band with primers IK01+IK25 in control => makes no sense, repeat
- same result
- transfer BAP1-pLF03 to new Cm+IPTG plate, colony 2 to LB w/o antibiotics, grow at 37°c#
2013-07-22
- no positive PCR of colony yet: run colony-PCR with primers IK01+IK03 (iTaq, 20 ml total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 180 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- primers RB43+RB44 (against sfp) iTaq, 20 µl total volume:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 12 | inf |
- unspecific bands in colony, no sfp bands at all
- re-run sfp PCR (primers RB43+RB44), include streaked BAP1 by Konrad
- sfp present in BAP1, not present in BAP1-pLF03, colony
- makes no sense, Konrad streaked his cells from the same batch of competent cells
- transfer BAP1, BAP1-pLF03, colony to liquid culture LB, grow at 37°C
2013-07-23

- run colony-PCR with primers RB43+RB44 (iTaq, 20 µl total volume, use 1 µl of liquid culture)
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- no sfp present
== Goal ==
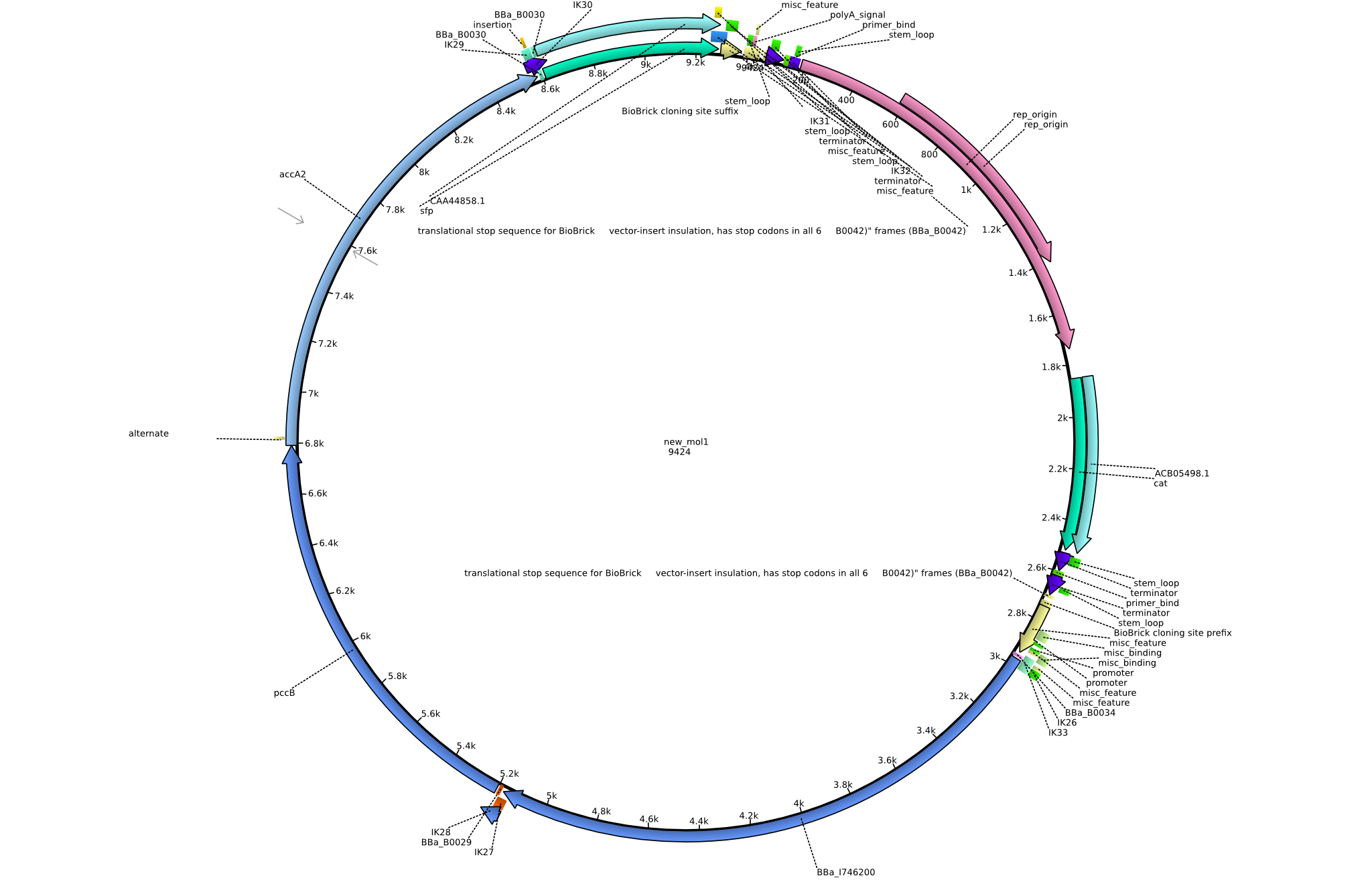
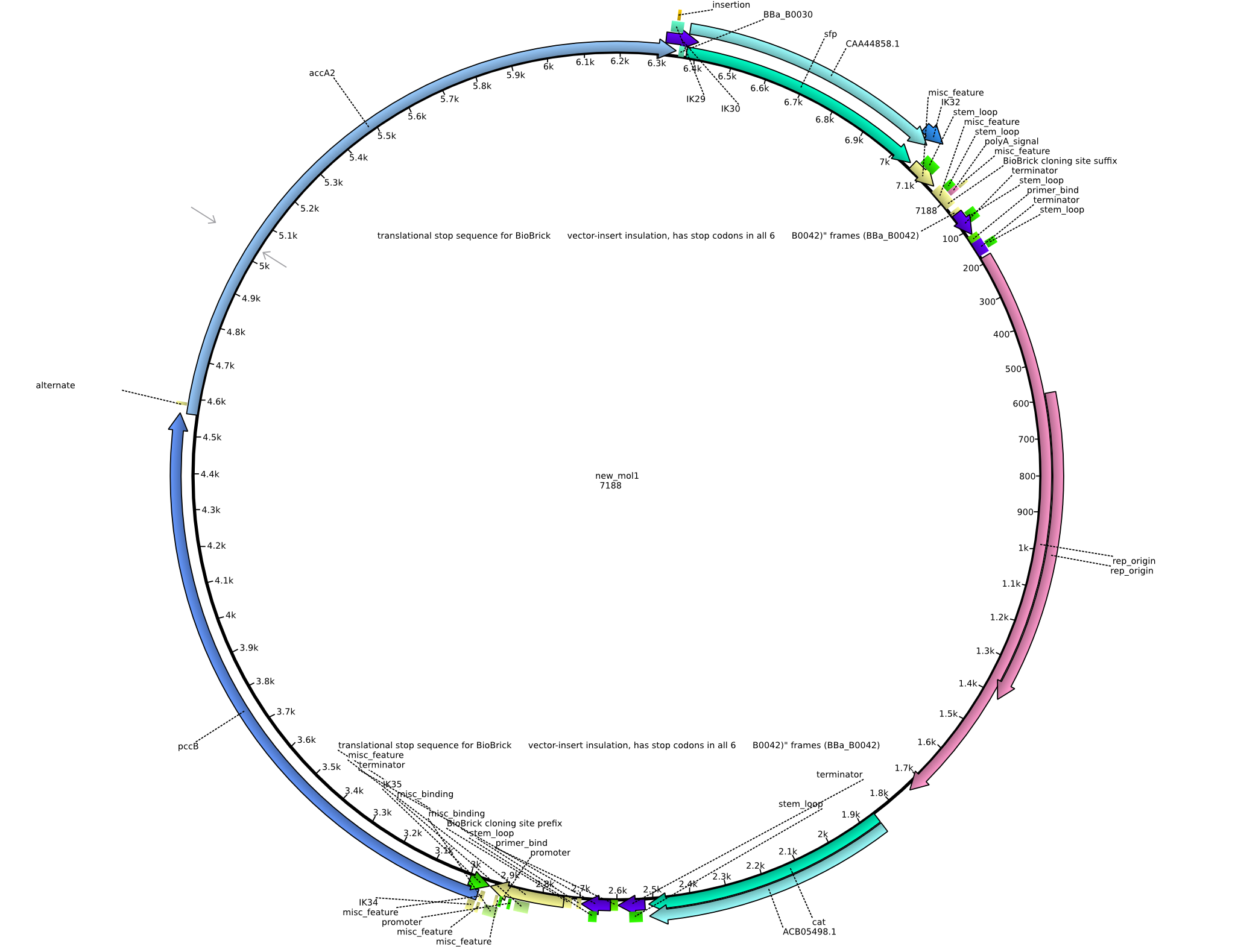
Back-up plan in case the genomic integration does not work. Two plasmids are to be constructed, pIK1 (containing BBa_I746200, pccB-accA, and sfp) and pIK2 (containing pccB-accA and sfp) in the pSB3C5 backbone. 6 fragments are to be amplified:
| fragment ID | source | primers | size [kb] |
|---|---|---|---|
| f1 | BBa_J04450 in pSB3C5 | IK32+IK33 | 3 |
| f2 | BBa_I746200 | IK26+IK27 | 2.3 |
| f3 | pccB-accA in pLF03 | IK28+IK29 | 3.4 |
| f4 | sfp in BAP1 | IK30+IK31 | 0.7 |
| f5 | BBa_J04450 in pSB3C5 | IK32+IK35 | 3 |
| f6 | pccB-accA in pLF03 | IK34+IK29 | 3.4 |
2013-07-22
- transform BBa_I746200 in pSB1AK3 (Spring 2012 Distribution Plate 2 Well 15E) in TOP10, plate on Amp
- transform BBa_J04450 in pSB3C5 (Spring 2012 Distribution Plate 1 Well 3C) in TOP10, plate on Cm
2013-07-23
- pick TOP10-BBa_I746200 colony, inoculate LB+Kan, grow at 37°C
- pick TOP10-BBa_J04450 colony, inoculate LB+Cm, grow at 37°C
- after 12 hours: add new antibiotics to medium, continue growing at 37°C
2013-07-24
- Prepare glycerol stocks
- make miniPreps: 11.1 ng/µl (BBa_J04450), 18 ng/µl (BBa_I746200)
2013-07-25
- run PCRs (using Q5, 20 µl total volume) using 4 ng of each template (pLF03 miniPrep from 2013-06-11, 21 ng/µl), sfp from Konrad's BAP1 plate:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 (f1,f2,f3,f5,f6) / 300 (f4) |
| 35 | 98 | 5 |
| 66 (f1,f2) / 60 (f4) / 61 (f3,f5,f6) | 10 | |
| 72 | 120 (f1,f2,f3,f5,f6) / 30(f4) | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- did not manage to get f3, f4, f6
- repeat: add 1 µl DMSO to reactions:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 35 | 98 | 5 |
| 56 (f4) / 57 (f3,f6) | 30 | |
| 72 | 150 (f3,f6) / 45(f4) | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- use pLF03 miniPrep from 2013-06-11, 27 ng/µl, repeat for f3, f6 (with 1 µl DMSO):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 12 | 98 | 10 |
| 62 ↓0.5°C | 10 | |
| 72 | 150 | |
| 23 | 98 | 10 |
| 58 | 10 | |
| 72 | 150 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
2013-07-26
- run PCR of f3, f6 with Phusion Flash (using pLF03 miniPrep from 2013-06-11, 21 ng/µl), without DMSO:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 65 | 5 | |
| 72 | 75 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 65 | 5 | |
| 72 | 75 | |
| 30 | 98 | 1 |
| 72 | 75 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- inoculate 10 ml LB+Amp with TOP10-pLF03, grow for 36h at 37°C, add Amp every 12 hours
2013-07-28
- make miniPrep -> very low yield (ca. 1-2 ng/µl)
- inoculate 6 ml LB+Amp with TOP10-pLF03, grow at 37°C
2013-07-29
- inoculate 10 ml 2xYT+Amp with TOP10-pKD46, grow at 30°C
2013-07-31
- miniPrep of pKD46 -> ca. 10 ng/µl
2013-08-03
- transform BAP1 with 5µl of pKD46 miniPrep form 2013-07-31, plate on Amp, grow at 30°C
2013-07-29
- make miniPrep -> very low yield (1-2 ng/µl)
- load 1µl of 97ng/µl pLF03 miniPrep from 2013-06-04 on gel -> ca. 20 ng/µl
- inoculate 50 ml LB+Amp with TOP10-pLF03, grow at 37°C
2013-07-30
- make midiPrep -> 1352 ng/µl in 66.5 µl
- medium was contaminated (also check with Fanny) -> low yields of previous miniPreps
2013-07-31
- send 20 µl of 1:15 dilution of midiPrep to sequencing with primers IK07, IK36, IK37: File:Heidelberg PLF03 seq 2013-07-31.zip
- perform multiple sequence alignment against assumed pLF03 sequence: IK07, IK36, IK37
- deletion at the end of accA2 -> primer can not bind
2013-08-05
- run colony-PCR against sfp on BAP1-pKD46 with primers RB43+RB44 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- inoculate 4 ml 2xYT+Amp with BAP1-pKD46, grow at 30°C
2013-08-06
- inoculate 50 ml 2xYT+Amp+Ara(0.5%) with BAP1-pKD46, grow at 30°C until OD=0.71
- run colony-PCR with primers RB43+RB44 from 50 ml culture:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- prepare electrocompetent BAP1-pKD46
2013-08-08
- electroporate BAP1-pKD46 with 500 ng DNA (5 µl of PCR amplificate from 2013-07-08)
- grow in SOC + IPTG(1mM) + Arabinose (0.5%) at 37°C, 400 rpm for 3h
- spin cells down (3 min at 8000 rcf)
- plate 10 µl, rest on Cm, grow at 37°C
2013-08-05
- received constructs from Matthew Mattozzi, Boston: pET21c-pcc-acca-mcce in NEB Turbo (Ampr), pWH8: mcc-pcs-pcc-acc-mmce in NEB Turbo (Kanr), E. coli MM17: MG1655 lambda(DE3) d(lacY) d(E14) mcr-pcs-pcc-acc::KanR integrated at phage site HK022 (Kanr)
- plates contaminated with Drosophila larvae
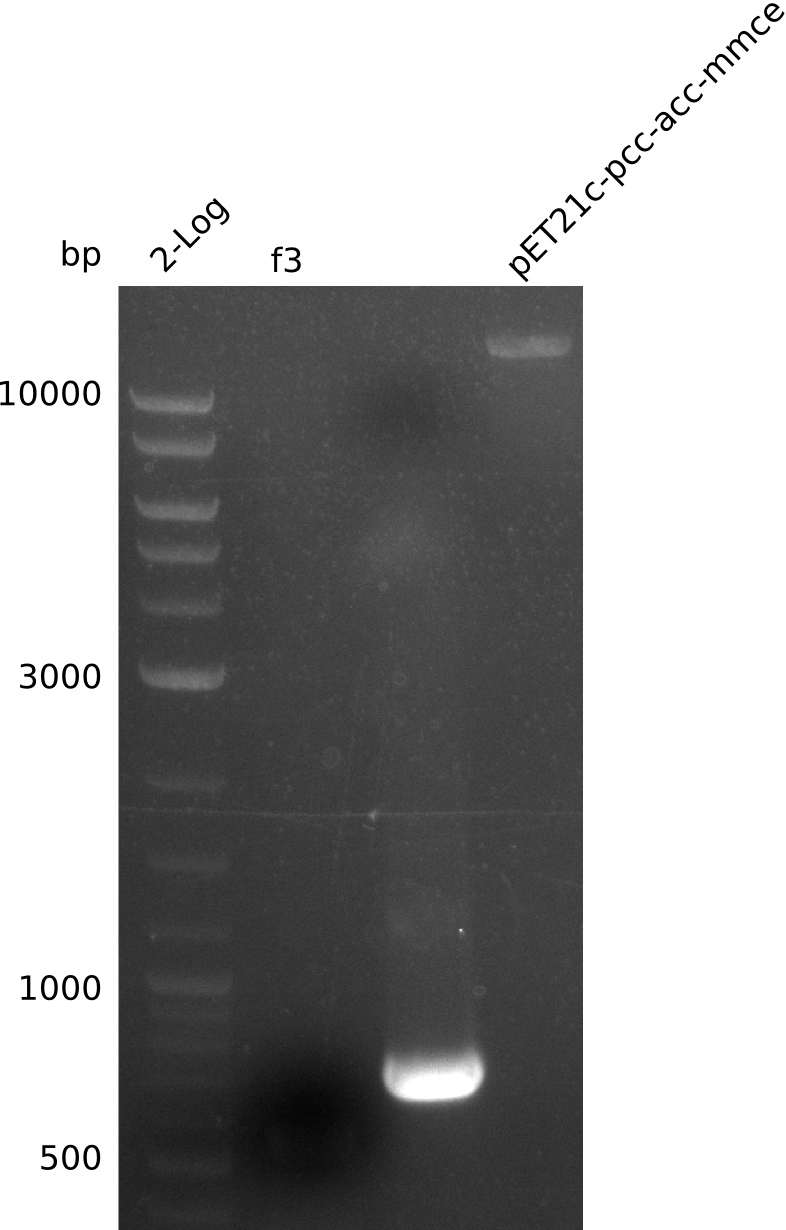
- re-streak on fresh plates, run colony-PCR for f3, f6 from pET21c-pcc-acca-mcce (Q5, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 5 |
| 61 | 10 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- no bands
2013-08-06
- run colony-PCR from pET21c-pcc-acca-mcce grown ON (Q5, 20 µl total volume) for fragments f3, f6:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 60 |
| 35 | 98 | 5 |
| 61 | 10 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- inoculate 5 ml 2xYT+Amp with colony from which colony-PCR was run, grow at 37°C
- no fragment for f3, re-run PCR (1 µl of liquid culture, 20 µl total, Phusion Flash):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 60 |
| 35 | 98 | 1 |
| 65 | 5 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- make miniPrep of liquid culture
- no PCR product, miniPrep runs above 10 kb supercoiled (whole plasmid is expected to have 9 kb) (miniPrep has ca. 10 ng/µl)
- re-run PCR for f3 from miniPrep (Q5, 0.2 µl DNA, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 60 |
| 35 | 98 | 5 |
| 61 | 10 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
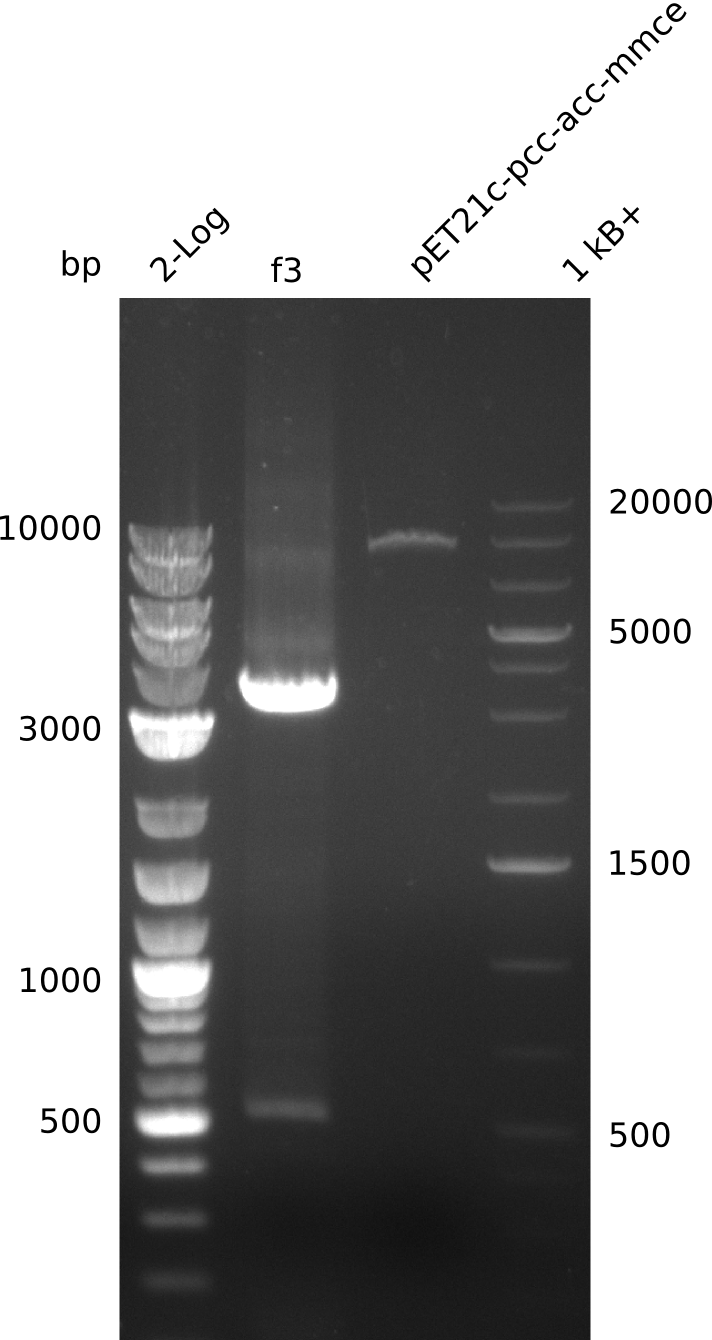
2013-08-07
- need to verify plasmid identity: linearize with BamHI (unique restriction site in both pET21c... and pWH8, use 5 µl of miniPrep, 30 µl total volume)
- linearized pET21c... plasmid runs at 9 kb -> right
- PCR of f3 worked -> hypothesis: Gibson overhang against permeability device bound to E. coli genome in colony-PCRs, as permeability device is mutated version of FepA, which is present in E. coli
- gel-purify all fragments
- f1: 54.2 ng/µl, f2: 64.4 ng/µl, f3: 24.4 ng/µl, f4: 14 ng/µl, f5: 47.6 ng/µl, f6: 25.2 ng/µl; all in 30 µl
- perform Gibson assembly:
- pIK1: 0.8 µl f1, 1.6 µl f2, 5 µl f3, 2 µl f4, 0.4 µl H2O, 10 µl NEB Gibson MM
- pIK2: 1 µl f5, 6 µl f6, 3 µl f4
- incubate at 50°C for 1h
- transform TOP10: 2 transformations per plasmid: 5 µl of Gibson assembly, 5 µl of 1:4 diluted Gibson assembly
- grow in 2xYT at 37°C, plate on Cm, grow at 37°C
2013-08-08
- pIK1: several colonies, some of them pink -> pick 10 white, grow in 2xYT+Cm at 37°C
- pIK2: 2 populations of colonies: large and small, several large colonies are pink -> pick 5 white large, 5 white small colonies, grow in 3 ml 2xYT+Cm at 37°C
2013-08-09
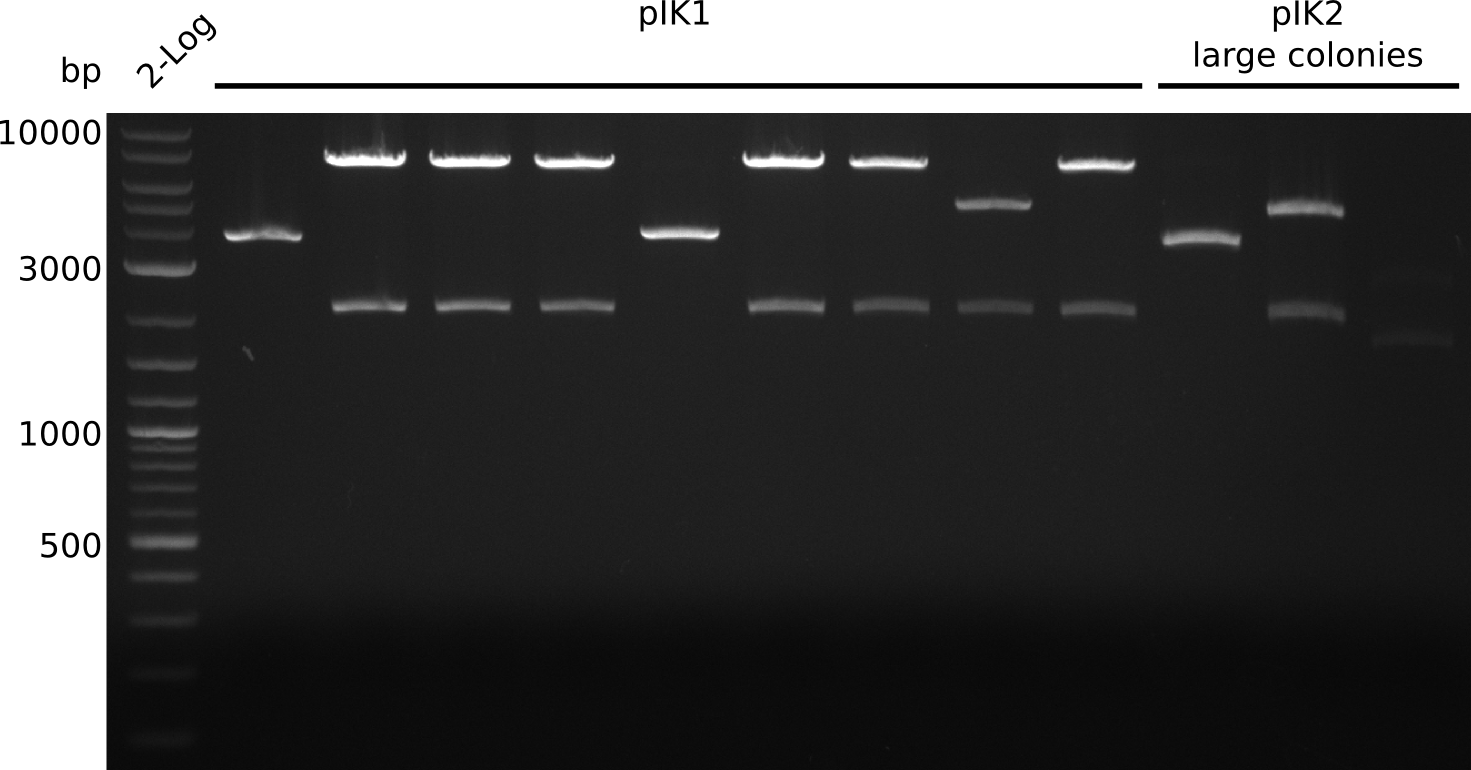
- colony 8 of pIK1 did not grow
- make miniPreps of 2 ml everything else (2 ml each) -> ca. 62 - 600 ng/µl
- miniPrep yields quite high, but consistent: pACYC with p15A origin (4 kb) [http://www.abgent.com.cn/downloads/BG0003-Plasmid-mini-kit.pdf yields 0.6 µg DNA / ml culture] in LB, culture in 2xYT/TB yields 2-5 times more cells (according to QiaGen) => ca. 10-15 µg expected for 9 kb plasmid
- digest with BamHI+HindIII (20 µl total volume, 0.5 µl enzyme, 1 µl DNA [except for sample with 62 ng/µl, there 2 µl DNA was used])
- expected fragments: 2151 bp + 7265 bp for pIK1, 2151 bp + 5029 bp for pIK2
- send pIK1.2 and pIK2.2 to sequencing with primers VF2 and VR
2013-08-12
- no colonies on 10 µl plate
- 2 large colonies, 1 small colony on rest plate
- run colony-PCR with primers IK01+IK02:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- genomic integration failed
== 2013-08-12 ==
2013-08-13
- sequences arrived: File:PIK1.2-pIK2.6 seq 2013-08-13.zip
- perform multiple sequence alignment against assumed sequences: pIK1.2 with IK25, pIK2.6 with VF2, pIK2.6 with VR
- sequences look good
- prepare glycerol stock of TOP10-pIK1.2
2013-08-14
- prepare glycerol stock of TOP10-pIK2.6
2013-08-19
- received BL21(DE3) from Gruss Lab, ZMBH
- grow in LB to OD=0.53, prepare competent cells
- co-transform with pIK1.2(0.2 µl = 120 ng)+pMM64(1 µl = 15 ng), pIK2.6(0.2 µl = 60 ng)+pMM64(1 µl = 15 ng), plate on Cm+Amp+IPTG, grow at 37°C
2013-08-20
- all colonies white
- pick 1 colony each, inoculate LB+Cm+Amp, grow at 37°C until cultures are opaque, add IPTG to 1mM, grow at 30°C
- no visible indigoidine production
- need to clone into a different backbone in order to be able to cotransform with pRB21
- inoculate LB+Kan with TOP10-pSB3K3-BBa_J04450, grow at 37°C
2013-08-21
- prepare glycerol stock of TOP10-pSB3K3-BBa_J04450
- perform miniPrep -> 10.1 ng/µl in 38 µl
- digest with EcoRI+SpeI: 380 ng of pSB3K3-BBa_J04450: use everything (50 µl total volume, 2x0.5 µl enzyme, expect 2.7kb & 1kb), 1200 ng of pIK1.2 (2 µl of miniPrep from 2013-08-09, 20 µl total volume, 2x0.5 µl enzyme, expect 6.7kb & 2.7kb), 900 ng of pIK2.6 (3 µl of miniPrep from 2013-08-09, 20 µl total volume, 2x0.5 µl enzyme, expect 4.4kb & 2.7kb)
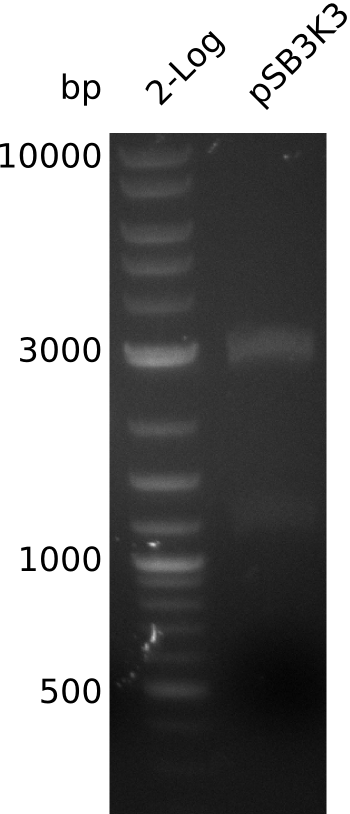
- pSB3K3-BBa_J04450 looks as though it was not cut: inoculate 50 ml TB+Kan with TOP10-pSB3K3-BBa_J04450, grow at 37°C
- as control for indigoidine production from pMM64: co-transform BL21(DE3) with pMM64(1 µl = 15 ng)+pMM65(0.2 µl = 13 ng), plate on Amp+Kan+IPTG
2013-08-22
- no colonies of BL21(DE3)-pMM64-pMM65: co-transform BL21(DE3) with pMM64(1 µl = 15 ng)+pMM65(1 µl = 75 ng), plate on Amp+Kan+IPTG
- prepare midiPrep of pSB3K3-BBa_J04450: 2952.1 ng/µl
- digest 0.4 µl of midiPrep with EcoRI+SpeI (20 µl total volume, 2x0.5 µl enzyme)
- band too weak: 2 possibilities:
- concentration measurement wrong
- mispipeted
- => load 1 µl of midiPrep on gel -> concentration measurement about right
- large pSB3K3 fragment over 3 kb, consistent with miniPrep from 2013-08-21 -> 2.7 kb expected, sequence wrong on parts.igem.org
- gel-purify pSB3K3, 6 kb fragment of pIK1, 4 kb fragment of pIK2
- 18 µl of pSB3K3 (not measured), 12.3 ng/µl pIK1 in 21 µl; 7.9 ng/µl pIK2 in 21 µl
- ligate at RT for 1h:
| what | µl |
|---|---|
| pSB3K3 | 9 |
| insert | 8 |
| T4 ligase | 1 µl |
| T4 ligase buffer | 2 µl |
- heat-inactivate: 75°C for 5 min
- transform 10 µl of each ligation into TOP10, plate on Kan, grow at 37°C (pSB3K3+pIK2 = pIK6)
2013-08-23
- pick 5 colonies each, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- no amplificate
- have another look at sequences (motivated by DelRest results): 1 bp missing in pIK1.2: frame shift in permeability device => send pIK1.3 to sequencing with primers VF2, VR, IK25
- pick 30 colonies of pSB3K3-pIK2, run colony-PCR with primers RB43+VR (pool 3 colonies in 1 PCR reaction, iTaq, 20 µl total volume)
- no positives => ligation did not work, most likely due to low DNA concentrations
- need to repeat: inoculate 2x5 ml 2xYT+Cm with TOP10-pIK2.6
2013-08-24
- make miniPrep of pIK2.6: 97 ng/µl in 37.5 µl
- digest 3 µg pSB3K3 (1 µl of midiPrep from 2013-08-22), 1.7 µg pIK2.6 (17 µl of miniPrep) with EcoRI+SpeI (20 µl total volume, 0.5 µl of each enzyme)
- treat pSB3K3 digest with antarctic phosphatase (37°C, 60 min)
- run gel, gel-purify -> 18 µl pSB3K3 (not measured), 15.0 ng/µl pIK2.6 in 20.5 µl
- ligate at RT for 1h:
| what | µl |
|---|---|
| pSB3K3 | 9 |
| pIK2.6 | 8 |
| T4 ligase | 1 µl |
| T4 ligase buffer | 2 µl |
- heat-inactivate: 75°C for 5 min
- transform 10 µl of ligation into TOP10, plate on Kan, grow at 37°C
2013-08-25
- pick 10 colonies, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- no amplificate
- VR primer might not be binding to pSB3K3: http://www.ccbi.cam.ac.uk/iGEM2006/index.php/Primer_List#BioBrick_verification_primers , https://2009.igem.org/Team:Groningen/Notebook/22_July_2009
- pick another 10 colonies, run colony-PCR with primers VF2+IK25 (expected: 1kb, iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- definite product, but at 1.3 kb => pSB3K3 was longer than expected in digest, might be a result from that
- unspecific bands probably due to melting temperature of IK25 being at 67°C
- inoculate 2xYT+Kan, grow at 37°C
- pSB3K3 used was from plate 5 => transform TOP10 with pSB3K3 from plate 2, well 6 F
2013-08-26
- make miniPreps of colonies 1 and 2: 71.4 ng/µl and 75.1 ng/µl in 38 µl
- digest with HindIII+BamHI (3 µl DNA, 0.5 µl of each enzyme, 20 µl total); expected: 4.1 kb, 2.1 kb, 0.9 kb
- no 0.9 kb fragment => no BamHI site in KanR gene, 2.1 fragment visible corresponds to acca-sfp => insert completely within backbone
- co-transform TOP10 with pIK6.1+pRB21
- sequences of pIK1.3 arrived: PIK1.3-2013-08-26.zip, PIK1.3-2013-08-26 VF2.clustal.txt, PIK1.3-2013-08-26 VR.clustal.txt
- point mutation in permeability device, interestingly, both the frameshift in pIK1.2 and this point mutation create a stop codon at the beginning of the permeability device
- send pIK1.4 to sequencing with primer VF2
2013-08-27
- TOP10-pIK6.1-pRB21 are not blue, but the indigoidine group is having problems with pRB21 -> pRB21 might not work
- sequence of pIK1.4 arrived: PIK1.4-2013-08-27.zip, PIK1.4-2013-08-27 VF2.clustal.txt
- again frame-shift in permeability device -> send pIK1.6, pIK1.7, pIK1.10 to sequencing with primer VF2
- make miniPrep of pSB3K3: 54.7 ng/µl in 38 µl
- digest pSB3K3 with EcoRI+SpeI (20 µl total volume, 5 µl DNA, 0.5 µl of each enzyme)
- gel-purify, ligate with pIK2.6 fragment from 2013-08-24 at RT for 1h -> pIK7:
| what | µl |
|---|---|
| pSB3K3 | 9 |
| pIK2.6 | 8 |
| T4 ligase | 1 µl |
| T4 ligase buffer | 2 µl |
- heat-inactivate: 75°C for 5 min
- transform 10 µl of ligation into TOP10, plate on Kan, grow at 37°C
2013-08-28
- pick colonies, run colony-PCR with primers VF2+IK25 (expected: 1kb, iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- grow in 2xYT+Kan at 37°C
2013-08-29
- sequences of pIK1 arrived: PIK1.6-pIK1.7-pIK1.10-2013-08-29.zip, PIK1.6-2013-08-29 VF2.clustal.txt, PIK1.7-2013-08-29 VF2.clustal.txt, PIK1.10-2013-08-29 VF2.clustal.txt
- frame-shift in pIK1.6, point mutations in pIK1.7, pIK1.10
- perform miniPreps of pIK7 -> ca. 50 ng/µl in 38 µl
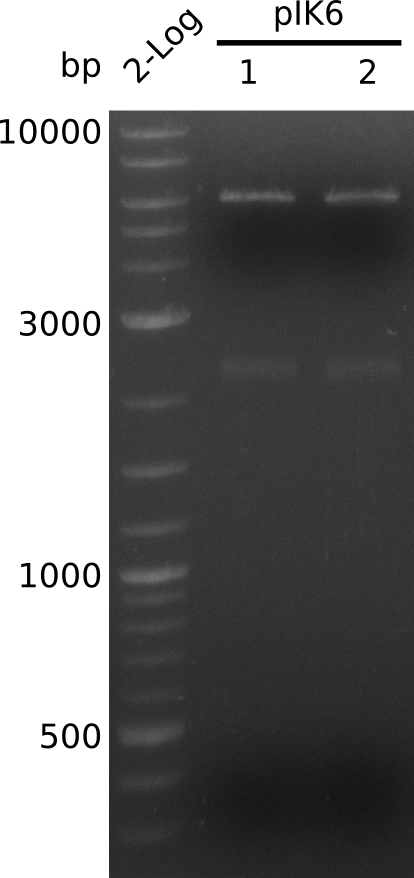
- digest 5 µl of pIK7.1, pIK7.2 with BamHI+HindIII (0.5 µl of each enzyme, 20 µl total volume)
- bands at right positions (expected: 4.1, 2.1, 0.8 kb)
- prepare glycerol stock of TOP10-pIK7.1
2013-08-30
- electroporate electrocompetent DH10ß with 1 µl, 14 µl of 1:4 Gibson mix for pIK1 from 2013-08-07
- plate 10 µl, rest of each electroporation on Cm, grow at 37°C
2013-08-31
- very large and small colonies present, after heat-schock transformation last time only large colonies present
- pick 5 small colonies from each plate, grow in 2xYT+Cm (no 2xYT left => last 3 colonies from 14 µl / rest plate grown in LB+Cm) at 37°C
2013-09-01
- pIK1.12 did not grow
- make miniPreps of all cultures -> concentrations about 200-300 ng/µl in 37.5 µl (except pIK1.14: 50 ng/µl)
- digest with BamHI+HindIII (20 µl total volume, 0.5 µl enzyme, 1 µl DNA)
- expected fragments: 2151 bp + 7265 bp
2013-09-02
- send pIK1.11, pIK1.13, pIK1.14 to sequencing with primer VF2
2013-09-03
- sequences arrived: PIK1.11-pIK1.13-pIK1.14-2013-09-03.zip, PIK1.11-2013-09-03 VF2.clustal.txt, PIK1.13-2013-09-03 VF2.clustal.txt, PIK1.14-2013-09-03 VF2.clustal.txt
- insertions leading to stop-codon in pIK1.11, pIK1.13; two bases of RBS missing in pIK1.14
- check wiki of Cambridge 2007 team, who constructed the permeability device: [http://www.ccbi.cam.ac.uk/iGEM2007/index.php/Detailed_background_information_-_signalling#Plan they tried different promoter strengths], [http://www.ccbi.cam.ac.uk/iGEM2007/index.php/Signalling_project_description#Standard_Construction_of_Permeability_Device only the weakest promoter gave colonies] => bacteria might be selecting against the permeability device
- put pIK1.12 in incubator again, maybe it has the right sequence and grows slowly
- pIK1.12 did grow, make miniPrep -> 67 ng/µl in 38 µl
- send pIK1.14 to sequencing with primers IK25, VR
2013-09-04
- sequences arrived: PIK1.14-2013-09-04.zip, PIK1.14-2013-09-04 IK25.clustal.txt, PIK1.14-2013-09-04 VR.clustal.txt
- sequences match
- prepare glycerol stock of DH10ß-pIK1.14
- digest pIK1.12 with BamHI+HindIII (20 µl total volume, 134 ng DNA [2 µl of miniPrep from 2013-09-03], 0.5 µl enzyme)
- digest looks about right, send to sequencing with primer VF2
2013-09-05
- sequence arrived: PIK1.12-2013-09-05.zip, PIK1.12-2013-09-05 VF2.clustal.txt
- ca. 1 kb missing from end of permeability device
2013-09-09
- co-transform TOP10 with pRB22.3 and pIK7.1, plate on Cm+Kan, grow at 37°C
2013-09-10
- after ca. 18h at 37°C: lots of tiny colonies, not blue
- leave at RT in drawer (no light)
2013-09-11
- colonies became blue
- start constructing pIK8: promoter even weaker than the one used by Cambridge 2007, weak RBS
- run PCRs of (2 ng DNA, 20 µl total volume, using Q5):
- pIK2.6 with primers IK40+IK43 -> f7
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 35 | 98 | 10 |
| 58 | 10 | |
| 72 | 300 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- BBa_I746200 with primers IK41+IK42 -> f8
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 35 | 98 | 10 |
| 60 | 10 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
2013-09-12
- run gel, gel-purify fragments -> 6.3 ng/µl in 20 µl (f7); 36.1 ng/µl in 20 µl (f8)
- perform Gibson assembly: 8 µl f7, 2 µl f8, 10 µl MM, incubate at 50°C for 1h
- dilute Gibson mix 1:4, electroporate electrocompetent DH10ß with 1 µl, 14 µl of dilution
- plate 10 µl, rest on Cm, grow at 37°C
2013-09-13
- very large and very small colonies present, pick starting with small colonies, run colony-PCR with primers IK26+VR (iTaq, 20 µl total volume) using 2 ng of BBa_I746200 in pSB1AK3 as positive control:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 240 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- clones grown in eppendorf tubes in 2xYT+Cm at 37°C
2013-09-14
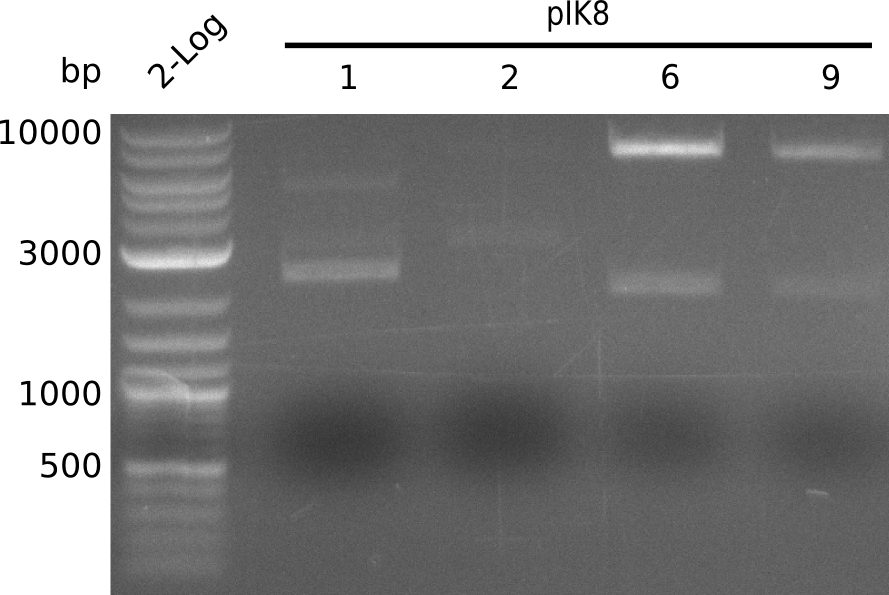
- run gel
- colonies designated pIK8.1, pIK8.2, pIK8.6, pIK8.9 positive
- transfer to baci falcons, add 2xYT+Cm, grow at 37°C
2013-09-15
- make miniPreps: 73.5 ng/µl (pIK8.1), 12.9 ng/µl (pIK8.2), 289.0 ng/µl (pIK8.6), 169.0 ng/µl (pIK8.9) in 37.5 µl each
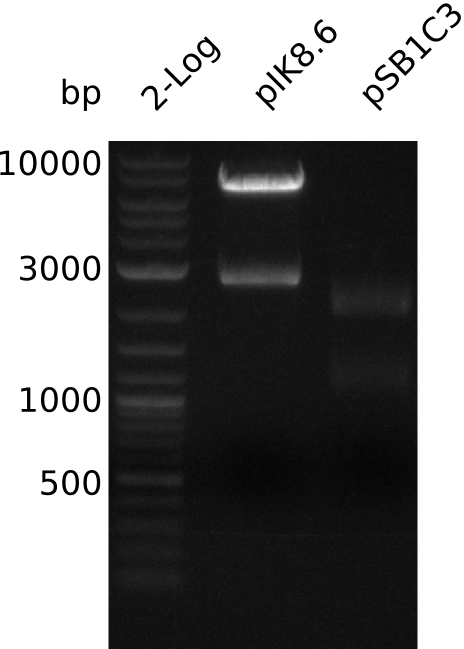
- digest with BamHI+HindIII using 2 µl (pIK8.1), 10 µl (pIK8.2), 1 µl (pIK8.6), 1 µl (pIK8.9) DNA (20 µl total volume)
- expected: 2.1 + 7.3 kb
2013-09-16
- send pIK8.6, pIK8.9 to sequencing with primer RB43
2013-09-17
- sequences arrived: PIK8.6-pIK8.9-2013-09-16.zip, PIK8.6-2013-09-16 RB43.clustal.txt, File:Heidelberg_PIK8.9-2013-09-16 RB43.clustal.txt
- no mutations at primer site
- sequence with VR primer
- inoculate 2x 6 ml 2xYT+Cm with DH10ß-pIK8.6
2013-09-18
- sequences arrived: PIK8.6-pIK8.9-2013-09-18.zip, PIK8.6-2013-09-18 VR.clustal.txt, File:Heidelberg_PIK8.9-2013-09-18 VR.clustal.txt
- sequences look good
- make miniPrep of pIK8.6 -> 269 ng/µl in 38 µl
- to verify functionality of permeability device: need to co-transform with indigoidine synthetase => need to clone into pSB3K3 -> pIK9
- need to clone into pSB1C3 for submission (pIK10, pIK11): digest 465 ng pSB1C3 (1 µl of Ralf's midiPrep) with EcoRI+SpeI, treat with antarctic phosphatase (37°C, 60 min)
- digest pIK8.6 with EcoRI+SpeI (3 µl DNA, 0.5 µl enzyme, 20 µl total volume)
- gel-purify pIK8.6 insert (20 ng / µl in 20 µl), pSB1C3 (18 µl, concentration not measured)
- ligate for 1 h at RT:
| pIK9 | pIK10 | pIK11 | |
|---|---|---|---|
| backbone | 9 µl pSB3K3 ES from 2013-09-27 | 9 µl pSB1C3 ES | 9 µl pSB1C3 ES |
| insert | 8 µl pIK8.6 ES | 8 µl pIK8.6 ES | 4.5 µl pIK2.6 ES (rest from 2013-08-24) |
| T4 ligase | 1 µl | 1 µl | 1 µl |
| T4 ligase buffer | 2 µl | 2 µl | 2 µl |
| H2O | 0 µl | 0 µl | 3.5 µl |
- transform TOP10 with 10 µl of each ligation, plate on LB+Kan(pIK9), LB+Cm(pIK10, pIK11)
2013-09-19
- colonies present for all ligations
- colonies for pIK10 extremely small -> permeability device toxic
- run colony-PCR using primers VF2+IK25 (expected: 1 kb, OneTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- grow in TB+Kan (pIK9) / TB+Cm (pIK10, pIK11)
- evening: make miniPreps of pIK9, pIK11 (pIK10 grew very slowly) -> ca. 50-60 ng/µl (pIK9), 100-300 ng / µl (pIK11) in 38 µl
- inoculate 2xYT+Cm with pIK8.1, pIK8.6
2013-09-20
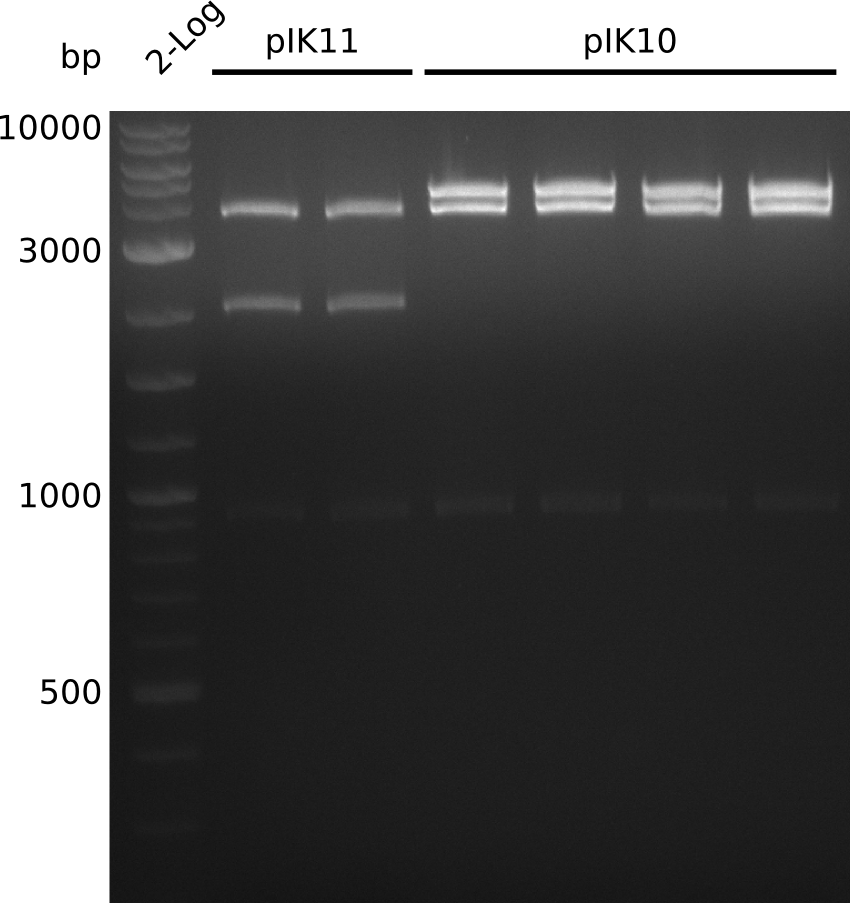
- make miniPreps of pIK10 -> 400-600 ng/µl in 38 µl
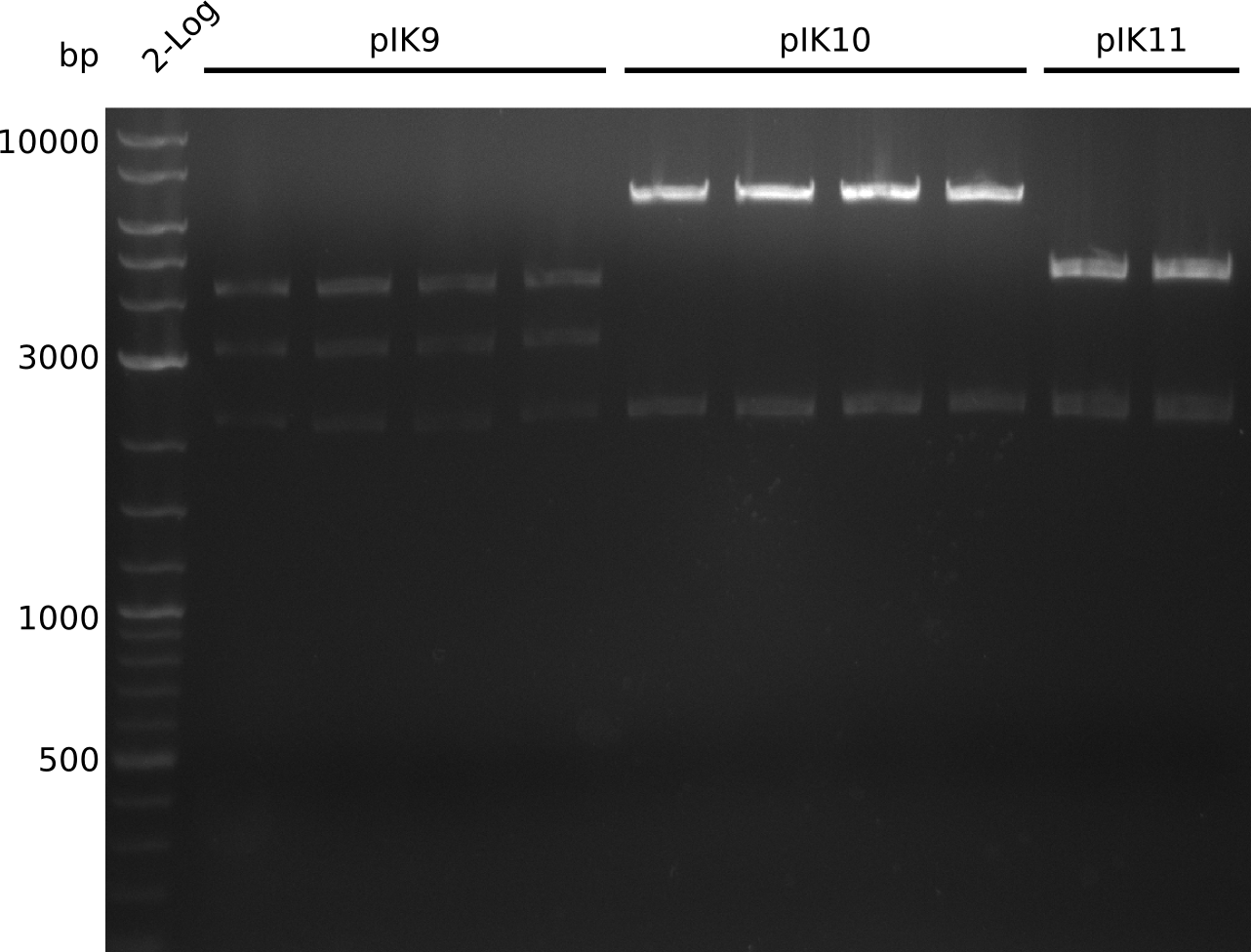
- digest everything with BamHI+HindIII, expected: 2.1 kb + 3.1 kb + 4.1 kb (pIK9), 2.1 kb + 6.6 kb (pIK10), 2.1 kb + 4.3 kb (pIK11) (20 µl total volume, 0.5 µl enzyme, using 0.5 µl (pIK10), 1 µl (pIK11), 2 µl (pIK9) DNA)
- to distinguish between pSB3C5 and pSB1C3: digest pIK10, pIK11 with XhoI (expected: 1 band for pSB3C5, 3 bands for pSB1C3)
- send pIK10.1, pIK10.3, pIK10.3, pIK11.1, pIK11.2 to sequencing with primers VF2, VR
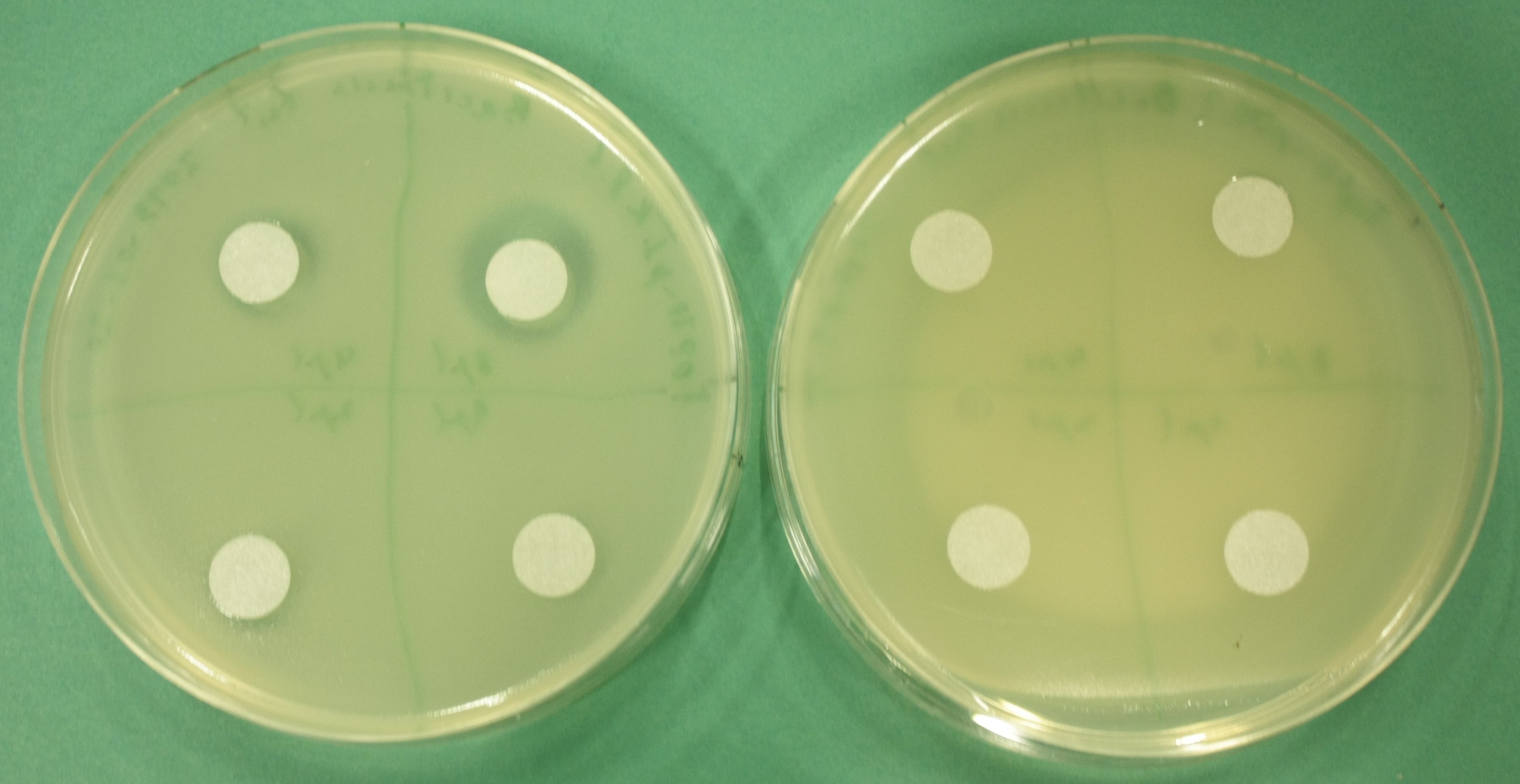
- test permeability device:
- inoculate 3 ml LB+Cm soft agar with 100 µl of pIK8.1, pIK8.6 ON culture, pour on LB+Cm plates
- add sterile filter paper, pipet 8 µl, 4 µl, 2 µl, 1 µl of Bacitracin on filter paper discs
- grow at 37°C
- Co-transform TOP10 with 25 ng pIK9.1, 10 ng pRB22.4, plate on Cm+Kan, grow at 37°C
2013-09-21
- co-transformats: lots of extremely small, blue-ish colonies, several big white colonies -> possibly mutated indigoidine synthetase, several big black colonies -> possibly mutated permeability device
- pick one little blue-ish colony, grow in 2xYT+Cm+Kan at 37°C
== 2013-09-23 ==
2013-09-27
- need to remove XbaI site for submission (for Boston) -> pIK12: amplify pccB-accA2 with primer introducing SpeI, ligate SpeI+Xba+ sites
- run PCR of pIK8.6 with primers VF2+IK44 (3 ng DNA, 20 µl total volume, using Q5) -> f9:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 35 | 98 | 10 |
| 55 | 10 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
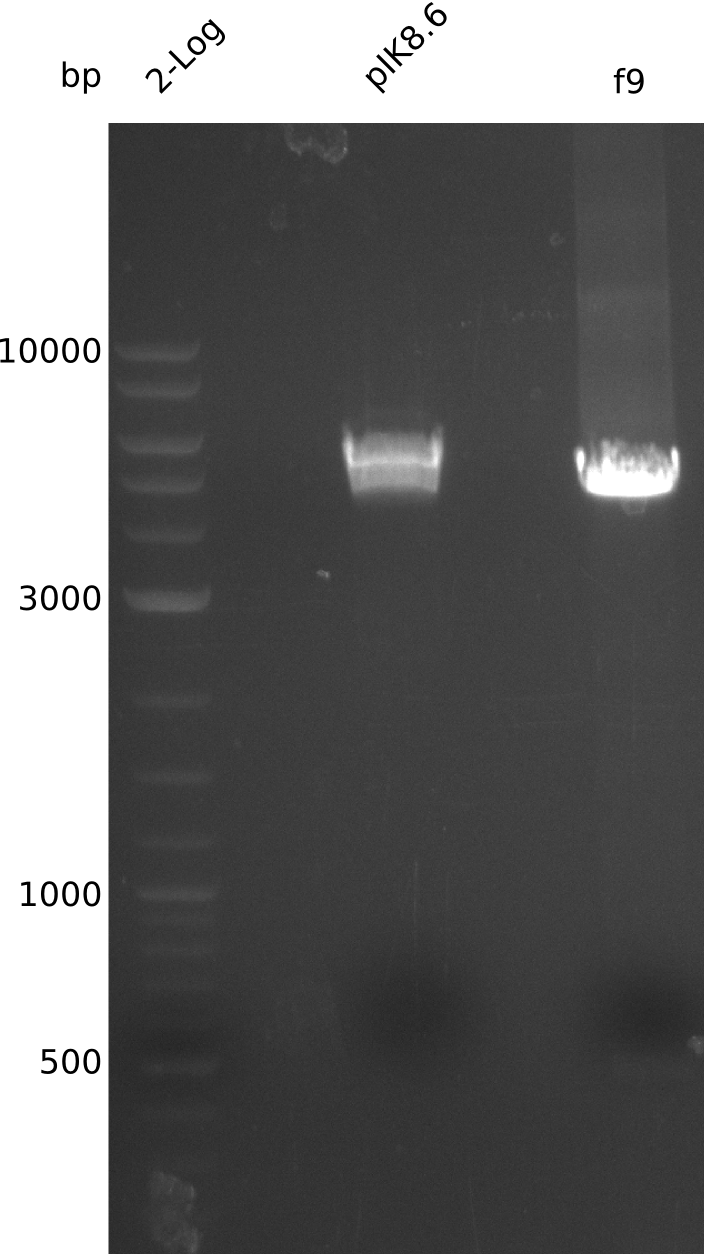
- digest 867 ng pIK8.6 (3 µl of miniPrep from 2013-09-15) with EcoRI+XbaI (20 µl total volume, using 0.5 µl enzyme), treat with antarctic phosphatase (37°C, 60 min)
- gel-purify large pIK8.6 fragment, f9 (elute f9 in 17 µl)
2013-09-28
- digest f9 with EcoRI+SpeI (0.5 µl enzyme, 20 µl total volume), purify with Nucleotide Removal Kit
- ligate at RT for 1h:
| what | µl |
|---|---|
| pIK8.6 | 8 |
| f9 | 9 |
| T4 ligase | 1 µl |
| T4 ligase buffer | 2 µl |
- heat-inactivate at 75°C for 5 min
- transform 10 µl into TOP10, plate 10 µl, rest on Cm, grow at 37°C
2013-09-28
- lots of small colonies on both plates, several large colonies on both plates
- pick 4 small, 1 large colonies from 10 µl plate, grow in 2xYT+Cm at 37°C
 "
"