Team:Heidelberg/Project/Indigoidine
From 2013.igem.org
m |
m |
||
| Line 24: | Line 24: | ||
<!--graphical abstract--> | <!--graphical abstract--> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | <a class="fancybox fancyGraphical" rel="group" href="Heidelberg_Indigoidine_graphical_abstract.png" > | + | <a class="fancybox fancyGraphical" rel="group" href="https://static.igem.org/mediawiki/2013/d/de/Heidelberg_Indigoidine_graphical_abstract.png" > |
| - | <img style="width:100%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 4px; border-color: grey;" src="https://static.igem.org/mediawiki/2013/ | + | <img style="width:100%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 4px; border-color: grey;" src="https://static.igem.org/mediawiki/2013/d/de/Heidelberg_Indigoidine_graphical_abstract.png"></img> |
</a> | </a> | ||
<div class="jumbotron"> | <div class="jumbotron"> | ||
| Line 61: | Line 61: | ||
<div class="item active"> | <div class="item active"> | ||
| - | <img src=" | + | <img src=""> |
<div class="container"> | <div class="container"> | ||
<div class="carousel-caption" data-offset="0"> | <div class="carousel-caption" data-offset="0"> | ||
| - | <p style="font-size:18px; color:#fff"> | + | <p style="font-size:18px; color:#fff"></p> |
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
<div class="item"> | <div class="item"> | ||
| - | <img src=" | + | <img src=""> |
<div class="container"> | <div class="container"> | ||
<div class="carousel-caption" data-offset="0"> | <div class="carousel-caption" data-offset="0"> | ||
| - | <p style="font-size:18px; color:#fff"> | + | <p style="font-size:18px; color:#fff"></p> |
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
<div class="item"> | <div class="item"> | ||
| - | <img src=" | + | <img src=""> |
<div class="container"> | <div class="container"> | ||
<div class="carousel-caption" data-offset="0"> | <div class="carousel-caption" data-offset="0"> | ||
| - | <p style="font-size:18px; color:#fff"> | + | <p style="font-size:18px; color:#fff"></p> |
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img src=""> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption" data-offset="0"> | ||
| + | <p style="font-size:18px; color:#fff"></p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img src=""> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption" data-offset="0"> | ||
| + | <p style="font-size:18px; color:#fff"></p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img src=""> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption" data-offset="0"> | ||
| + | <p style="font-size:18px; color:#fff"></p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img src=""> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption" data-offset="0"> | ||
| + | <p style="font-size:18px; color:#fff"></p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="item"> | ||
| + | <img src=""> | ||
| + | <div class="container"> | ||
| + | <div class="carousel-caption" data-offset="0"> | ||
| + | <p style="font-size:18px; color:#fff"></p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 236: | Line 276: | ||
</script> | </script> | ||
</html> | </html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:Heidelberg/Templates/Footer-Nav}} | {{:Team:Heidelberg/Templates/Footer-Nav}} | ||
{{:Team:Heidelberg/Templates/Fancybox}} | {{:Team:Heidelberg/Templates/Fancybox}} | ||
Revision as of 00:46, 5 October 2013


Indigoidine. Proving Modularity of NRPS by Shuffling Domains.

Highlights
- Harvest of delftibactin, a gold-precipitating peptide, from its native, cultured host Delftia acidovorans.
- Enrichment of pure gold from electronic waste with purified delftibactin.
- Optimization of the Gibson-Assembly method for the creation of large plasmids (> 30 kbp) with high GC content.
- Amplification and cloning of all components required for delftibactin production.
- Transfer of the entire pathway from D. acidovorans for the synthesis of delftibactin to E. coli.
Abstract
An integral characteristic of synthetic biology yet often undermined is the ability to learn fundamental knowledge by systematically perturbing a biological system. Non-ribosomal peptide synthetases (NRPS) are predestinated for such a trial and error approach. Their hierarchical organization into modules and domains offer a unique opportunity to spin around their inherent logical assembly and observe if their functionality is preserved. Following this idea, we prove the interchangeability of NRPS domains at the example of indC from Photorhabdus luminescens laumondii TT01 (DSM15139). The native NRPS domains have been replaced with domains from other bacterial organisms and fully synthetic domains. To quantify the NRPS efficiency we established an indigoidine assay based on OD measurement of the blue-colored pigment. Interestingly, we find that our data points out the dependence on the T-domain and the 4'-Phosphopanthetheinyl-transferases (PPTases), resulting in different levels of indigoidine synthesis. Furthermore, we introduce HiCT - High throughput protocols for circular polymerase extension Cloning and Transformation - a new standard for the assembly of combinatorial gene libraries (RFC 99).
Introduction
Most modules of non-ribosomal peptide synthetase (NRPS) pathways consist of three domain types: condensation, adenylation and thiolation domain (see Figure 1a), also called peptidyl-carrier-protein domain (PCP)-domain (reviewed in
For example, a single module of P. luminescens laumondii TT01 (DSM15139) contains an internal oxidation domain (Ox-domain) in its A-domain and a special TE-domain (Figure 2a). This enzyme first adenylates L-glutamine (A-domain), which is then attached to the T-domain. The TE-domain cleaves and catalyzes the cyclization of the substrate, which is further oxidized by the Ox-domain. The oxidation of two cyclic glutamines results in the formation of an insoluble small molecule (Figure 2b)[Reference].
The small molecule produced by the pathway described above is a blue-colored pigment called indigoidine. Accordingly, the catalytic NRPS is referred to as indigoidine synthetase or blue pigment synthetase encoded by various bacterial strains such as S. lavendulae subsp. lavendulae (ATCC11924) or P. luminescens (
Aim
We want to find the best approach to determine the linker structure between the A-, T- and TE-domain of indC by exchanging the native T-domain with the T-domain of bpsA. We want to proof that it is possible to exchange the indC T-domain with T-domains from other NRPS modules that are not related to indigoidine synthetases and/or the P. luminescens strain We want to point out the great potential of NRPS for synthetic approaches by designing synthetic T-domains that will result in a functional indigoidine synthetase when introduced to indC We want to show that the activity of NRPS is dependant on the PPTase used to activate them and that the right combination of PPTase and T-domain plays a crucial role when trying to increase the overall product yield. We want to exhibit the potential of NRPS modularity by converting native NRPS modules with glutamine specificity into an indigoidine synthetase by introducing an oxidation domain into the Adenylation-domain. Additionally we will try out various domain combinations to assemble and configure a indigoidine synthetase comprised of different NRPS modules.Results
Engineered indigoidine synthetases retain functionality
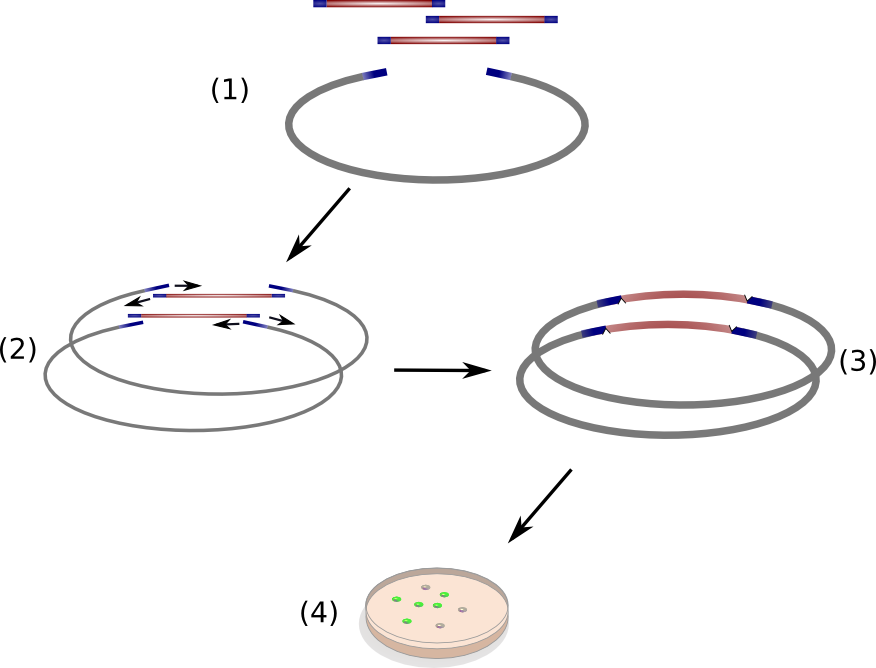
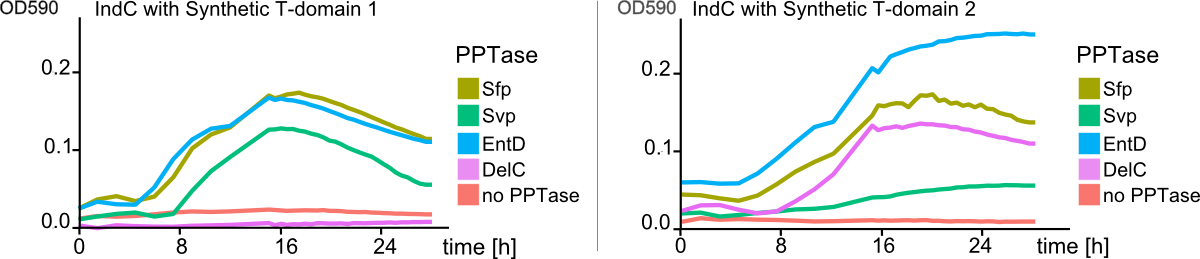
We successfully engineered the nonribosomal peptide synthetase indC from P. luminescens subsp. Laumondii TT01 by replacing its native T-domain with both T-domains of other NRPS modules from different bacterial strains and synthetic T-domains of own design, thus creating a library of 58 engineered variants of the indC indigoidine synthetase. So far only the exchange of the T-domain in the related indigoidine synthetase bpsA from S. lavendulae ATCC11924 with the T-domain of the E. coli entF NRPS module was reported, postulating that the T-domain cannot be replaced by other native T-domains retaining the overall protein function, since the engineered variants of bpsA were incapable of producing the blue pigment indigoidine (However, our data shows that the T-domain of indC can be replaced by the T-domain of bpsA only if specific T-domain border combinations are used (see Fig. x) to define the replaced fragment (Figure xa). We proved that indC retained its functionality when the T-domain of the native indC gene was replaced with T-domains of other NRPS modules (see Figure X), applying the T-domain border combination "A2" (see Fig. x). We also tried the T-domain border combination "B2" with the same T-domains, as these T-domain borders were proposed by the Pfam domain prediction tool (pfam.sanger.ac.uk). In this approach less transformants were capable of producing the plue pigment (data not shown), suggesting that the T-domain border combination "A2" can be seen as an improved variant compared to the Pfam prediction.

Discussion
We showed that it is possible to replace the T-domain of an indigoidine synthetase with T-domains of from other NRPS modules to proof that one can in principle also replace the T-domain of a given NRPS module, preserving its function. We propose a method to design synthetic NRPS domains and proof their functionality by introducing them into the indigoidine synthetase indC, thus resulting in a blue phenotype of positive transformants. Moreover, we submitted the RFC [99], describing "high throughput protocols for circular polymerase extension cloning and transformation" (Hi-CT). To further establish the indigoidine synthetase indC as a model for NRPS research we created a library of 58 engineered indC variants – giving insights in how synthetic buiding blocks should be designed, where the domain borders should be set, which domains from respective NRPS pathways and bacterial strains can be used, when creating novel engineered NRPS pathways. Together with our software "NRPS-Designer" and the labelling of engineered NRPS products, we pioneered the research on high-throughput methods for creation of novel NRPS systems, which could lead to a faster development in drug research, recycling, cell signaling and other applications one could think of when creating customized peptides with hundreds of possible monomers.







 "
"









