Team:Heidelberg/Project/Delftibactin
From 2013.igem.org
| Line 245: | Line 245: | ||
<p>We developed a cloning strategy which allowed us to clone all necessary genes encoding the delftibactin-producing non-ribosomal peptide synthetases and polyketide synthetases from the del cluster (about 59 kb in total) and express them in <i>E. coli</i>. In addition, introducing the methylmalonyl-CoA pathway into <i>E. coli</i> provided one of the basic substrates for the del pathway which is endogenously not present in <i>E. coli</i>.</p> | <p>We developed a cloning strategy which allowed us to clone all necessary genes encoding the delftibactin-producing non-ribosomal peptide synthetases and polyketide synthetases from the del cluster (about 59 kb in total) and express them in <i>E. coli</i>. In addition, introducing the methylmalonyl-CoA pathway into <i>E. coli</i> provided one of the basic substrates for the del pathway which is endogenously not present in <i>E. coli</i>.</p> | ||
| - | Therefore, our initial aim was the genomic integration of the genes encoding for the methylmalonyl-CoA pathway into <i>E. coli</i> using the lambda red system established by [Dazenka and wanna].banane This pathway is required for sufficient delftibactin production, as it supplies the substrate methylmalonyl-CoA for DelF, the PKS of the delftibactin cluster. Because this genomic integration turned out to be more challenging than expected a new strategy was developed. Two plasmids were created: pIK2 containing the mm-CoA pathway amplified from <i>Streptomyces coelicolor</i> as well as the PPTase sfp, amplified from <i>Bacillus subtilis</i> in the BioBrick backbone pSB3C5. The permeability device (<a href='http://parts.igem.org/wiki/index.php?title=Part:BBa_I746200'>BBa_I746200</a>) for the outer membrane of <i>E. coli</i> was cloned into another plasmid (pIK1). Since the iGEM Team Cambridge 2007 showed that <a href='http://parts.igem.org/wiki/index.php?title=Part:BBa_I746200'>BBa_I746200</a> is toxic if produced in higher quantities we inserted it into pIK2 between the two terminators driven by a weak promoter ( | + | Therefore, our initial aim was the genomic integration of the genes encoding for the methylmalonyl-CoA pathway into <i>E. coli</i> using the lambda red system established by [Dazenka and wanna].banane This pathway is required for sufficient delftibactin production, as it supplies the substrate methylmalonyl-CoA for DelF, the PKS of the delftibactin cluster. Because this genomic integration turned out to be more challenging than expected a new strategy was developed. Two plasmids were created: pIK2 containing the mm-CoA pathway amplified from <i>Streptomyces coelicolor</i> as well as the PPTase sfp, amplified from <i>Bacillus subtilis</i> in the BioBrick backbone pSB3C5. The permeability device (<a href='http://parts.igem.org/wiki/index.php?title=Part:BBa_I746200'>BBa_I746200</a>) for the outer membrane of <i>E. coli</i> was cloned into another plasmid (pIK1). Since the iGEM Team Cambridge 2007 showed that <a href='http://parts.igem.org/wiki/index.php?title=Part:BBa_I746200'>BBa_I746200</a> is toxic if produced in higher quantities we inserted it into pIK2 between the two terminators driven by a weak promoter (<a href="http://parts.igem.org/Part:BBa_J23114">BBa_J23114</a>) and a weak RBS (<a href="http://parts.igem.org/Part:BBa_B0030">BBa_B0030</a>), resulting in the final plasmids pIK8 with a total size of 9,467 bp, which was transformed into the <i>E.coli</i> strains DH10ß and BL21 DE3 via electroporation. |
Revision as of 10:07, 28 October 2013


Gold Recycling. Using Delftibactin to Recycle Gold from Electronic Waste.
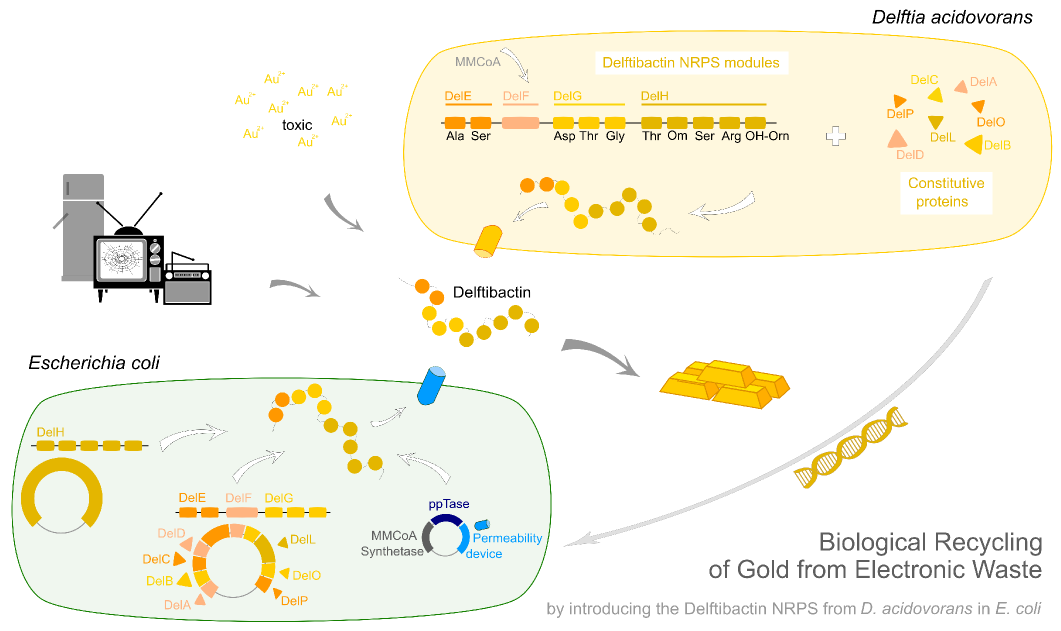
Highlights
- Production and purification of delftibactin, a gold-precipitating NRP, from its native, cultured host Delftia acidovorans.
- Recovery of pure gold from electronic waste by Delftia acidovorans and purified delftibactin.
- Optimization of the Gibson assembly method for the creation of large plasmids (> 30 kbp) with high GC content.
- Amplification and cloning of all components required for recombinant delftibactin production.
- Transfer of the entire pathway from D. acidovorans for the synthesis of delftibactin to E. coli.
Abstract
Efficient recycling of gold from electronic waste using delftibactin
Undoubtedly, gold is one of the most precious materials on earth. Besides its common use in art and jewelry, gold is also an essential component of our modern computers and cell-phones. Due to the fast turn-over of today’s high-tech equipment, millions of tons of electronic waste accumulate each year containing tons of this valuable metal. The main approach nowadays to recycle gold from electronic waste is by electrolysis. Unfortunately, this is a highly inefficient and expensive procedure, preventing most of the gold from being recovered.
Earlier this year, a publication in Nature Chemical Biology reported the existence of a non-ribosomal peptide – delftibactin - which has the astonishing property to specifically precipitate elemental gold from gold-ion containing solutions. Naturally, delftibactin is produced by Delftia acidovorans, an extremophile bacterium, which secretes delftibactin to complexate and dispose of toxic gold ions present in its environment. Although the exact delftibactin production pathway is not known, bioinformatic predictions claim a non-ribosomal peptide synthesis pathway encoded on a large, 59 kb gene cluster (the del-cluster) to be responsible for delftibactin production.
In this subproject we want to demonstrate that the natural secondary metabolite delftibactin can be efficiently produced in E. coli and used for the recycling of gold from electronic waste. To this end, we developed a cloning strategy based on an optimized Gibson Assembly protocol, enabling the cloning of large, GC-rich genomic regions onto regular low-copy plasmids. We thereby engineered three different plasmids (about 70 kb in total size) enabling the expression of the predicted del-cluster from regular E. coli promoters along with the methylmalonyl-CoA pathway providing the basic delftibactin building blocks and a NRPS activating PPTase, Sfp from Bacillus subtilis.
We want to show that these large constructs can be potentially inserted and expressed by E. coli with the promising perspective that delftibactin could readily be used as an efficient way of gold recycling from electronic waste.
Introduction
The quest for a magical substance to generate gold from inferior metals stirred the imagination of generations. However, this substance, the Philosopher’s Stone, stands for more than just the desire to produce gold. There was a time when the fabled Philosopher’s Stone also represented wisdom, rejuvenation and health. Nowadays, gold is still of great importance for us as it is needed for most of our electronic devices.
In 2007, more than two tons of gold, worth $92 million, were discarded hidden in electronic waste in Germany Quellebanane. Most of the precious element ends up on waste disposal sites as only a minor fraction of 10-15% [Gesine Kauffmann, Elektroschrott – die neue Schürfstelle für Gold, 08.12.11, welt, http://www.welt.de/wissenschaft/umwelt/article13755992/Elektroschrott-die-neue-Schuerfstelle-fuer-Gold.html]banane of the gold is recycled also due to the small amounts per device. Since our planet’s gold supplies are limited, the metal is more and more depleted and the value of gold continuously reaches all-time highs. In order to satisfy our society’s need for gold, we have to develop heavy mining techniques involving strong acids, causing devastating impact on humans and environment [1] [2] [3].
Besides economical usage of the resource gold, one way to reduce global demands for gold is elevation of gold recovery [4]. Intriguingly, nature itself offers a structure that has been reported to efficiently extract pure gold from solutions containing gold ions. This fascinating molecule is called delftibactin and is in fact a small peptide secreted by a metal-tolerant bacterium called Delftia acidovorans Quelle.banane
This extremophile has the incredible ability to withstand toxic amounts of gold ions in contaminated soil. If one could culture these bacteria and produce delftibactin in large scales, could one potentially recover gold from electronic waste in a cost- and energy-efficient way? But what is the special feature of delfibactin to precipitate gold that efficiently?
Delftibactin is a non-ribosomal peptide (NRP) [5] [6]. The efficient and non-pollutant large-scale production of this NRP in E. coli could revolutionize the recovery of gold from electronic waste and additionally highlight the plethora of versatile applications for non-ribosomal peptide synthetases (NRPSs). The most striking feature of these non-ribosomal synthetases is their ability to incorporate far more than the 21 common amino acids into peptides. They make use of numerous modified and even non-proteinogenic amino acids to assemble peptides of diverse functions [7].
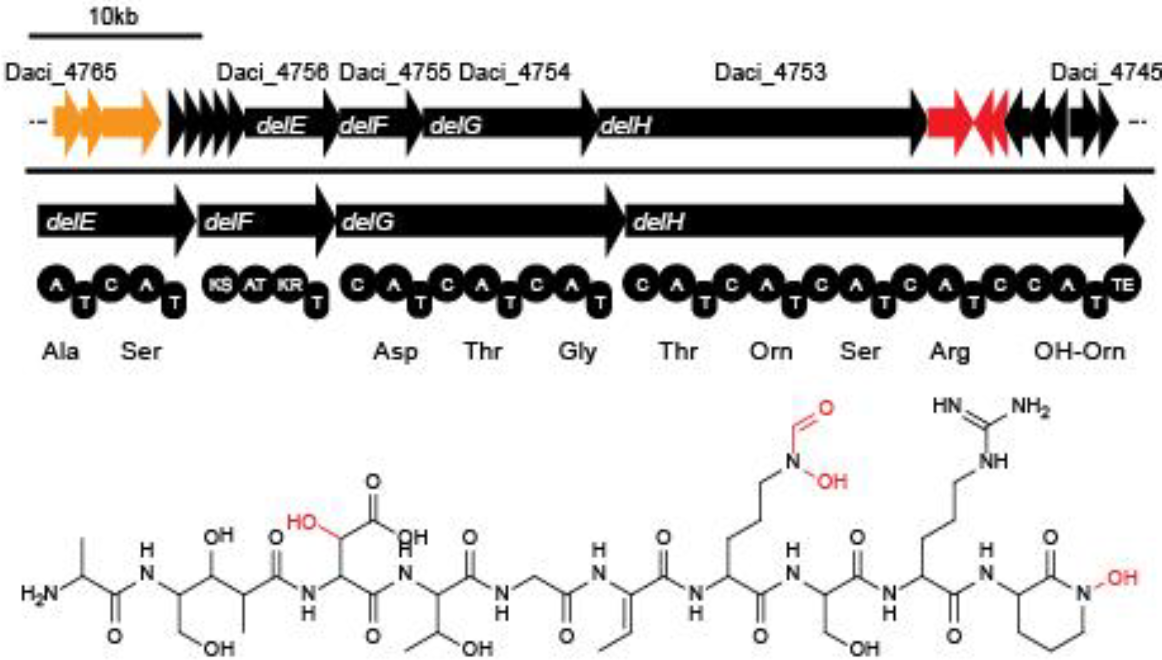
Delftibactin is a NRP produced by a hybrid NRPS/polyketide synthase (PKS) system. In their recent publication, Johnston and colleagues [5] predicted that the enzymes responsible for producing delftibactin are encoded on a single gene cluster, hereafter referred to as del cluster. It comprises 59 kbp encoding for 21 genes. DelE, DelF, DelG and DelH constitute the hybrid NRPS/ PKS system producing delftibactin, with DelE, DelG and DelH comprising the NRPS and DelF the PKS. The remaining enzymes involved in the delftibactin synthesis pathway are required for maturation or post-synthesis modification of delftibactin. The predicted activities [5] of the assumed proteins are:
- DelA: MbtH-like protein, most likely required for efficient delftibactin synthesis [8]
- DelB: thioesterase
- DelC: 4’-phosphopanteinyl transferase: required for maturation of ACP/PCP subunits
- DelD: taurine dioxygenase
- DelL: Ornithine N-monooxygenase
- DelP: N5-hydroxyornithine formyltransferase
We introduced the large del cluster into the commonly used, easy-to-culture model organism E. coli with the aim of recombinant delftibactin expression. Although the del cluster contains the native PPTase of D. Acidovorans we additionally introduced the sfp phosphopanteinyl transferase from Bacillus subtilis as this PPTase is able to activate a wide variety of PKSs including those from Saccharomyces cerevisiae. Importantly, it has been proven to work in E. coli by the indigoidine project of our own team [9]. Additionally, DelF, the polyketide synthetase of the del cluster requires methylmalonyl-CoA as substrate. This metabolite, from now on abbreviated as mmCoA is not produced by E. coli. Therefore, we also transfer the mmCoA synthesis pathway from B. subtilis into E. Coli. This should allow for efficient production of recombinant delftibactin.
As the del cluster starts with Daci_4760 (DelA; Daci IDs are NCBI Gene gene symbols, Del* gene names as referred to in [10]) and promoters within the Del-cluster were bioinformatically predicted upstream of Daci_4750 (DelK), Daci_4760 (DelA) and Daci_4746 (DelO) we assumed that the entire sequence from Daci_4760 (DelA) to Daci_4753 (DelH) is transcribed as a single polycistronic mRNA of approximately 40 kbp in size [10].
Facing these challenges, we decided to approach the project by cultivation of D. acidovorans and the isolation of native delftibactin to reproduce the findings of Johnston and colleagues [5].
.In order to achieve recombinant expression of delftibactin, we decided to introduce constructs coding for the delftibactin-cluster [File:Del cluster.gb] the methylmalonyl-CoA pathway and the PPTase sfp. In addition, we transformed the permeability device BBa_I746200 from the parts registry for the export of recombinant delftibactin out of the target organism E. coli. The desired genes from the del cluster were subdivided onto two different plasmids in order to decrease plasmid size and thereby avoid the intricacies expected for cloning of a single 59 kbp plasmid as well as to allow for faster trouble shooting in case issues with the cloning of particular genes occur:
- Plasmid 1: methylmalonyl-CoA pathway, PPTase sfp & permeability device BBa_I746200, transcription regulated by inducible lac promoter, chloramphenicol resistance;
- Plasmid 2: DelH, transcription regulated by inducible lac promoter, ampicillin resistance;
- Plasmid 3: DelA-P, genes of the del cluster required for production of delftibactin, transcription regulated by inducible lac promoter, kanamycin resistance.
Here we show successful amplification, cloning and transformation of plasmids above 30 kbp in size as well as expression of the desired genes of the del cluster from its natural host D. Acidovorans. Furthermore, we demonstrate efficient recovery of purified gold from electronic waste using the non-ribosomal peptide delftibactin. Additionally, we proved toxicity of DelH for our target organism E. coli when expressed as the only gene of the del cluster: cloning of DelH into a construct without promoter lead to depletion of DelH from our target system which previously had been selecting for mutated versions of DelH.

Results
Efficient Recycling of Gold from Electronic Waste Using Endogenously-Derived Delftibactin
As a first step into the direction of an environmentally friendly procedure for recycling gold from gold-containing waste, we wanted to show that the non-ribosomal peptide delftibactin can be used to precipitate gold from gold ion-containing solutions.
We obtained D. acidovorans DSM-39 from the DSMZ and successfully reproduced the paper by Johnston and colleagues [5]. What they had been able to show was that delftibactin selectively precipitates gold from gold solution. In our experiments, precipitation on agar plates worked even better than described by Johnston et al. (Fig. 1). D. acidovorans is capable to precipitate solid gold from gold chloride solution as purple-black nanoparticles.
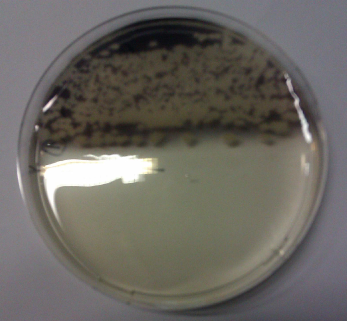
We also showed that another strain, Delftia acidovorans SPH-1, is also able to precipitate gold ions to gold nanoparticles. When using the supernatant of a culture gold nanoparticles were precipitated at an amount which caused a color change to black already at low concentrations of 0.35 µg/ml of gold chloride. With increasing concentration of gold chloride more nanoparticles formed, even though the process became slower above a certain concentration (Fig. 5a). Furthermore, we melted the purple-black nanoparticles to shiny, solid gold as shown in Fig. 5b,c,d.

Next, we established a purification protocol for delftibactin using HP20 resins. Additionally, we proved precipitation of gold by the purified delftibactin (Fig. 6) and detected it by Micro-TOF(Fig.7).


Moreover we were interested in the potential of delftibactin for the recycling of gold from electronic waste. An important prerequisite for efficient gold recycling from electronic waste is the feasibility for gold precipitation from gold solutions of low concentration. Undoubtedly, this is likely the case for most gold solutions derived from electronic waste.Therefore, we used an old, broken CPU and established a protocol for dissolving gold from gold-containing metal waste. We incubated gold covered CPU pins in aqua regia resulting in a gold-ion containing solution (figures 8 to 10). We showed precipitation of dissolved gold recovered from electronic waste by D. acidovorans. Adding this solution to D. acidovorans SPH-1 agar plates resulted in the formation of solid gold nanoparticles.
Taken together, we have successfully established a method enabling the recycling of pure gold from electronic waste using delftibactin produced by D. acidovorans.

Although the recycling was working efficiently in our hands, the approach of using the natural D. acidovorans bacterial strain for delftibactin production on a larger scale has several disadvantages:
Therefore, we wanted to engineer an E. coli strain producing delftibactin in high yields, thereby circumventing the above mentioned limitations.
Expression of Del Cluster Genes in E. coli
We developed a cloning strategy which allowed us to clone all necessary genes encoding the delftibactin-producing non-ribosomal peptide synthetases and polyketide synthetases from the del cluster (about 59 kb in total) and express them in E. coli. In addition, introducing the methylmalonyl-CoA pathway into E. coli provided one of the basic substrates for the del pathway which is endogenously not present in E. coli.
Therefore, our initial aim was the genomic integration of the genes encoding for the methylmalonyl-CoA pathway into E. coli using the lambda red system established by [Dazenka and wanna].banane This pathway is required for sufficient delftibactin production, as it supplies the substrate methylmalonyl-CoA for DelF, the PKS of the delftibactin cluster. Because this genomic integration turned out to be more challenging than expected a new strategy was developed. Two plasmids were created: pIK2 containing the mm-CoA pathway amplified from Streptomyces coelicolor as well as the PPTase sfp, amplified from Bacillus subtilis in the BioBrick backbone pSB3C5. The permeability device (BBa_I746200) for the outer membrane of E. coli was cloned into another plasmid (pIK1). Since the iGEM Team Cambridge 2007 showed that BBa_I746200 is toxic if produced in higher quantities we inserted it into pIK2 between the two terminators driven by a weak promoter (BBa_J23114) and a weak RBS (BBa_B0030), resulting in the final plasmids pIK8 with a total size of 9,467 bp, which was transformed into the E.coli strains DH10ß and BL21 DE3 via electroporation.
Details of our cloning strategy are shown in Fig. 14. Notably, the three plasmids we wanted to assemble are of large size (23, 32 and 10 kbp in size) and cloning was further complicated due to the high GC content and presence of repetitive elements within the del genes.
Gibson Assembly & Transformation
Assembly of plasmids above 30 kb in size composing of multiple fragments is challenging when using conventional restriction enzyme based cloning. Thus, we decided to use Gibson Assembly, a method which was introduced to the iGEM community by Cambridge in iGEM 2010 as a powerful alternative to common cloning procedures. The assembled constructs of up to 32 kbp in size were transformed into E. coli via electroporation. Correct assembly of the fragments was tested by analytical restriction digests. The exemplary restriction digests shown above (Fig. 16) confirmed the correct assembly of the three desired constructs as it displays the expected band pattern. For cloning of the mm-CoA pathway, clones (6 and 9) show the expected restriction pattern (Fig. 16a). Sequencing of the assembly sites of these constructs confirmed the restriction digest results.

Cloning of the DelRest plasmid was validated by different suitable analytic restriction digests (Fig. 16b) and also confirmed by sequencing. The sequence was compared with the available D. acidovorans SPH-1 reference sequence obtained from NCBI (for further information please visit our labjournal). The high quality of the alignment shows that Gibson assembly is a suitable cloning approach for rapid assembly of large NRPS and PKS expression constructs.
Analytical restriction digest of DelH (Fig.16c) also gave rise to a number of positive clones. However, the sequencing results derived from all DelH showed various mutations, which were exclusively located within the region of the first DelH forward primer. Most of these mutations are deletions present in the first 30 bp of the DelH coding region thereby resulting in frameshifts and formation of a stop codon disposing E. Coli of expression of DelH. As there was no clone without mutations, we proceeded with DelH clone C5, as this clone did not have any bp deletion but only harbored a substitution mutation at bp position 28 of the ORF, leading to the conversion of Alanine at position 10 to Threonine. Two representative sequences compared to Delftia acidovorans SPH-1 are listed below (Tab. 1) and display two of the observed mutations in different DelH clones.
Tab.1 DelH 5’ sequence, in which most mutations were observed. The ATG start codon is depicted in bold. The table shows the sequence comparison between the DelH reference strand of D. acidovorans and two different exemplary E. coli clones transformed with the plasmid pHM04 (assembled DelH expression vector). The second line shows the accumulated deletion and the third line shows the clone containing 'only' single base pair substitution. Deletions appeared quite frequently while a substitution was only found in a single clone C5. The substitution changes the corresponding Alanine codon to Threonine.| Organism | Plasmid containing | DNA -Sequence | Conclusion |
|---|---|---|---|
| D. acidovorans | none | ATG GACCGTGGC CGCCTGCGC CAAATCGC | correct |
| E. coli DH10ß | pHM04 | ATG GACCGTG-C CGCCTGCGC CAAATCGC | deletion |
| E. coli DH10ß C5 | pHM04 | ATG GACCGTGGC CGCCTGCGC CAAATCAC | substitution |
Due to these observations, we hypothesize that expression of DelH is toxic for E. coli. Therefore natural selection only leads to survival of those clones which incorporate constructs giving rise to unfunctional, truncated DelH proteins or avoiding any expression. This phenomenon of frequent mutations within the primer binding site was also observed when we started cloning of the permeability device used in the pIK8 construct. The sequenced plasmids displayed a high accumulation of mutations compared to other constructs. In case of the methylmalonyl-CoA plasmid (pIK8), the problem was solved by the usage of a weak promoter and a weak ribosome binding site from the parts registry for driving the expression of the permeability device (see above). Based on this knowledge, DelH is currently being assembled into the desired backbone pSB6A1 containing a weak promoter BBa_J23114 and the ribosome binding site BBa_B0032, also reducing expression. While the new plasmid is constructed, the following experiments were performed with the C5 clone as we hypothesized that DelH C5 bearing the single nucleotide exchange at position 28 might still show expression of functional DelH when transformed into E. coli (the corresponding amino acid exchange is located at the N-terminus).
Expression of the Delftibactin NRPSs & Associated Genes
For further characterization of our constructs, we analyzed expression of the delftibactin NRPS/PKS pathway in E. coli. For expression of DelH and DelRest, SDS-PAGEs were conducted followed by Coomassie staining. Native cells as well as cells transformed with the plasmid backbones obtained from the parts registry containing the used antibiotic resistance markers served as control groups. The proteins DelE, DelG and DelH are significantly larger than any protein that is expressed by our host E. coli. Therefore, the expression of the introduced genes was detectable on the SDS-PAGE (Fig. 17) without specific labeling of the proteins. Although expression of these large proteins was weak, clear distinct bands of the sizes predicted for DelE and DelG were detected. Accordingly a band at the predicted size of DelH for the clone transformed with DelH C5 was visible. As the lac promoter regulating expression of DelE and DelG also drives expression of DelA, DelB, DelC, DelD and DelF, one can conclude simultaneous expression of these del proteins. This is also in accordance with the predicted distribution of promoters within the delfticbatin cluster.

Furthermore, the expression of the PPTase was verified by performing an IndC activity assay established by the indigoidine subproject. The indigoidine synthetase IndC activity is dependent on the presence of a functional PPTase. Upon activation, indC produces the blue pigment indigoidine. Co-transformation of the corresponding plasmid pIK8 construct (enabling PPTase expression) with an IndC indigoidine synthetase expression construct lacking a PPTase expression cassette (pRB22 was conducted. The transformed E. coli displayed inhibited growth and developed the expected blue phenotype. From these results, we conclude that the PPTase on pIK8 is functionally expressed (note: decelerated growth kinetics of E. coli results from the metabolic burden that is caused by the synthesis of the indigoidine).
For proving the production of the permeability device which is needed for export of delftibactin, a zone of inhibition test with bactracin was performed. Bacitracin is an antibiotic not able to pass the bacterial cell wall by passive transport[Sampson, B. A., Misra, R., & Benson, S. A. (1989). Identification and Characterization of a New Gene, (1972)].banane Growth of bacteria containing the permeability device was inhibited upon application of bacitracin confirms the expression of the transporter. Growth of control cells without the device was not impaired by application of bacitracin (Fig. 18).

In conclusion, we successfully expressed the recombinant delftibactin NRPS/ PKS pathway as well as the required methylmalonyl-CoA pathway, the PPTase and permeability device in E. coli.
Furthermore, we showed, that it is not only possible to assemble large plasmids (in sum these were 67 kpb in total size in our case) and transform them into E. coli, but demonstrated successful expression of large NRPS/PKS modules in our host strain.
Discussion and outlook
Motivation
Delftibactin is a secondary metabolite naturally produced by Delftia acidovorans and has the ability to specifically precipitate gold from solutions. Today, electronic waste has become an immense environmental problem not only in developed nations, but also third-world countries through the increasing export of waste and disposal challenges.
Therefore, an efficient method to recycle electronic waste is urgently needed. From our work we concluded, that delftibactin could be used as an efficient substance to recycle gold. In fact, many different electronic scraps contain considerable gold amounts such as PC mainboards (566 ppm in terms of weight) and mobile phones (350 ppm). And those electronic devices are more and more demanded by society though their average lifetime seems to decrease steadily. To date, existing methods for gold recycling and safe disposal of electronic waste are still very energy-consuming and harmful to the environment.
Nowadays, harsh substances, such as strong acids, are conventionally used for leaching gold from electronic waste. Applied chemicals pose a potential thread to the environment as they could contaminate the biosphere. Biological agents could become an alternative to chemical clearance of metals from electronic waste. Nature's gold-altering microbes are non-pollutive and they are not even genetically modified. While bacteria such as Cupriavidus metallidurans convert the dissolved element to its metallic form inside the cell [11], species like Chromobacterium violaceum [4] or Delftia acidovorans [5] secrete substances to the surrounding medium for gold precipitation. These microorganisms can therefore be engineered to increase the yield of bioleaching substances.
Achievements
In our experiments, we successfully managed to dissolve electronic waste in aqua regia (i.e. nitro-hydrochloric acid) and neutralize the solution to receive gold chloride. When we added the supernatant of a D. acidovorans liquid culture to the gold chloride solution, pure gold was precipitated. Furthermore, we were able to melt the precipitated gold resulting in little gold flakes. The procedure already worked on a small scale in our project. With increased efforts, yields of Delftibactine for industrial applications are conceivable. In those dimensions, gold could easily be recycled. With Delftibactin there is no further need for chemical reducing reagents to purify gold from solution.
Nevertheless, the efficiency of our approach still has to be improved. We were able to apply Delftibactin for the extraction of gold from electronic waste but still had to expose the CPUs with aqua regis to bring gold in solution. Ideally, one should get rid of the use of this highly corrosive mixture.
To further increase the efficiency of Delftibactin production, we aimed at transferring the entire synthesis machinery for Delftibactin into E. coli. As the NRPS producing delftibactin is a very large enzyme complex consisting of many modules, amplification, cloning and transformation of the constructs was very challenging. Nonetheless, we successfully managed to amplify, assemble and transform all of the genes (in sum 54 kbp) required for production of Delftibactin. In this process, we established protocols, including Gibson assembly of large fragments, and transformation of those constructs into E. coli via electroporation. Optimized procedures will ease the cloning of large customized NRPSs in future.
Challenges
A hurdle to overcome is that the DelH displayed an above-average mutation rate when present in E. coli without the rest of the Delftibactin pathway genes. DelH could potentially be toxic to the E. coli. Therefore, cells select for mutations that render the clones unable to express a funtional DelH protein [12]. In our experiments, the putative selection pressure towards nonfunctional DelH was manifested in deletions, insertions or substitutions in the respective ribsosome binding side or at the beginning of DelH. However, we managed to obtain a single clone, which only possessed a point mutation that potentially does not interfere with protein expression since no false stop codon was created and no frame shift was caused. In this clone, DelH was successfully expressed, yet with an amino acid exchange (Alanine to Threonine) at the N terminus of the protein. At the moment, we are aiming to obtain the gene without any mutation by lowering protein expression. To this end, the most-promising approach is to design a DelH construct containing a weak promoter and weak ribosome binding side. This would decrease the selection pressure caused by DelH. An alternative strategy would be the construction of DelH without any promoter or ribosome binding site. As soon as we would get a mutation-free DelH fragment,,it could be ligated into a plasmid backbone via conventional restriction enzyme-based cloning. A weak promoter and ribosome binding side would keep the detrimental impact of DelH as low as possible. Despite the point mutation, we were able to show that DelH is expressed in E. coli. Our prediction of the tertiary structure of DelH indicated that the.substituted amino acid in the mutant DelH is located at the outer side of the protein. Nevertheless it could still have an important function. Therefore it is not possible to foresee whether the mutation would have any effect on the function of DelH. However alanine is an unpolar amino acid whereas threonine is polar. If the mutation was in a functional region, this could have significant effects rendering DelH nonfunctional but keeping E. coli alive.

1. Eisler R, Wiemeyer SN (2004) Cyanide hazards to plants and animals from gold mining and related water issues. Reviews of environmental contamination and toxicology: 21–54.
2. Eisler R (2004) Arsenic hazards to humans, plants, and animals from gold mining. In:. Reviews of environmental contamination and toxicology. Springer. pp. 133–165.
3. Donato DB, Nichols O, Possingham H, Moore M, Ricci PF, et al. (2007) A critical review of the effects of gold cyanide-bearing tailings solutions on wildlife. Environment international 33: 974–984.
4. Tay SB, Natarajan G, bin Abdul Rahim MN, Tan HT, Chung MCM, et al. (2013) Enhancing gold recovery from electronic waste via lixiviant metabolic engineering in Chromobacterium violaceum. Scientific reports 3.
5. Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, et al. (2013) Gold biomineralization by a metallophore from a gold-associated microbe. Nature chemical biology 9: 241–243.
6. Strieker M, Tanović A, Marahiel MA (2010) Nonribosomal peptide synthetases: structures and dynamics. Current opinion in structural biology 20: 234–240.
7. Caboche S, Leclère V, Pupin M, Kucherov G, Jacques P (2010) Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. Journal of bacteriology 192: 5143–5150.
8. Baltz RH (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. Journal of industrial microbiology & biotechnology 38: 1747–1760.
9. Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, et al. (1998) Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37: 1585–1595.
10. de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J (2012) PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC genomics 13: 299.
11. Reith F, Etschmann B, Grosse C, Moors H, Benotmane MA, et al. (2009) Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proceedings of the National Academy of Sciences 106: 17757–17762.
12. Cárcamo J, Ravera MW, Brissette R, Dedova O, Beasley JR, et al. (1998) Unexpected frameshifts from gene to expressed protein in a phage-displayed peptide library. Proceedings of the National Academy of Sciences 95: 11146–11151.
 "
"















