Team:Heidelberg/Project/Indigoidine
From 2013.igem.org


Indigoidine. Proving Modularity of NRPS by Shuffling Domains.
Abstract
An integral characteristic of synthetic biology yet often undermined is the ability to learn fundamental knowledge by systematically perturbing a biological system. Non-ribosomal peptide synthetases (NRPS) are predestinated for such a trial and error approach. Their hierarchical organization into modules and domains offer a unique opportunity to spin around their inherent logical assembly and observe if their functionality is preserved. Following this idea, we prove the interchangeability of NRPS domains at the example of indC from Photorhabdus luminescens laumondii TT01 (DSM15139). The native NRPS domains have been replaced with domains from other bacterial organisms and fully synthetic domains. To quantify the NRPS efficiency we established an indigoidine assay based on OD measurement of the blue-colored pigment. Interestingly, we find that our data points out the dependence on the T-domain and the 4'-Phosphopanthetheinyl-transferases (PPTases), resulting in different levels of indigoidine synthesis. Furthermore, we introduce HiCT - High throughput protocols for circular polymerase extension Cloning and Transformation - a new standard for the assembly of combinatorial gene libraries (RFC 99).
Introduction
Most modules of non-ribosomal peptide synthetase (NRPS) pathways consist of three domain types: condensation, adenylation and thiolation domain (see Figure 1a), also called peptidyl-carrier-protein domain (PCP)-domain (reviewed in

Besides these fundamental domains (C-domain, A-domain and T-domain), some NRPS modules incorporate additional domains enlarging the amount of potential catalytic reactions, such as cyclization, epimerization or oxidation of the amino acid [Reference].
For example, a single module of P. luminescens laumondii TT01 (DSM15139) contains an internal oxidation domain (Ox-domain) in its A-domain and a special TE-domain (Figure 2a). This enzyme first adenylates L-glutamine (A-domain), which is then attached to the T-domain. The TE-domain cleaves and catalyzes the cyclization of the substrate, which is further oxidized by the Ox-domain. The oxidation of two cyclic glutamines results in the formation of an insoluble small molecule (Figure 2b)[Reference].

Results
Expression of functional indigoidine synthetase indC derived from ''P. lumninescens'' in five substrains of ''E. coli''
The open reading frame of the native indigoidine synthetase indC was amplified from genomic DNA of ''P. lumninescens'' and cloned into a plasmid under the control of an lac-inducible promoter. This indC expression cassette was transformed into different substrains of ''E. coli'', namely DH5alpha, MG1655, BAP1, TOP10 and NEB Turbo. All of these host strains express the endogenous PPTAse entD which is responsible for the transfer of the 4'-phosphopantetheine residue from coenzyme A to the apo-domain of EntF, a T-domain in the enterobactin pathway(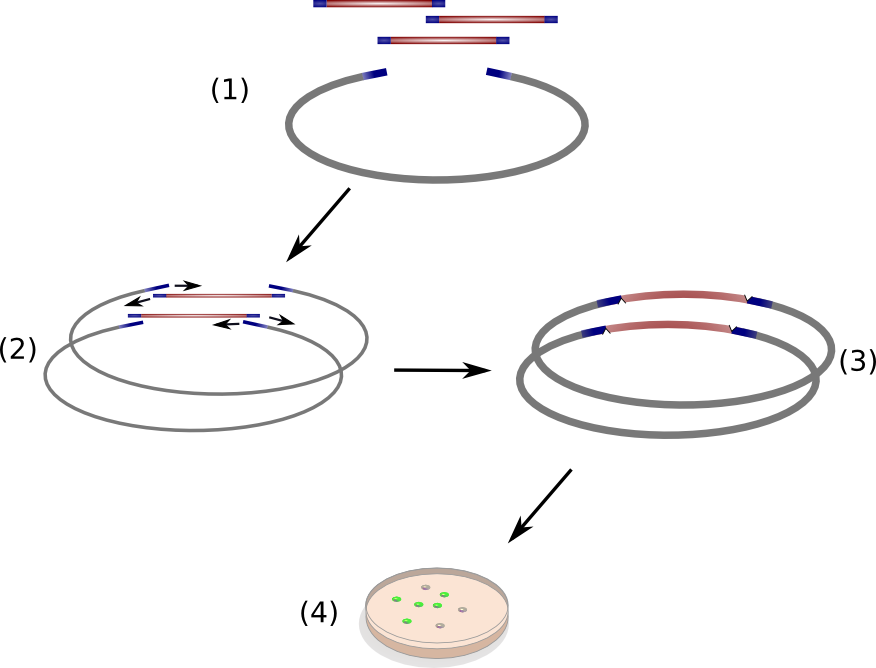
Improved Production of Indigoidine by Co-transformation of Host Strain with Supplementary PPTases
The expression of indC under activation by endC was sufficient for easy detection of indigoidine production on plates harboring indC-carrying cells. In order to determine, whether the amount of indigoidine production in the ''E. coli''TOP10 cells is dependent on the quality of the interacting of indC with the PPTase, four PPTase dervied from varying origins were selected and amplified from the genome of the hosts of origin. ''E. coli''TOP10 cells were co-transformed with plasmids coding for the different PPTases and the plasmid containing the expression cassette for indC. As reference for the endogenous PPTase activity served cells only transformed with the indC plasmid. Irrespective of the PPTase, growth of colonies was retarded. Remarkably however, colonies co-transformed with the PPTase plasmid remained of smaller size than the ones only carrying the indC construct. On the other side, indigoidine production was more diffuse in the latter cells with secretion of the blue pigment into the agar (Figure 6b, indC) and only slight blue-greenish coloring of the colonies. The four PPTases additionally introduced into the TOP10 cells were all shown to be functional (blue phenotype of the transformants, Figure 6b), but lead to the retention of most of the indigoidine within the cells. Colonies of cells transformed with thess constructs, were of convex shape and of distinct, dark blue color. Overall, cells carrying an additional PPTase showed increased indigoidine production compared to the cells relying on the endogenous entD.Synthetic T-Domains Generated by Consensus and Guided Random Design Method are Functional
The main structurel characteristic of NRPSs is their modular composition on different levels. The indogoidine synthetase indC is a one-module NRPS comprised of the three domains, namely AoxA, T and TE. Since the functionality of this NRPS is detectable by the bare eye, it offers a perfect and simple experimental set-up for proof of principle experiments regarding the interchangeability of domains from different NRPS. We choose a two-pronged domain shuffling approach: First, we transferred native T-domains derived from either different host species and/or NRPSs of entirely different function into the indC indigoidine synthetase. Second, we deviced three methods for the generation of synthetic T-domains based on different NRPS libraries generated by BLAST search against either specific subranges of host organisms or restricting the query sequence to be BLASTed.(link) For the libraries, where the query sequence was restricted to the T-domain of indC, prior knowledge about the T-domain boundaries was required. We used PFAM, a web-tool which allows -amongst other functions- for the prediction of NRPS's module and domain boundaries, to define the T-domain of indC. As depicted in Figure 7, both approaches lead principally to fully functional indCs. The synthetic T-domains 1, 3 and 4 showed the same retarded growth and indigoidin production on plates as did the native T-domain derived from ''P. lumninescens''. The colonies obtained after co-transformation with supplementary PPTase plasmid were small in size and of dark blue color. Compared to synthetic T-domain 5, indigoidin production started earlier (approximately after 24-30 hours). In contrast to the synthetic domains 1,3 and 4 which were designed by the consensus method and showed medium to high similarity to the sequence of origin, synthetic domain 5 was generated by the guided random method. Remarkably, even though 39 out of the 62 amino acids of the original T-domain were exchange, the indigoidine synthetase with this T-domain was still functional. Closer analysis of the sequence compared to the original indC T-domain sequence showed, that the characteristics of the amino acid sequence, i. e. for instance polar or charged amino acids, were retained in 72% of the sequence. Also, the GGXS core sequence of the T-domain at which the activation by the PPTas occurs was conserved.Domain Shuffling Works across Modules Derived from different Pathways and host organisms
Multiple web-tools exist which offer the prediction of NRPS module and domain boundaries. One of the most common used prediction tools is PFAM which we used as a starting point to determine the best method for defining domain boundaries. PFAM predicted large linker structures between the end of the A and the beginning of the T-domain. Using these domain boundaries for the native T-domains did only yield one functional native T-domain. We tried to improve this yield by defining new T-domain boundaries based on the predictions of PFAM and multiple sequence alingments with the respective homology libraris at the predicted linker regions. Boundaries were set closer to the preceeding A-domain, at regions were less sequence conservation was observed. Figure 9 depicts the success of this strategy as two additional native T-domains derived from delH4 and bpsA (indigoidine synthetase) led to functional indCs and the production of indigoidine. In addition, the native T-domain from plu2642 which was already shown to be functional(compare Figure 7) showed faster and increased indigoidine production (deep blue agar plate, lower right panel on Figure 9). The results obtained from this experiments proofed two concepts. First, domain shuffling is possible across different species as the T-domains of delH4 and bpsA were derived from ''D. acidovorans'' and ''S. lavendulae lavendulae'', respectively and were functional in ''E. coli''. Also, shuffling of domains from modules of different substrate specificity has been proofen herein. Second, manually adjusting the boundaries predicted by PFAM based on MSA is a functional method to predict functional T-domain boundaries.Discussion
In this subproject, we wanted to set the basis for engineering entirely synthetic NRPS modules composed of user-defined domains. As model system, we used the unimodular indigoidine synthetase NRPS from P. luminescens subsp. Laumondii TT01. We predicted the modular composition and domain borders of IndC using our own NRPS-Designer software. We then started by replacing the native IndC T-domain with T-domains derived from different NRPS pathways from different bacterial strains, among those the T-domain from the BpsA indigoidine synthetase from S. lavendulae ATCC11924. Constructs were transformed into E. coli alongside with an PPTase expression cassette in order to screen for functional IndC variants. As hoped, a subset of the natural T-domains were functioning in context of the IndC scaffold module, leading to indigoidine production and thereby blue coloring of colonies and corresponding liquid cultures. We then engineered a variety of synthetic T-domains derived from consensus sequences of different natural T-domains. Again, a subset of these T-domains successfully maintained indigoidine production (Fig. 9). Notably, one of our engineered IndC construcats showed an indigoidine production even higher compared to the wild-type IndC (T-domain Plu2642; Figure 3). This is particularly remarkably as our results contradict to previous studies of NRPS domains that reported the native T-domain of the indigoidine synthetase BpsA to be absolutely essential for protein function (and therefore not replaceable by other T-domains). ([Owen2012]). However, to our surprise, the BpsA T-domain-containing IndC construct did not yield any detectable indigoidine production, although BpsA shares strong sequence homology with IndC. We hypothesized, that the selection of the exact border could be critical for maintaining domain functionality when introduced into a novel NRPS module scaffold. Therefore, we amplified different BpsA T-domain variants differing in their domain border and introduced them into the IndC scaffold. Remarkably, a subset of the resulting IndC variants showed successful indigoidine production. We thus revaluated all native and synthetic T-domains in light of this finding and performed a second screening round in which we were able to rescue even more functional IndC variants, proofing our abovementioned hypothesis. We also co-transformed all engineered IndC construct bearing different natural and synthetic T-domains with four different PPTase expression constructs. To our surprise, the T-domains used not only determined general efficiency of indigoidine production, but also the efficiency of NRPS activation by the different activating PPTases. In conclusion we were able to demonstrate, that it is indeed possible to replace single Domains from NRPS modules, while preserving or even enhancing its functionality. In addition, we established an approach for the design of synthetic T-domains and proved their functionality by introducing them into the indigoidine synthetase indC scaffold. Moreover, we established a high throughput protocol for circular polymerase extension cloning and transformation (Hi-CT) (BBF RFC 99), which we applied for our domain shuffling approach. In summary, we created a library of 58 engineered indC variants. In addition we perforemd measurement of blue pigement production over time, which gave us novel insights in how NRPS domains should be designed, where the domain borders between different domains in a single NRPS module have to be set and which domains from respective NRPS pathways and bacterial strains can be used, when creating novel engineered NRPS pathways. We implemented our findings into the "NRPS-Designer" Software, so that the underlying algorithm for NRPS design takes into consideration the abovementioned findings (e.g. domain border setting) which are certainly crucial for successful in silico prediction of functional NRPSs. Thereby, our project pioneers the research on high-throughput methods for creation of synthetic NRPS modules composed of user-defined domains. We believe that our findings will highly contribute to future development of custom NRPSs.





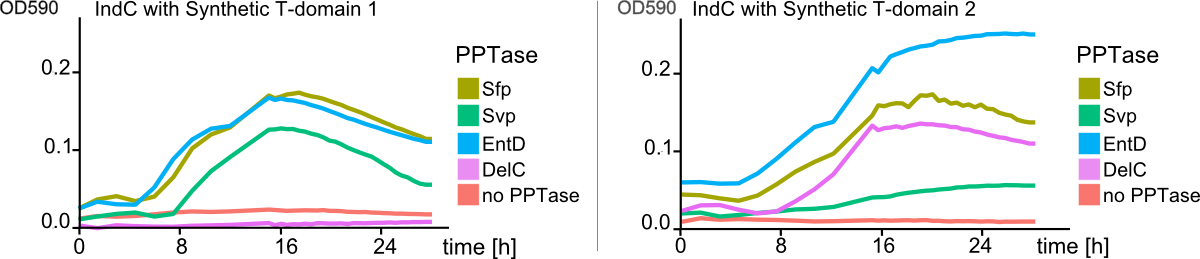


 "
"


















