Team:Heidelberg/Delftibactin/MMCoA
From 2013.igem.org


Methylmalonyl-CoA Pathway. Making the whole thing work.
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet.
Week 6
This week the project DelRest was launched. In order to clone the Delftibactin cluster from D. Acidovorans into E.coli we decided to use Gibson cloning. Therefore Gibson primers were designed to amplify our target backbone pSB4K5 with an overlap to DelA. Furthermore the Gibson primer connecting DelOP and DelAG introduces the ribosome binding site BBa_BNILS. Accordingly the Gibson primer for joining DelL with DelOP includes the ribosome binding site BBa_BNILS. The very last gibson primer used to amplify DelL consequently has an overlap to the beginning of the mRFP reporter gene connected to pSB4K5. Check out our vectormap if you are curious about the detailed primer design. The goal of this week was to amplify our previously assembled backbone with the intended overlaps as well as the desired genes from our host organism D. Acidovorans .
Week 7
This week the project DelRest was launched. In order to clone the Delftibactin cluster from D. Acidovorans into E.coli we decided to use Gibson cloning. Therefore Gibson primers were designed to amplify our target backbone pSB4K5 with an overlap to DelA. Furthermore the Gibson primer connecting DelOP and DelAG introduces the ribosome binding site BBa_BNILS. Accordingly the Gibson primer for joining DelL with DelOP includes the ribosome binding site BBa_BNILS. The very last gibson primer used to amplify DelL consequently has an overlap to the beginning of the mRFP reporter gene connected to pSB4K5. Check out our vectormap if you are curious about the detailed primer design. The goal of this week was to amplify our previously assembled backbone with the intended overlaps as well as the desired genes from our host organism D. Acidovorans .
Week 8
This week the project DelRest was launched. In order to clone the Delftibactin cluster from D. Acidovorans into E.coli we decided to use Gibson cloning. Therefore Gibson primers were designed to amplify our target backbone pSB4K5 with an overlap to DelA. Furthermore the Gibson primer connecting DelOP and DelAG introduces the ribosome binding site BBa_BNILS. Accordingly the Gibson primer for joining DelL with DelOP includes the ribosome binding site BBa_BNILS. The very last gibson primer used to amplify DelL consequently has an overlap to the beginning of the mRFP reporter gene connected to pSB4K5. Check out our vectormap if you are curious about the detailed primer design. The goal of this week was to amplify our previously assembled backbone with the intended overlaps as well as the desired genes from our host organism D. Acidovorans .
Week 9
This week the project DelRest was launched. In order to clone the Delftibactin cluster from D. Acidovorans into E.coli we decided to use Gibson cloning. Therefore Gibson primers were designed to amplify our target backbone pSB4K5 with an overlap to DelA. Furthermore the Gibson primer connecting DelOP and DelAG introduces the ribosome binding site BBa_BNILS. Accordingly the Gibson primer for joining DelL with DelOP includes the ribosome binding site BBa_BNILS. The very last gibson primer used to amplify DelL consequently has an overlap to the beginning of the mRFP reporter gene connected to pSB4K5. Check out our vectormap if you are curious about the detailed primer design. The goal of this week was to amplify our previously assembled backbone with the intended overlaps as well as the desired genes from our host organism D. Acidovorans .
Week 10
This week the project DelRest was launched. In order to clone the Delftibactin cluster from D. Acidovorans into E.coli we decided to use Gibson cloning. Therefore Gibson primers were designed to amplify our target backbone pSB4K5 with an overlap to DelA. Furthermore the Gibson primer connecting DelOP and DelAG introduces the ribosome binding site BBa_BNILS. Accordingly the Gibson primer for joining DelL with DelOP includes the ribosome binding site BBa_BNILS. The very last gibson primer used to amplify DelL consequently has an overlap to the beginning of the mRFP reporter gene connected to pSB4K5. Check out our vectormap if you are curious about the detailed primer design. The goal of this week was to amplify our previously assembled backbone with the intended overlaps as well as the desired genes from our host organism D. Acidovorans .
Week 11
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
At the beginning of the week, we could verify that the Gibson Assembly for Tripeptide I was indeed positive, however, the other Gibson Assemblies did not work properly. Instead of picking new colonies, we decided to optimize the Gibson recipe instead, as backbone religations were the most common problem. With these improved protocols, we used Gibson Assembly for the Dipeptide, Tripeptide II and Tetrapeptide I, later that week, Tetrapeptide II followed. After the Transformation to DH10β cells and screening by restriction digest we could send samples for the Dipeptide and Tetrapeptide I to sequencing and obtained a positive alignment. Hence we transformed BAP I cells with the positive constructs. The same was...
Week 12
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
At the beginning of the week, we could verify that the Gibson Assembly for Tripeptide I was indeed positive, however, the other Gibson Assemblies did not work properly. Instead of picking new colonies, we decided to optimize the Gibson recipe instead, as backbone religations were the most common problem. With these improved protocols, we used Gibson Assembly for the Dipeptide, Tripeptide II and Tetrapeptide I, later that week, Tetrapeptide II followed. After the Transformation to DH10β cells and screening by restriction digest we could send samples for the Dipeptide and Tetrapeptide I to sequencing and obtained a positive alignment. Hence we transformed BAP I cells with the positive constructs. The same was...
Week 13
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 14
At the beginning of the week, we could verify that the Gibson Assembly for Tripeptide I was indeed positive, however, the other Gibson Assemblies did not work properly. Instead of picking new colonies, we decided to optimize the Gibson recipe instead, as backbone religations were the most common problem. With these improved protocols, we used Gibson Assembly for the Dipeptide, Tripeptide II and Tetrapeptide I, later that week, Tetrapeptide II followed.
Week 15
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 16
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 17
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 18
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 19
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 20
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 21
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 22
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Week 23
Cras justo odio, dapibus ac facilisis in, egestas eget quam. Donec id elit non mi porta gravida at eget metus. Nullam id dolor id nibh ultricies vehicula ut id elit.
Methods:
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet.Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. .
Goal
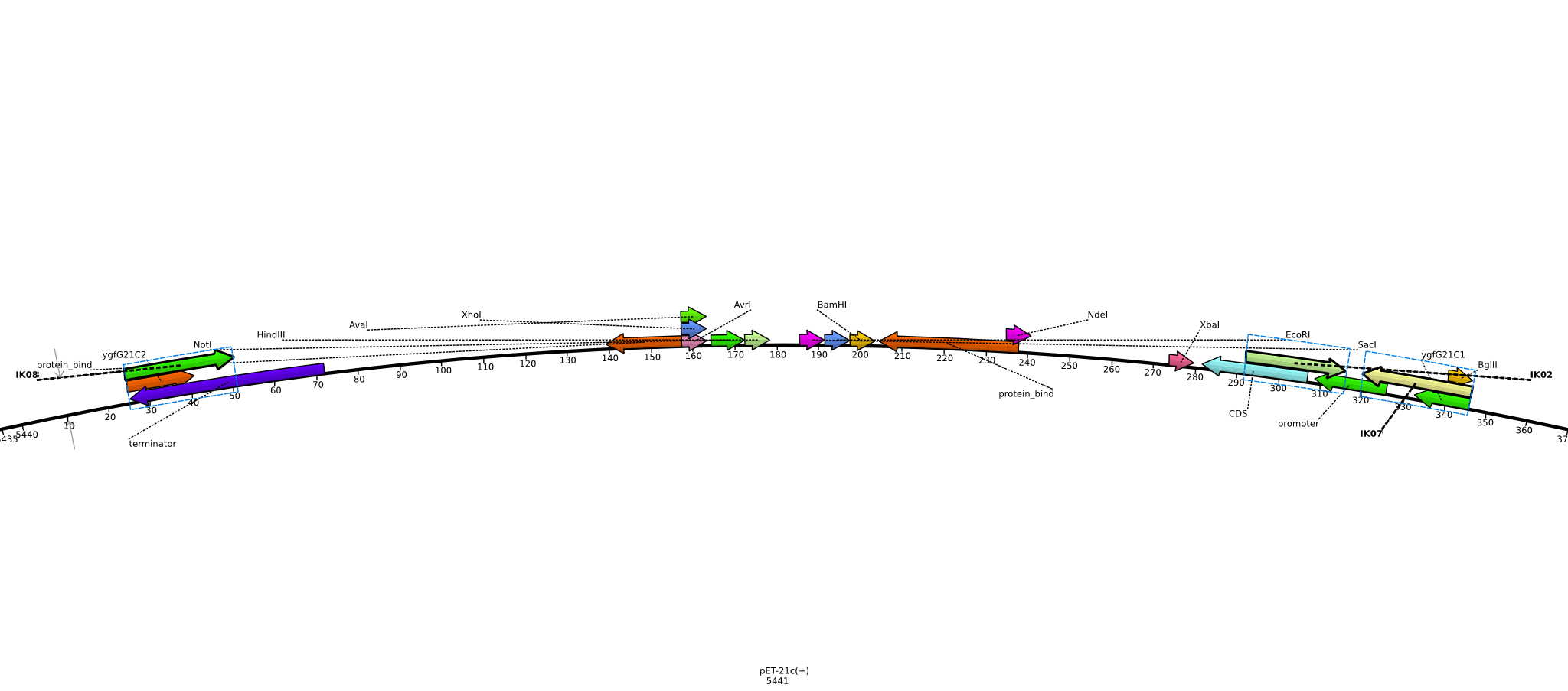
Integration of the methylmalonyl-CoA sythesis pathway into the BAP1 genome. This is a reproduction of a part of <bib id="pmid17959404"/>. For the integration, the λ-red protocol established by Datsenko and Wanner<bib id="pmid10829079"/> is used. pKD20/pKD46 plasmids contain the λ-red genes begind an arabinose inducible promoter, a temperature-sensitive origin, and an ampicillin selection marker. The part to be integrated is on the pLF03 plasmid, which is identical to pYW201<bib id="pmid17959404"/> in every way except for the selection marker in the insert (pYW201 has kanamycin resistance, pLF03 chloramphenicol). pLF03 contains an ampicillin marker that is expressed constitutively, chloramphenicol is only used to test for successful integration, as it is behind an IPTG-inducible T7 promoter (as are the other two genes, pccB and accA1). pLF03 is derived from pET-21c. The expected amplificate size for PCR of pLF03 is 4kb. pccB-accA will replace the ygfG gene in the BAP1 genome.
2013-06-03
- inoculate 50 ml LB with BAP1, grow at 37°C until OD=0.57
- prepare BAP1 glycerol stocks
- prepare competent BAP1
- recover pKD46, pCP20, pLF03
- transform TOP10 with pKD46, plate on Amp, grow at 30°C
- transform TOP10 with pLF03, plate on Amp, grow at 37°C
- transform TOP10 with pCP20, plate on Amp, grow at 30°C
2013-06-04
- inoculate 4 ml LB+Amp with TOP10-pLF03, grow at 37°C
- make miniPrep of pLF03 => 97 ng/µl in 27 µl
- transform BAP1 with pKD46, plate on Amp, grow at 30°C
2013-06-05
- prepare 50 ml LB + Amp + Arabinose(0.1%), inoculate with BAP1-pKD46
- prepare 4 ml LB + Amp, inoculate with TOP10-pKD46 (for miniPrep)
- no colonies with pCP20 => repeat transformation with three times the amount of plasmid (30 µl)
- BAP1-pKD46 reached OD=0.68 at 22:30 => put in fridge over night
2013-06-06
- inoculate 50 ml LB + Amp + Arabinose(0.1%) with 500 µl BAP1-pKD46 from ON culture (fridge)
- at OD=0.49: make glycerol stocks, prepare electrocompetent BAP1-pKD46
- make miniPrep of TOP10-pKD46 ON culture -> 31 ng/µl
- low yield of TOP10 miniPrep: make 2 miniPreps of BAP1-pKD46 ON culture -> 45.5 ng/µl and 46.0 ng/µl
- evening: no colonies with pCP20 -> write Lei Fang (Pfeifer lab) -> their stock is bad, we should get pCP20 somewhere else
2013-06-07
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) of with primers ygfG21C1 and ygfG21C2 using Phusion polymerase:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 10 |
| 66 | 30 | |
| 72 | 90 | |
| 1 | 72 | 450 |
| 1 | 4 | inf |
- using hot start at 98°C
- two PCR samples were run, in one the annealing step at 66°C was left out
- => no amplificate
2013-06-08
- Looking in more detail at the primers, the parts binding to the template have melting temperatures of 40 and 45°C (the rest is homologous to E. coli genome for recombination)
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion polymerase:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 5 | 98 | 10 |
| 39 | 45 | |
| 72 | 90 | |
| 5 | 98 | 10 |
| 45 | 45 | |
| 72 | 90 | |
| 20 | 98 | 10 |
| 58 | 45 | |
| 72 | 90 | |
| 1 | 4 | inf |
- using hot start at 98°C
- two PCR samples were run, in one the annealing step at 58°C was left out
- => no amplificate
2013-06-09
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion polymerase:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 35 | 98 | 10 |
| 40 | 60 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- using hot start at 98°C
- amplificate has 1.3kb -> too short, but specific (one distinct band)
2013-06-10
- need to verify plasmid identity: digest pLF03 with EcoRI (for linearization) and XhoI+NdeI (verify insert size); 388 ng (4 µl of miniPrep from 2013-06-04) each
- insert only 3.3 kb in length, expected >4 kb ([http://www.uniprot.org/uniprot/Q9RGQ6 AccA1]: 590AS * 3 = 1770 bp, [http://www.uniprot.org/uniprot/Q9X4K7 PccB]: 530AS * 3 = 1590 bp, [http://www.uniprot.org/uniprot/P62577 CatR]: 219 AS = 657 bp; adds up to 4017 bp, not counting RBS and spacers between the individual genes)
- contacted Lei Fang (Pfeifer Lab), there are NdeI and EcoRI sites in catR (they were not indicated in his plasmid map)
- to verify presence of chloramphenicol acetyl transferase in insert: transform BAP1 with 97 ng pLF03 (1 µl of miniPrep from 2013-06-04), plate on Amp + IPTG
- evening: pick BAP1-pLF03 colonies, transfer to Cm + IPTG
- inoculate 4 and 5 ml of liquid culture (Amp) with BAP1-pLF03
2013-06-11
- BAP1-pLF03 grew on Cm + IPTG
- try to optimize PCR:
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion Flash F548 polymerase (performed by Dominik):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 45 | 5 | |
| 72 | 120 | |
| 25 | 98 | 1 |
| 55 | 5 | |
| 72 | 120 | |
| 1 | 72 | 120 |
| 1 | 4 | inf |
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Taq polymerase (cheaper):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 94 | 180 |
| 35 | 94 | 20 |
| 41.5/43 | 60 | |
| 72 | 480 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
- prepare glycerol stock of BAP1-pLF03
- make miniPreps of BAP1-pLF03 -> 21 ng / µl and 27 ng / µl in 27.5 µl
- received just plated DH5α-pCP20 from Sourjik Lab, grow at 30°C
2013-06-12
- purify Phusion Flash PCR product with Qiagen PCR purification kit -> 120 ng / µl in 27.5 µl
- inoculate 2 x 5 ml LB + Amp with DH5α-pCP20, grow at 30°C
2013-06-13
- miniPreps of DH5α-pCP20 -> 7.8 ng / µl and 14.6 ng / µl in 27.5 µl, respectively
- low yield of miniPreps: inoculate 2 x 5 ml LB + Amp with DH5α-pCP20, grow at 30°C
2013-06-14
- miniPreps of DH5α-pCP20 -> 25 ng / µl and 29 ng / µl in 27.5 µl, respectively
- prepare glycerol stocks of DH5α-pCP20
- to avoid electroporation with intact pLF03: add 10 µl of PCR amplificate (1.2 mg) on gel, perform gel extraction (QiaGen gel extraction kit) -> 3.2 ng / µl
- add 240 ng (2 µl) raw PCR product to 100 µl electrocompetent BAP1-pKD46
- add 16 ng (5 µl) gel-extracted PCR product to 100 µl electrocompetent BAP1-pKD46
- electroporate 50 µl each, add to 1 ml SOC with 1 mM IPTG
- shake at 300 rpm, 37°C for 1 h
- spin cells down (3 min 10 000 rpm = 8000 g), decant supernatant (leave about 200 µl supernatant in eppendorf tube)
- resuspend pellet, plate 100 µl on Cm-IPTG, leave rest at RT
- grow at 37°C
2013-06-15
- one colony on plate with gel-extracted PCR product
- many colonies on plate with PCR-purified PCR product
- pick colonies from plate with PCR-purified PCR product, run colony-PCR (Taq, 25 µl total volume) with primers IK01 and IK02 (expected product: 465 bp):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 180 |
| 30 | 95 | 30 |
| 61 | 30 | |
| 72 | 30 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product -> pick colony from gel-extracted plate, pick colonies from PCR-purified plate, run colony-PCR (Taq, 25 µl total volume) with primers IK01 and IK02:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 240 |
| 30 | 95 | 30 |
| 61 | 30 | |
| 72 | 30 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product -> 2 possibilities:
- all colonies bear complete pLF03 plasmid (PCR was run with 5 ng template, DNA concentration in PCR product: 120 ng / µl, electroporated with 2 µl -> 1/6 ng pL03 vs. 240 ng amplificate)
- PCR did not work
- for colony from gel-extracted plate + 3 colonies from PCR-purified plate: add up to 3 ml medium, add IPTG (1 mM) + Cm, grow at 37°C
- plate rest of bacteria electroporated with gel-extracted PCR product on Cm + IPTG
2013-06-16
- no colonies on gel-extracted plate
- make miniPreps of ON cultures, run gel
- pLF03: 9.2 kb, bands above 10 kb, bands smear, low intensity (applied all 30 µl of miniPrep to gel) -> fragments of genomic DNA?
- => PCR went wrong?
2013-06-17

- repeat colony PCR, use 1 µl of liquid culture from gel-extracted plate, pick colonies from PCR-purified plate (Taq, 25 µl total volume)
- 2 conditions:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 600 |
| 30 | 95 | 240 |
| 64 | 30 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 600 |
| 12 | 95 | 240 |
| 66°C ↓0.5°C | 30 | |
| 72 | 60 | |
| 18 | 95 | 240 |
| 60°C | 30 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no positives
- plate samples of liquid cultures from 2013-06-15 on Amp, grow at 37°C
2013-06-18
- colonies grew on Amp
2013-06-19
- prepare for repetition of experiment: run 4 PCRs of 1 ng pLF03 (miniPrep from 2013-06-04) with primers ygfG21C1 and ygfG21C2 using Phusion Flash (performed by Dominik), one reaction = 50 µl total volume
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 45 | 5 | |
| 72 | 130 | |
| 25 | 98 | 1 |
| 55 | 5 | |
| 72 | 120 | |
| 1 | 72 | 120 |
| 1 | 4 | inf |
- pool products, purify (digestion only 3 h)
- nanoDrop: 2088 ng/µl -> remeasure: 532 ng/µl
- load 1 µl purified PCR product on gel -> very weak band -> nanoDrop wrong
- to eliminate error: make fresh electrocompetent cells:
- pick colony from BAP1-pKD46 plate from 2013-06-04
- inoculate 1 ml LB + Amp
- grow at 30°C
2013-06-20
- too little cells: use aliquot from 2013-06-06
- use all available DNA (14 µl) and 100 µl cells (complete aliquot)
- grow in SOC + IPTG(1mM) at 300 rpm for 3h
- spin cells down, decant supernatant, resuspend in remainder of medium
- plate 10 µl, rest on Cm + IPTG
2013-06-24

- no colonies on 10 µl plate, 2 colonies on rest plate
- colony-PCR using primer pairs IK01+IK02, IK01+IK03 (positive control), IK05+IK06 (negative control) (Taq, 20 µl total volume)
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 30 | 95 | 60 |
| 62 | 30 | |
| 72 | 60 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- inconclusive results: no band for IK05+IK06 in BAP1-pLF03 (negative control)
- grow liquid cultures at 37°C
2013-06-25
- repeat PCR with 1 µl of ON cultures:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 (IK01+IK02; IK01+IK03;IK05+IK06) / 240 (IK01+IK06) | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 (IK01+IK02; IK01+IK03;IK05+IK06) / 240 (IK01+IK06) | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- genomic integration failed
- pick colony from BAP1-pKD46 plate from 2013-06-04, grow in 3 ml LB+Amp at 30°C
2013-06-26
- inoculate 50 ml LB + Amp + Ara(0.5%) with 1.1 ml of ON culture
- grow at 30°C to OD=0.73, prepare electrocompetent BAP1-pKD46
2013-06-27
- run PCR of 5 ng pLF03 (1 µl 1:20 dilution of miniPrep from 2013-06-04) with primers IK07 and IK08 using Phusion polymerase (20 µl total volume) on cycler 1 (does not cool down to 4°C) (using hot start):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product -> repeat PCR with cycler 2 (fully functional)
- no product -> run 2-step PCR with 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Phusion polymerase (20 µl total volume, using hot start) on cycler 2:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 72 | 150 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no product
2013-06-28
- run PCR of 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Q5 polymerase (20 µl total volume, using hot start) on cycler 2:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 1800 (30 min) |
| 1 | 4 | inf |
- perfect PCR => Phusion is crappy
- run 4 PCRs of 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Q5 polymerase (50 µl total volume, using hot start) on cycler 2:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 45 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 1800 (30 min) |
| 1 | 4 | inf |
2013-06-29
- load 1 µl of PCR product on gel -> specific amplificate
- pool products (also include amplificate from 2013-06-28), purify (only first step, as no DpnI available)
2013-06-30
- inoculate 1 ml LB+Amp with 10 µl of ON culture from 2013-06-25, grow at 30°C
2013-07-01
- finish purification (digest for 5h, dissolve in 50 µl H2O) -> 1162 ng/µl (performed by Fanny)
- prepare electrocompetent cells from ON culture (add 0.5% arabinose at dilution) (performed by Fanny)
- electroporate fresh electrocompetent cells, 1 aliquot from 2013-06-26 (23 µl DNA each) (performed by Fanny)
- grow in SOC at 30°C for 30 min, transfer to falcons, add 1 ml LB, add IPTG (1 mM), grow at 37°C for 4h
- spin cell down (8000 rcf for 3 min), decant supernatant, resuspend pellet in remaining medium
- plate on Cm+IPTG (2 plates for each sample: 10µl + rest, plates marked F: from -80°C, marked H: newly prepared cells), grow at 37°C
2013-07-02

- bacteria lawn on rest-plate from -80°C, possible colonies on other plates (barely visible, may be bubbles)
- pick 2 colonies from F-rest, sample of lawn, 2 colonies from H-10µl, colony-PCR with primers IK01+IK02, IK01+IK03 (positive control) (20µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- very evening: colonies on F-10µl, H-rest, H-10µl plates appear -> leave at 37°C ON
2013-07-03

Lane 1: NEB 2-log
Top: lanes 2-4: negative control (BAP1-pLF03); lanes 5-13: colony-PCR F-10µl
bottom:lanes 2-4: colony-PCR of F-10µl; lanes 5-13: colony-PCR of H-10µl; lanes 2,5,11: primers IK01+IK02; lanes 3,6,12: primers IK01+IK03 (positive control); lanes 4,7,13: primers IK05+IK06 (negative control)
- colonies on F-rest plate appeared (plate left ON at RT)
- pick 4 colonies from F-10µl, 3 colonies from H-10µl, colony-PCR with primers IK01+IK02, IK01+IK03 (positive control), IK05+IK06 (negative control):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- F-10µl 1,2,4 show no negative control -> grow at 37°C
- continue growing plates at 37°C until evening, leave at RT ON
2013-07-04

Lane 1: NEB 2-log
Top: lanes 2-10: cultures from 2013-07-03; lanes 2,5,8: primers IK01+IK02; lanes 3,6,9: primers IK01+IK03; lanes 4,7,10: primers IK05+IK06; lane 11: BAP1-pLF03 (negative control); lanes 12-13: colonies from F-10µl plate; lanes 11-13: primers IK05+IK06
bottom: lanes 2-13: primers IK05+IK06; lanes 2,3: colonies from F-10µl plate; lanes 4-6: colonies from F-rest plate; lanes 7-10: colonies from H-10µl plate; lanes 11-13: colonies from H-rest plate
- repeat colony-PCR for 3 ambiguous cultures form 2013-07-03 (use 1 µl of culture), pick 14 new colonies (only primers IK05+IK06); 20 µl total volume:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no positives
2013-07-05
- digest 450 ng pKD46 (10 µl of 45.5 and 46 ng/µl miniPreps from 2013-06-06) with EcoRI (expected: 4816+1505 bp), BamHI+NotI (expected: 3675+2646 bp), PstI+NotI (expected: 3711+2363+243 bp); 30 µl total volume
- right plasmid
- inoculate 2 x 7 ml LB+Cm+IPTG(1mM) with colonies from F-10µl plate, grow at 30°C
2013-07-06
- add Cm (1:3000), IPTG (1:1000) to liquid cultures, grow for 8h at 30°C
- make miniPreps -> 25 ng/µl and 14.4 ng/µl in 27.5 µl
- digest 25 ng/µl miniPrep with EcoRI (use all of miniPrep)
- load digest completely, 10 µl of 14.4 ng/µl miniPrep on gel
- no bands -> nanoDrop crappy, no plasmid
- wrong strain? Streak BAP1 on Cm, grow at 37°C
2013-07-08
- no colonies of BAP1 on Cm
- repeat PCR of pLF03:
- run 4 PCRs of 4 ng pLF03 (0.2 µl of 21 ng/µl miniPrep from 2013-06-11) with primers IK07 and IK08 using Q5 polymerase (50 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 45 | 98 | 5 |
| 66 | 30 | |
| 72 | 150 | |
| 1 | 72 | 1800 (30 min) |
| 1 | 4 | inf |
- extract amplificate from gel (4 lanes pooled, extracted by Dominik -> 119 ng/µl; 4 lanes pooled, extracted by Ilia -> 111.5 ng/µl)
- load 1 µl of each extract on gel
2013-07-09
- electroporate electrocompetent BAP1-pKD46 with 1µl, 5µl, 25µl DNA
- resuspend in 1 ml SOC + IPTG (1mM) + Ara (0.1%)
- grow for 90 min at 37°C, 400 rpm
- spin cells down, plate 10µl, rest of each sample on Cm+IPTG
2013-07-10
- on all plates: large + small colonies
- small colonies too small to pick => pick 10 large colonies from 25µl DNA / 10 µl bacteria plate, run colony-PCR with primers IK05+IK06 (Taq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no integration
- continue growing plates at 37°C

Top: lane 1: NEB 2-log; lanes 2,4,6,8,10,12: primers IK05+IK06; lanes 3,5,7,9,11,13: primers IK01+IK03 (positive control); lanes 2,3: BAP1-pLF03 (control); lanes 4-7: colonies from 1µl/10µl plate; lanes 8-11: 1µl/rest; lanes 12-13: 5µl/10µl
Bottom: lane 11: NEB 2-log; lanes 1,3,5,7,9: primers IK05+IK06; lanes 2,4,6,8,10: primers IK01+IK03 (positive control); lanes 1-2: 5µl/10µl; lanes 3-6: 5µl/rest; lanes 7-8: 25µl/10µl; lanes 9-10: 25µl/rest
- when small colonies large enough: pick, run colony-PCR (OneTaq, 20µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- 1st colony from 5µl/rest plate (marked 7) seems promising => grow at 37°C in 1 ml LB
2013-07-11
- run full colony-PCR of liquid culture (OneTaq, 20 µl total volume, use 1 µl of culture):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 18 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no integration
- 3rd population of colonies appeared on plates (stored at RT): very small, probably satellites

- run colony-PCR of liquid with primers IK07+IK08 (test for integration in wrong place) (OneTaq, 20µl total volume, use 1 µl liquid culture):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 30 | 95 | 60 |
| 66 | 30 | |
| 72 | 600 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- no integration at all

Top: lane 1: NEB 2-log; lane 2: BAP1-pLF03(control); lanes 3-9: colonies from 1µl/10µl plate; lanes 10-13: colonies from 1µl/rest plate
Bottom: lane 1: NEB 2-log; lanes 2-10: colonies from 1µl/rest plate (lane 5: swiped 3rd population of extremely small colonies)
- pick 20 new colonies, run colony-PCR with primers IK05+IK06 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- unspecific bands (also present in colony-PCR from 2013-07-10)
- unspecific bands not present in BAP1-pLF03 PCR (control) and bottom lane 5 (satellite colonies) => not due to iTaq
- assume: some amount of satellites was picked together with colonies from plates -> give band at 500 bp; partially amplified ygfG fragments prime integrated insert
- do multiple sequence alignment of ygfG, [http://www.ncbi.nlm.nih.gov/nuccore/AF113605 pccB], [http://www.ncbi.nlm.nih.gov/nuccore/AF113603 accA1], and [http://www.ncbi.nlm.nih.gov/nuccore/AY048742 catR] using ClustalO => long enough stretches of partially identical sequences present
- add 1mM IPTG to cultures with colonies from 1µl/10µl plate picked today (grew in 500 µl LB at 37°C for several hours), after 2h at 37°C: streak on Cm+IPTG
- pick 10 colonies each from 1µl/10µl and 1µl/rest plates, transfer to new Cm+IPTG plate, grow at 37°C
- pick 1 colony from 1µl/10µl plate, inoculate 1 ml LB + Cm + IPTG (1mM)
2013-07-12

Top: lane 1: NEB 2-log; lane 2: BAP1-pLF03(control); lanes 3-12: colonies liquid culture that were plated on Cm+IPTG; lane 13: colony from 1µl/10µl plate that was transferred to new Cm+IPTG plate
Bottom: lane 1: NEB 2-log; lanes 2-6: colonies from 1µl/10µl plate that were transferred to new Cm+IPTG plate; lanes 7-12: colonies from 1µl/rest plate that were transferred to new Cm+IPTG plate; lane 13: liquid culture (Cm+IPTG(1mM)) from 1µl/10µl plate
- colonies grew slowly
- run colony-PCR with primers IK01+IK02 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- no bands
- run colony-PCR with primers IK05+IK06 of 2 colonies from liquid culture that were transferred to Cm + IPTG plate (iTaq, 20µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 120 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- band at 1.2 kb is now specific -> WTF?!?
2013-07-13

- need to determine whether 1.2 kb band is specific: run gradient PCR of colony 2 (iTaq, 20 µl total volume) with primers IK05+IK06:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 63-68 (ΔT = 1) | 30 | |
| 72 | 120 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- intensity of 1.2kb band decreases, intensity of 0.5 kb band (control) does not -> 1.2 kb band is unspecific product
- check for presence of genomically integrated insert: run colony-PCR of colony 2 with primers IK07+IK08 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 66 | 30 | |
| 72 | 600 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- transfer some of colony 2 on Amp (check for presence of pLF03 as a whole), grow at 37°C
- transfer some of colony 2 to liquid culture: LB + Cm + IPTG (1mM), grow at 37°C
2013-07-14
- bacteria grew on Amp => 3 possibilities:
- Amp plates not selective
- bacteria have complete pLF03 plasmid
- bacteria still have pKD46, although grown at 37°C
- plate BAP1, BAP1-pKD46 on Amp, grow at 37°C
- colony-PCR did not work for the control -> repeat with OneTaq (use 1 µl of liquid culture)
- no amplificate for colony: possibly, Taq has problems amplifying 4.8 kb from genomic DNA

- run colony-PCR with primers IK01+IK06 (OneTaq, 20 µl total volume, use 1 µl of liquid culture):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 240 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 240 | |
| 1 | 72 | 600 |
| 1 | 4 | inf |
- control shows bright band where expected, colony does not => something is integrated, unknown what
2013-07-15

- BAP1 did not grow on Amp, BAP1-pKD46 did grow on Amp at 37°C => no discrimination between pLF03 and pKD46 possible
- Taq might have problems amplifying 4.8 kb from genomic DNA => use Phusion Flash (primers IK07+IK08, 20 µl total volume, use 1 µl of liquid culture, old BioRad cycler):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 100 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- control did not work -> repeat in new BioRad cycler (pick from plate)
- same result
2013-07-18


- run colony-PCR with primers IK24, IK25 (OneTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 180 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- 1.2 kb band with primers IK01+IK25 in control => makes no sense, repeat
- same result
- transfer BAP1-pLF03 to new Cm+IPTG plate, colony 2 to LB w/o antibiotics, grow at 37°c#
2013-07-22
- no positive PCR of colony yet: run colony-PCR with primers IK01+IK03 (iTaq, 20 ml total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 12 | 95 | 60 |
| 68 ↓0.5°C | 30 | |
| 72 | 180 | |
| 23 | 95 | 60 |
| 62 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- primers RB43+RB44 (against sfp) iTaq, 20 µl total volume:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 12 | inf |
- unspecific bands in colony, no sfp bands at all
- re-run sfp PCR (primers RB43+RB44), include streaked BAP1 by Konrad
- sfp present in BAP1, not present in BAP1-pLF03, colony
- makes no sense, Konrad streaked his cells from the same batch of competent cells
- transfer BAP1, BAP1-pLF03, colony to liquid culture LB, grow at 37°C
2013-07-23

- run colony-PCR with primers RB43+RB44 (iTaq, 20 µl total volume, use 1 µl of liquid culture)
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 12 | inf |
- no sfp present
2013-07-29
- inoculate 10 ml 2xYT+Amp with TOP10-pKD46, grow at 30°C
2013-07-31
- miniPrep of pKD46 -> ca. 10 ng/µl
2013-08-03
- transform BAP1 with 5µl of pKD46 miniPrep form 2013-07-31, plate on Amp, grow at 30°C
29-07-2013
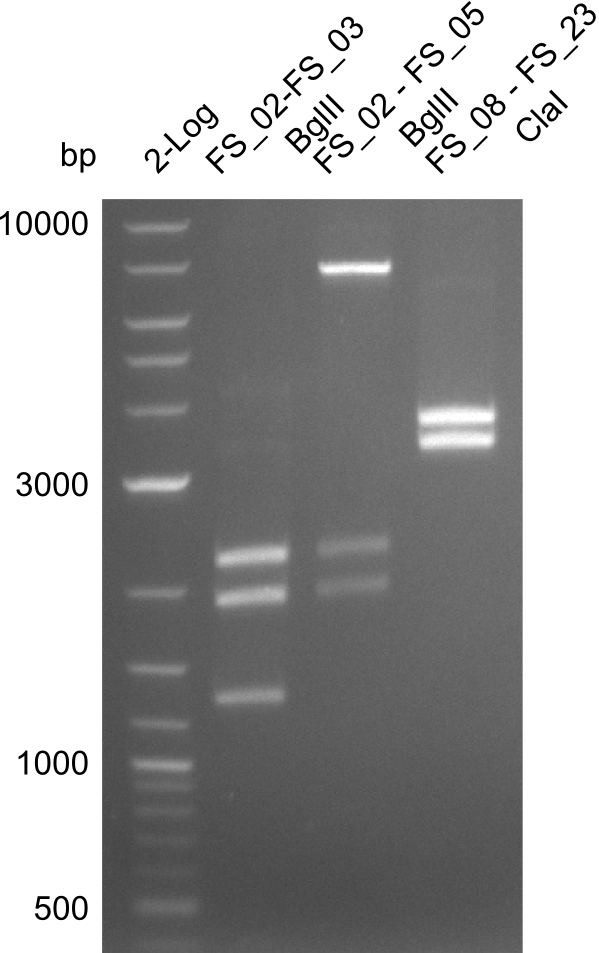
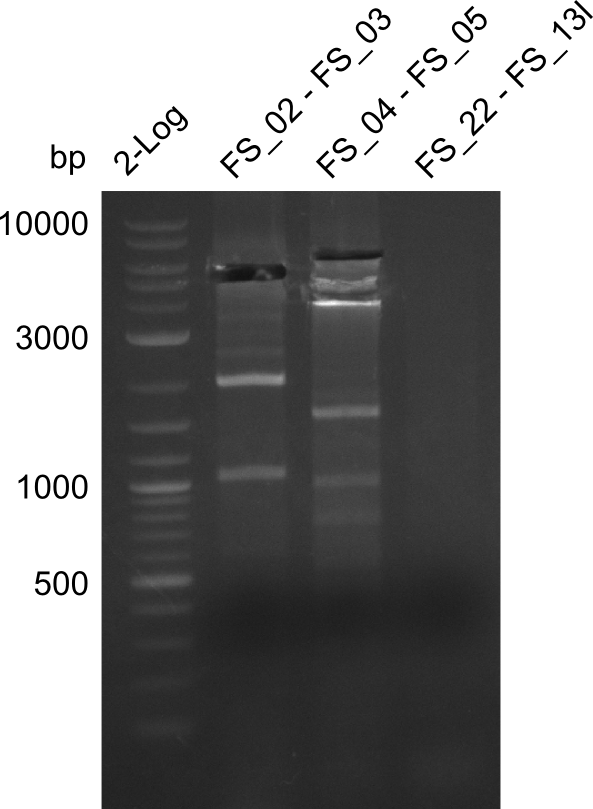
Restriction digest of fragment FS_02 to FS_03 (5.3 kb; 27-07-2013; II) with BglII
Incubation at 37°C for about 3 h
| what | µL |
|---|---|
| FS_02 to FS_03 (27-07-2013; II) | 15 |
| BglII | 1 |
| Buffer 3.1 | 2 |
| dd H2O | 2 |
| Expected fragment lengths [bp] | 2146, 1862, 1306 |
Results:
- Restriction digest shows the expected product sizes
- indicator for correct amplicon but to be sure, PCR product will be prepared for single read sequencing by GATC
2013-08-05
- run colony-PCR against sfp on BAP1-pKD46 with primers RB43+RB44 (iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- inoculate 4 ml 2xYT+Amp with BAP1-pKD46, grow at 30°C
2013-08-06
- inoculate 50 ml 2xYT+Amp+Ara(0.5%) with BAP1-pKD46, grow at 30°C until OD=0.71
- run colony-PCR with primers RB43+RB44 from 50 ml culture:
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 60 |
| 55 | 30 | |
| 72 | 180 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- prepare electrocompetent BAP1-pKD46
2013-08-08
- electroporate BAP1-pKD46 with 500 ng DNA (5 µl of PCR amplificate from 2013-07-08)
- grow in SOC + IPTG(1mM) + Arabinose (0.5%) at 37°C, 400 rpm for 3h
- spin cells down (3 min at 8000 rcf)
- plate 10 µl, rest on Cm, grow at 37°C
07-08-2013
Sequencing Results
We send the following samples (amplified fragments of D. acidovorans DSM-39 and backbone pSB4K5 from the partsregistry) to GATC for sequencing:
- AE (FS_02-FS_03) with Primer FS_02
- AE (FS_02-FS_03) with Primer FS_03
08-08-2013
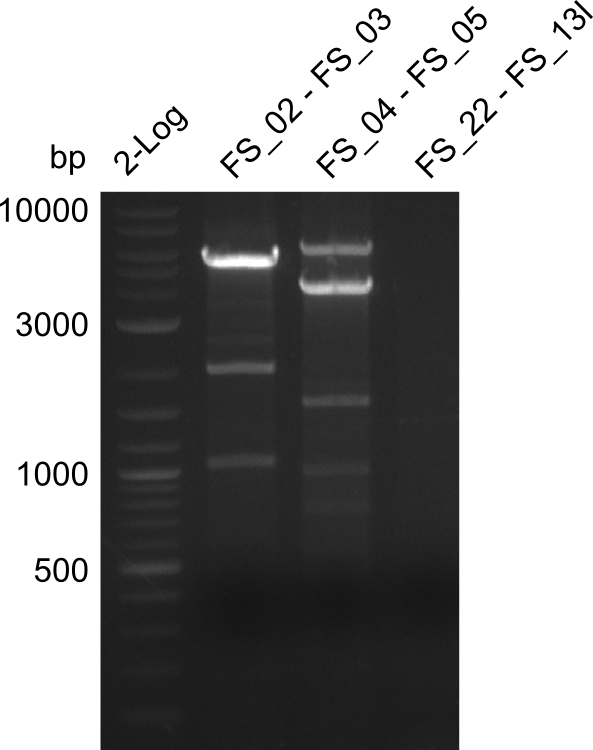
Amplification from FS_02 to FS_03; 5.3kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02: (1/10) | 4 |
| FS_03: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:30 | |
| 1 | 72 | 7min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE was repeated successfully with the new strain SPH-1
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- if fragment is used in Gibson assembly the amplification has to be repeated to increase the amount of product
12-08-2013
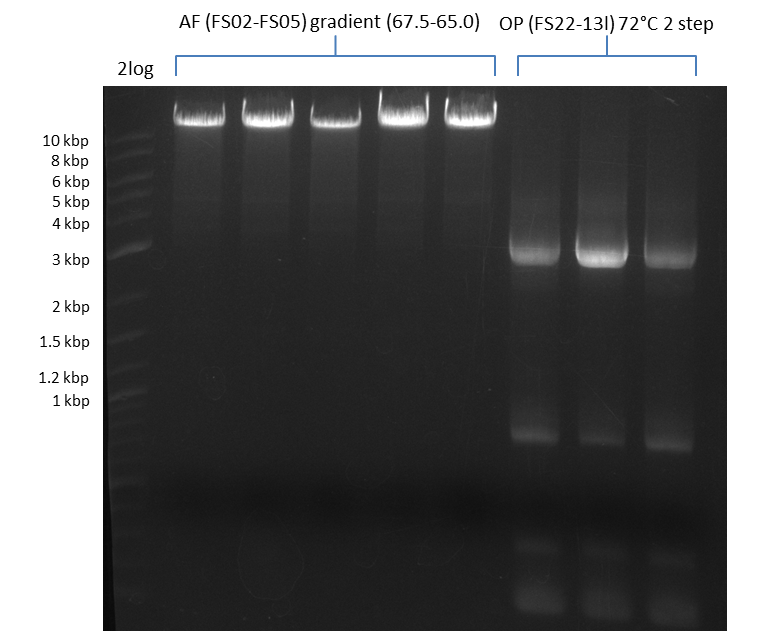
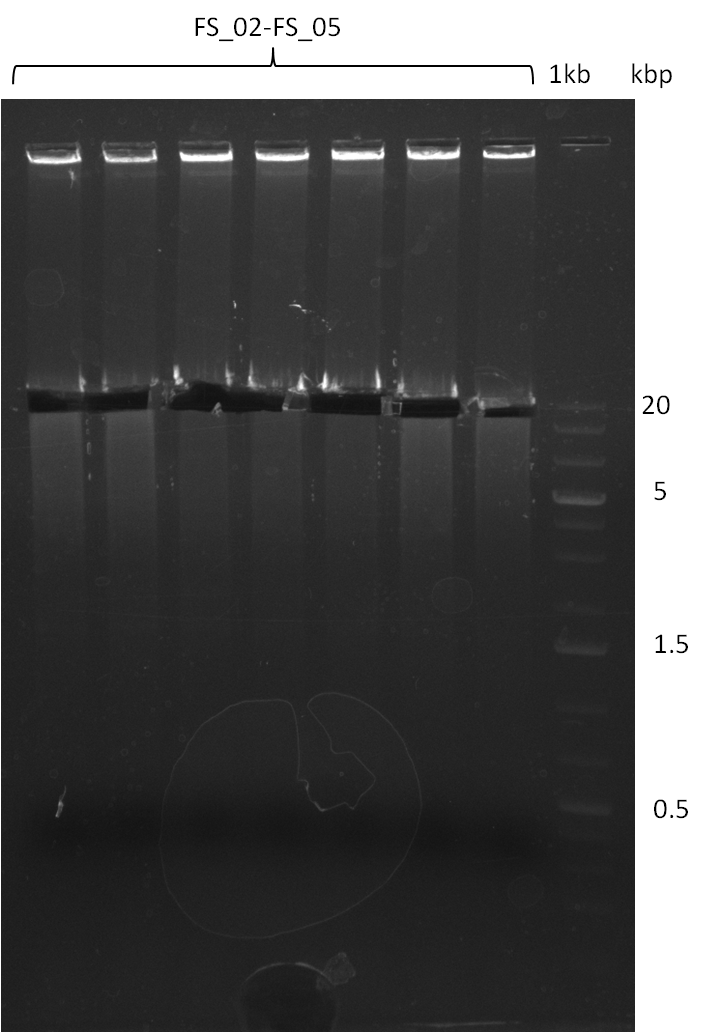
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4.5 µL FS_02 | |||||||||
| Primer rev | 4.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 67.5 - 65.0 (ΔT = 0.5) ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 65.5 - 63.0 (ΔT = 0.5) | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked well, gradient displays an optimal annealing temperature of 65.5°C, which will be used for further amplifications
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
13-08-2013
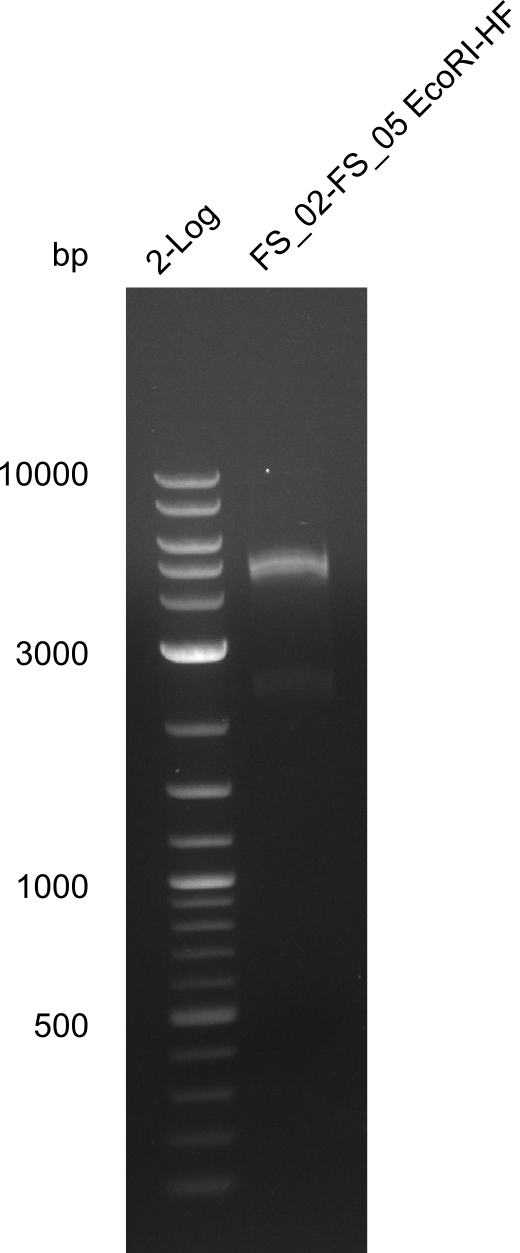
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 12-08-2013 with EcoRI-HF
Incubation at 37°C for 2 hours
| what | µL |
|---|---|
| FS_02 to FS_05 (12-08-2013) | 20 |
| EcoRI-HF | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 1.5 |
Expected fragment sizes: 2.26kbp; 4.62kbp; 4.32kbp
Results:
- Weak bands of about 4.6kbp visible, as well as on of about 2.6kbp
- digest will be repeated using higher concentrations of DNA to clearify results of restriction digest
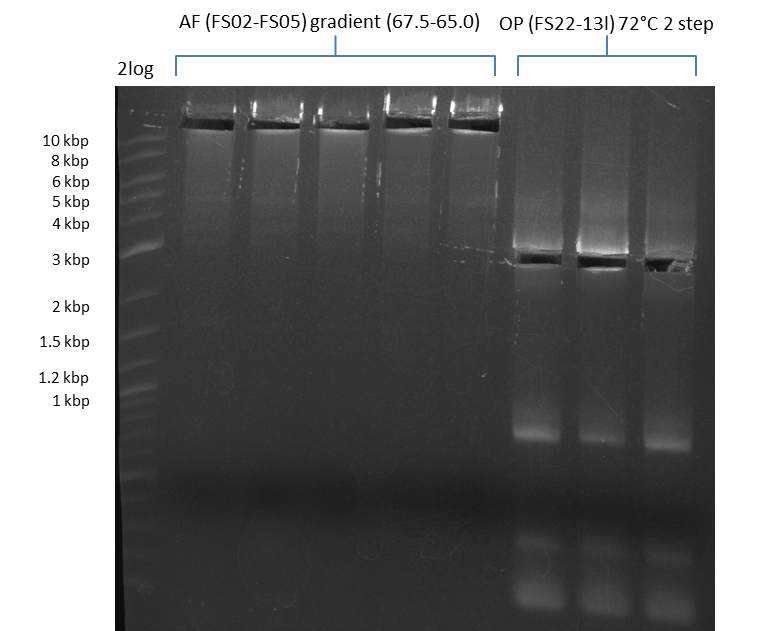
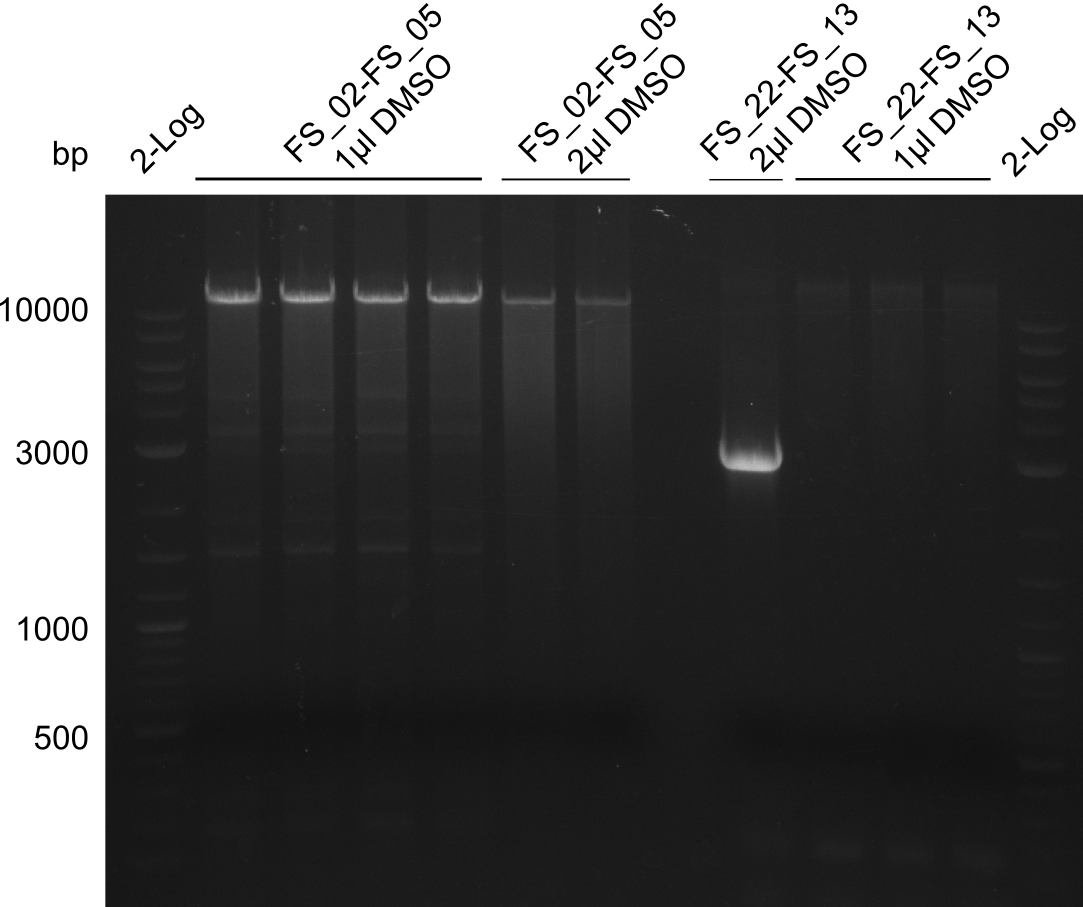
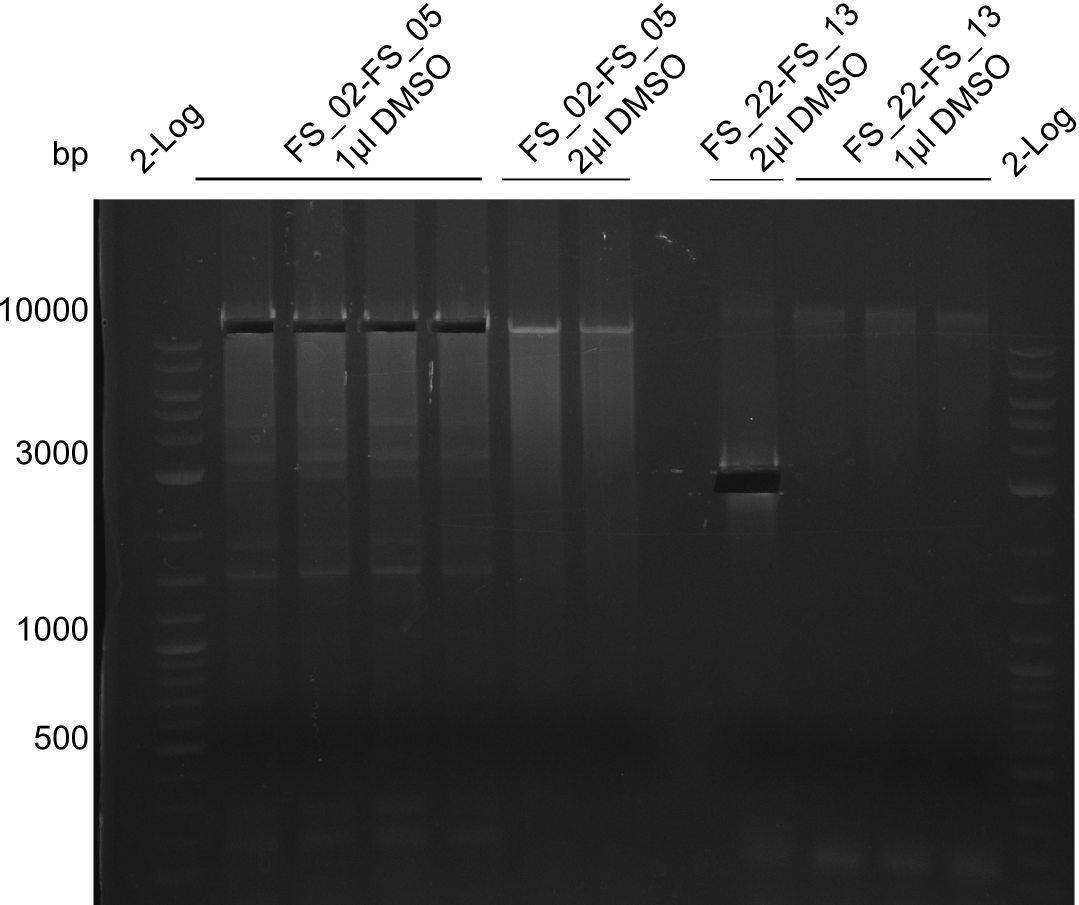
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction
1 sample contains 2µL of DMSO
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 4.5 µL FS_02 | |||||||||
| Primer rev | 4.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65.5 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 63.5 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked though a slight smear occured, therefore PCR will be repeated on the more precise Biometra TProfessional Basic cylcer again
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
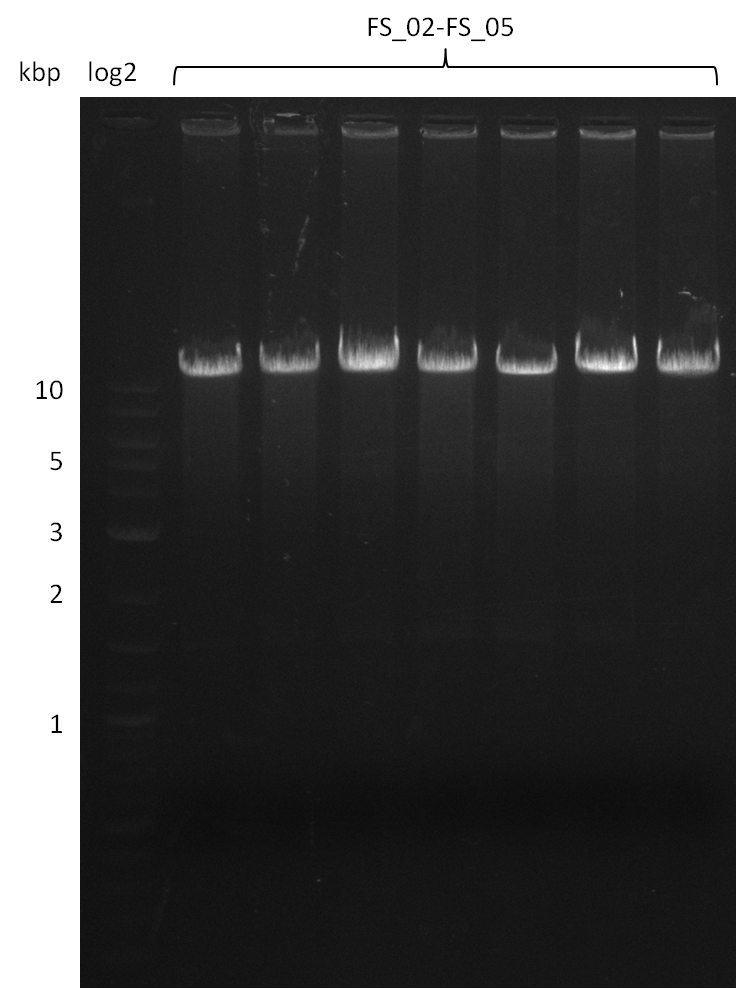
14-08-2013
Concentration measurement (FS_02 to FS_05; 11.2 kb)
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelAF | FS02-FS05 | 11-08-2013 | ~10 ng/µL |
| DelAF | FS02-FS05 | 12-08-2013 | 0 ng/µL |
| DelAF | FS02-FS05 | 12-08-2013 | 0 ng/µL |
Results:
- concentrations are not sufficient for gibson assembly
- PCR will be repeated and gel slices of different reactions will be pooled for one gel extraction using QIAquick Gel Extraction Kit to obtain the concentrations needed for gibson assembly
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 11-08-2013 with EcoRI-HF
Incubation at 37°C for 2 hours 45min
| what | µL |
|---|---|
| FS_02 to FS_05 (11-08-2013) | 18 |
| EcoRI-HF | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 3.5 |
Expected fragment sizes: 2.26kbp; 4.62kbp; 4.32kbp
Results:
- One band of about 5kbp and one of 2.5kbp
- no clear result, digest will be repeated with another enzyme (ClaI), as the enzyme used was beyond expiration date
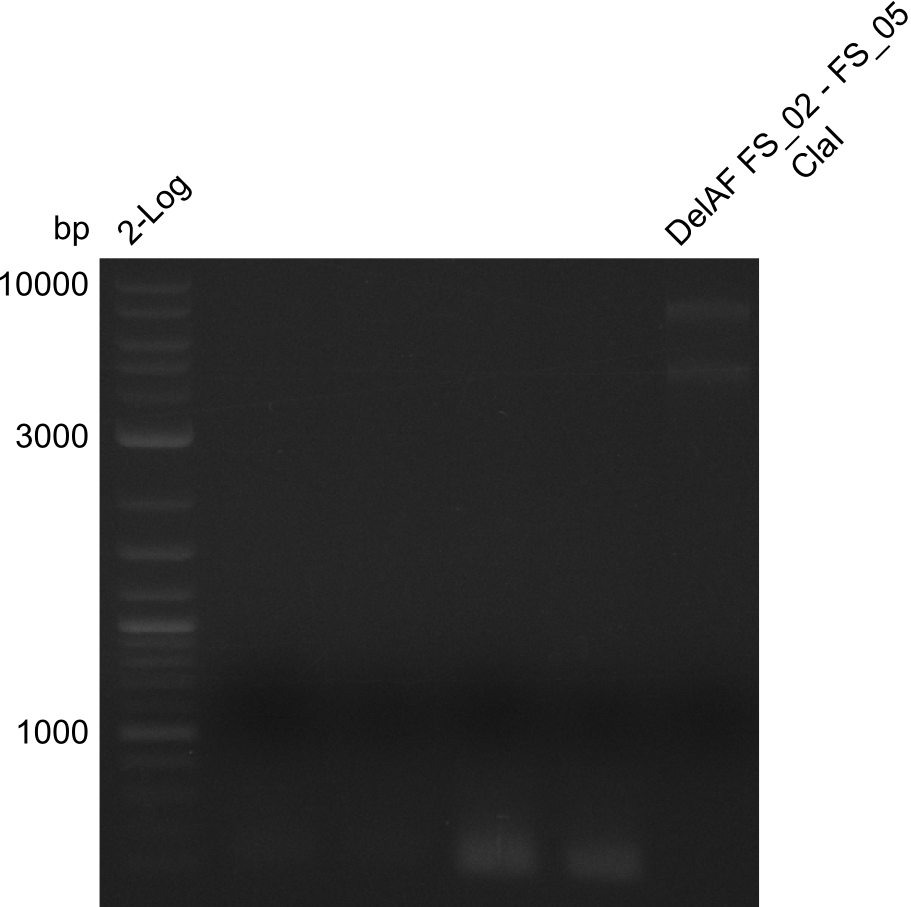
Restriction digest of fragment FS_02 to FS_05; 11.2 kb; 10-08-2013 with ClaI
Incubation at 37°C for 2 hours
| what | µL |
|---|---|
| FS_02 to FS_05 (10-08-2013) | 20 |
| ClaI | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 1.5 |
Expected fragment sizes: 6.9kbp, 4.3kbp
Results:
- restriction digest displays the expected fragments, therefore amplification of the desired fragment can be assumed
- PCR product will be prepared for sequencing by GATC to proof amplification of the desired DNA sequence before Gibson Assembly
17-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction of DelAF
6x20 µL
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 2.5 µL FS_02 | |||||||||
| Primer rev | 2.5 µL FS_05 | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
| DMSO | 1 µL | |||||||||
| dd H2O | 4 µL | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65.5 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 63.5 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked
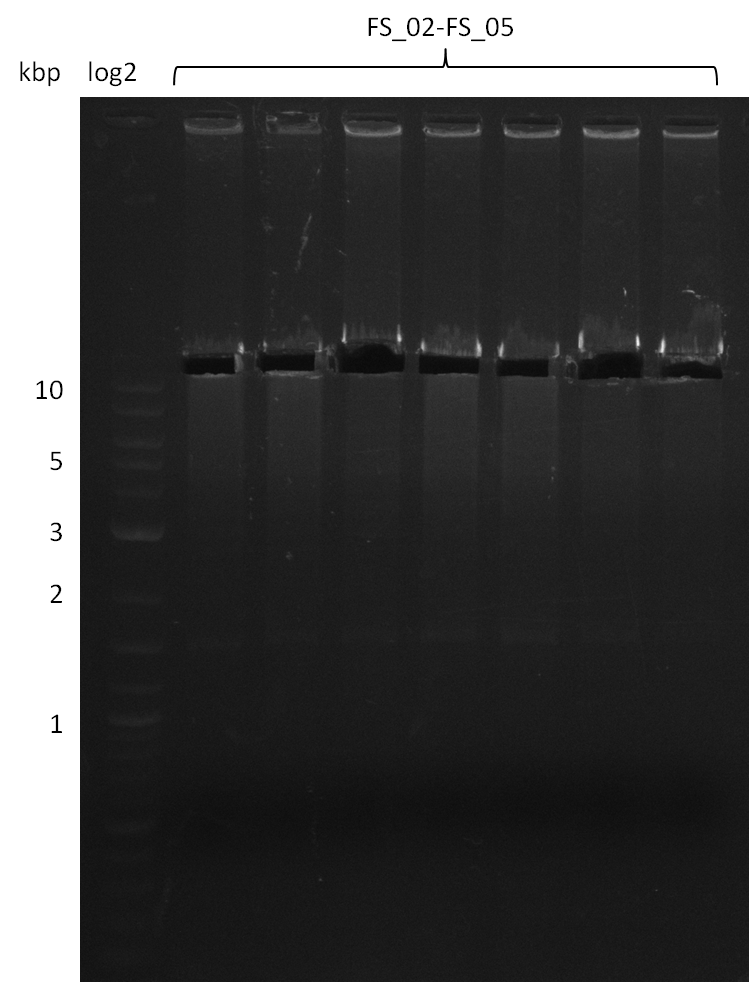
- bands were cut out, pooled and DNA purified using QIAquick Gel Extraction Kit to obtain concentrations needed for gibson assembly
18-08-2013
Amplification from FS_02 to FS_05; 11.2 kb
- Reaction of DelAF
6x20 µL
| Reagent | DelAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Template | D.acidovorans SPH-1 colony | |||||||||
| Primer fw | 2.5 µL FS_02 | |||||||||
| Primer rev | 2.5 µL FS_05 | |||||||||
| DMSO | 1 µL | |||||||||
| Phusion Ready Mix | 10 µL | |||||||||
| dd H2O | 4 µL | |||||||||
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65.5 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 63.5 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- Amplification of DelAF worked
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
12-08-2013
Amplification from FS_02 to FS_11; 22.8 kb
- Reaction
| Reagent | DelAG |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.5 µl FS_02 |
| Primer rev | 4.5 µl FS_11 |
| DMSO | 1 µl |
| Phusion Ready Mix | 10 µl |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 72.0 - 67.0 (ΔT = 0.5) ↓ 0.5 | 5 | |
| 72 | 7:20 | |
| 18 | 98 | 1 |
| 70.0 - 65.0 (ΔT = 0.5) | 5 | |
| 72 | 7:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
Results:
- none of the amplifications led to yields of DNA sufficient for Gibson Assembly, therefore the used combination of primers seems not to be sufficient for amplification of 22.0 kbp
13-08-2013
Amplification from FS_02 to FS_11_short; 22.8 kb
- Reaction
| Reagent | DelAG |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.0 µl FS_02 |
| Primer rev | 4.0 µl FS_11_short |
| DMSO | 2 µl |
| Phusion Ready Mix | 10 µl |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 72.0 - 67.0 (ΔT = 0.5) ↓ 0.5 | 5 | |
| 72 | 7:20 | |
| 18 | 98 | 1 |
| 70.0 - 65.0 (ΔT = 0.5) | 5 | |
| 72 | 7:20 | |
| 1 | 72 | 10min |
| 1 | 10 | inf |
15-08-2013
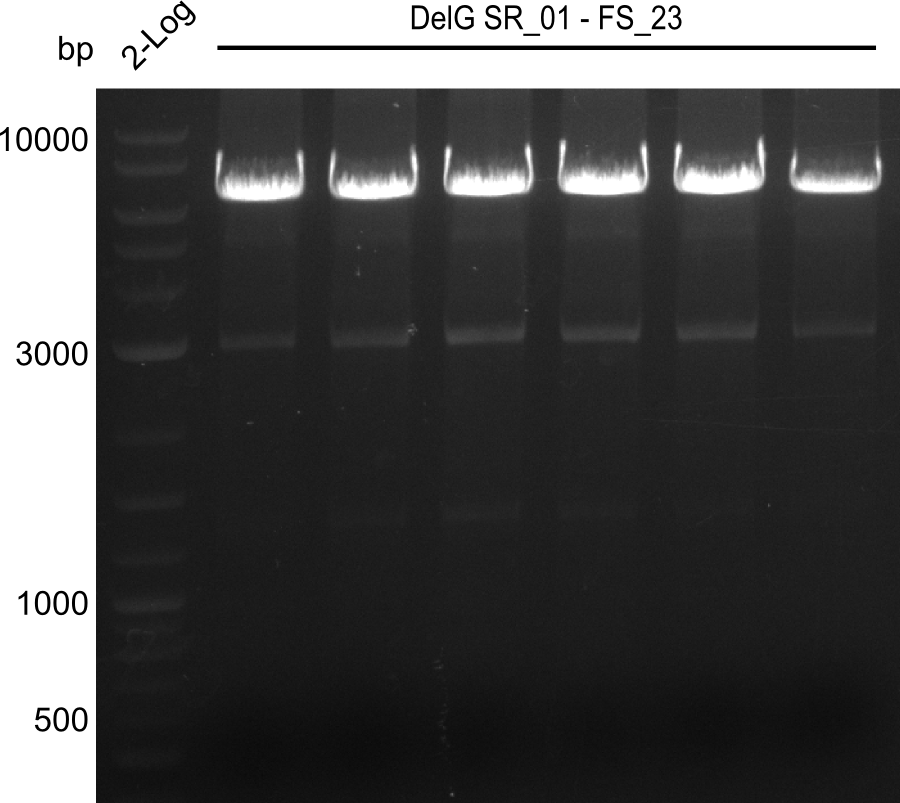
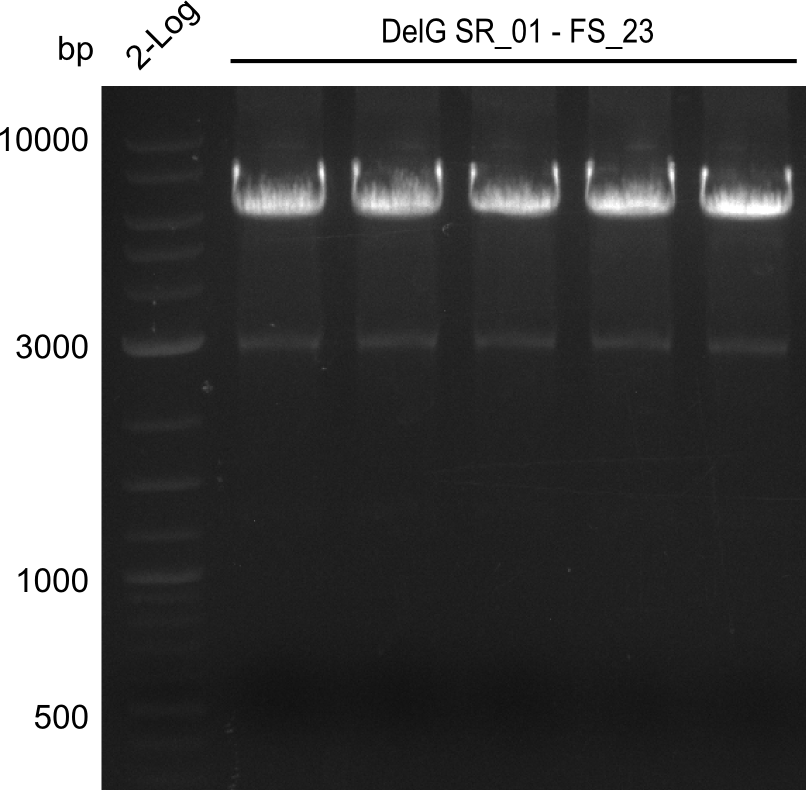
Amplification from SR_01 to FS_23; 6.2 kb
- Reaction (5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| SR_01: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- Bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- Amplification will be repeated to increase the concentration for the Gibson Assembly
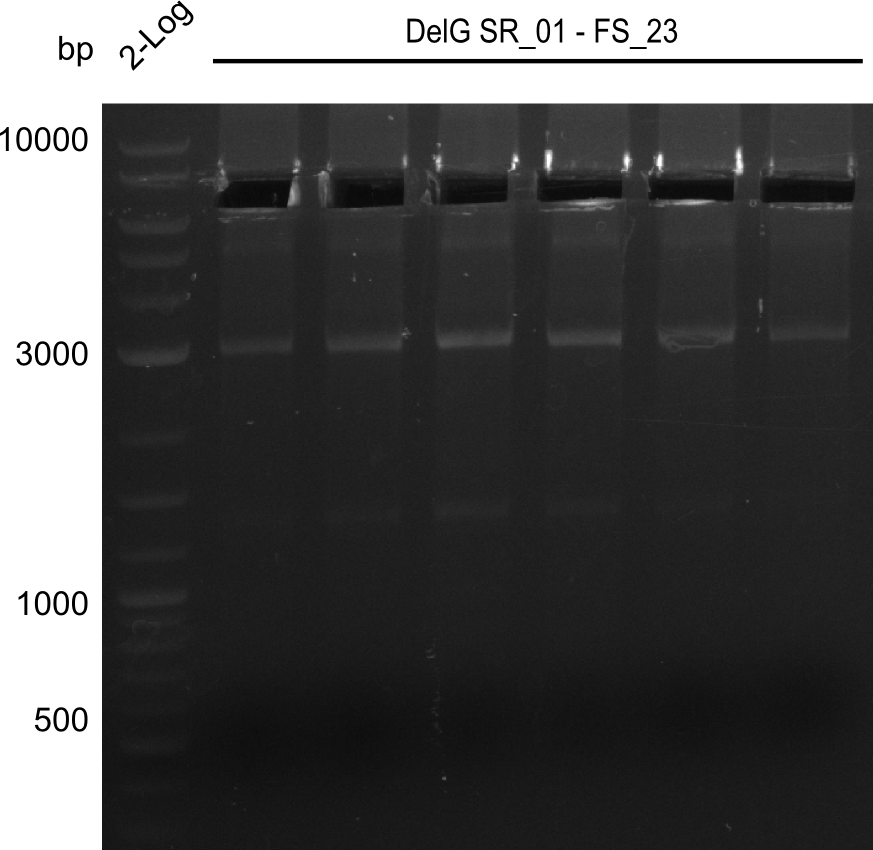
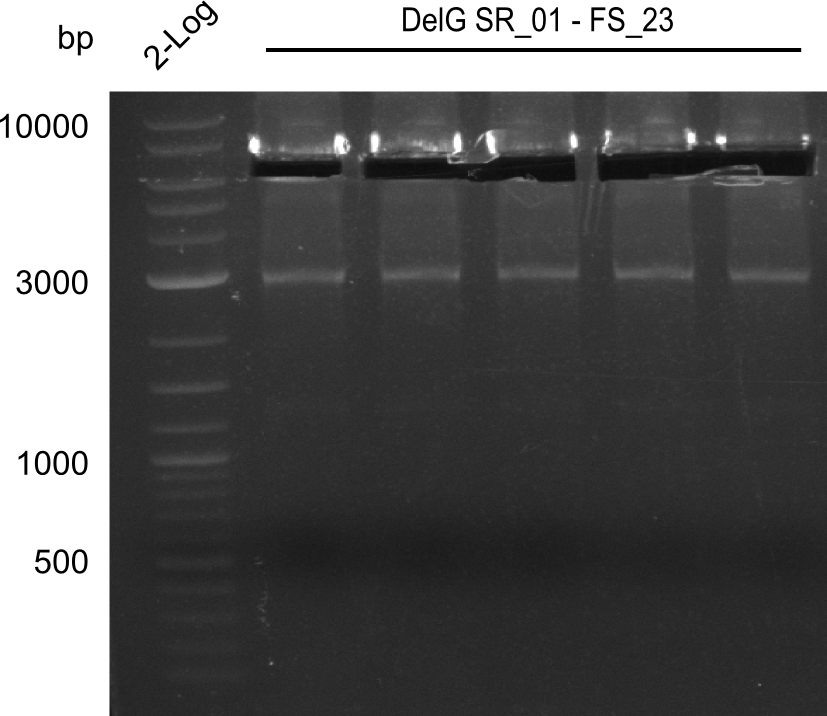
16-08-2013
Concentration measurement
the fragment was gel purified
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelG | SR01-FS23 | 15-08-2013 | 52.6 ng/µl |
| DelG after nucleotide removal | SR01-FS23 | 15-08-2013 | 30 ng/µl |
Amplification from SR_01 to FS_23; 6.2 kb
- Reaction (5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| SR_01: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 3:20 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 3:20 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelG was successful
- Bands were cut out and DNA purified using QIAquick Gel Extraction Kit
12-08-2013
Amplification I from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP | |
|---|---|---|
| Template | D.acidovorans SPH-1 colony | 1µl FS_22-FS_13_short 10-08 |
| Primer fw | 2 µl FS_22 | |
| Primer rev | 2 µl FS_13 | |
| Phusion Ready Mix | 10 µl | |
| dd H2O | 5 µl | |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP did not work
- PCR will be repeated at very high annealing temperatures to ensure that primers with very complex secondary structure such as the Gibson-Primer FS_13long are able to bind
Amplification II and III from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP |
|---|---|
| Template | 1µl FS_22-FS_13_short 10.08 |
| Primer fw | 4 µl FS_22 |
| Primer rev | 4 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
- Conditions
| Biorad MyCycler | ||||||
|---|---|---|---|---|---|---|
| Cycles | temperature [°C] DelOP II | Time | Cycles | temperature [°C] DelOP III | Time | |
| 1 | 98 | 10 s | 1 | 98 | 10 s | |
| 30 | 98 | 1 s | 12 | 98 | 1 s | |
| 74.0 - 70.0 (ΔT = 2.0) ↓ 0.5 | 5 s | |||||
| 72 | 1:00 min | 72 | 3:15 min | |||
| 18 | 98 | 1 s | ||||
| 66 | 5 s | |||||
| 68 / 70 / 72 | 1:00 min | |||||
| 1 | 72 | 10 min | 1 | 72 | 10 min | |
| 1 | 10 | inf | 1 | 10 | inf | |
Results:
- Amplification of DelOP was successful, 2-step PCR with FS_22-FS_13s as template led to expected bands as well as a slight smear and an unintended product
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be repeated to obtain more sample
Re-PCR from FS_22 to FS_13; 2.7 kb; 10-08-2013)
3x 20µl
- Reaction
| Reagent | DelOP |
|---|---|
| Template | 1µl FS_22-FS_13_short 10-08-2013 |
| Primer fw | 4 µl FS_22 |
| Primer rev | 4 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:05 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successfull
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- gradient PCR with annealing at 74°C, 72°C and 70°C will be carried out to increase yield
Amplification V from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4.5 µl FS_22 |
| Primer rev | 4.5 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 75.0 - 72.0 (ΔT = 0.5) | 1:05 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
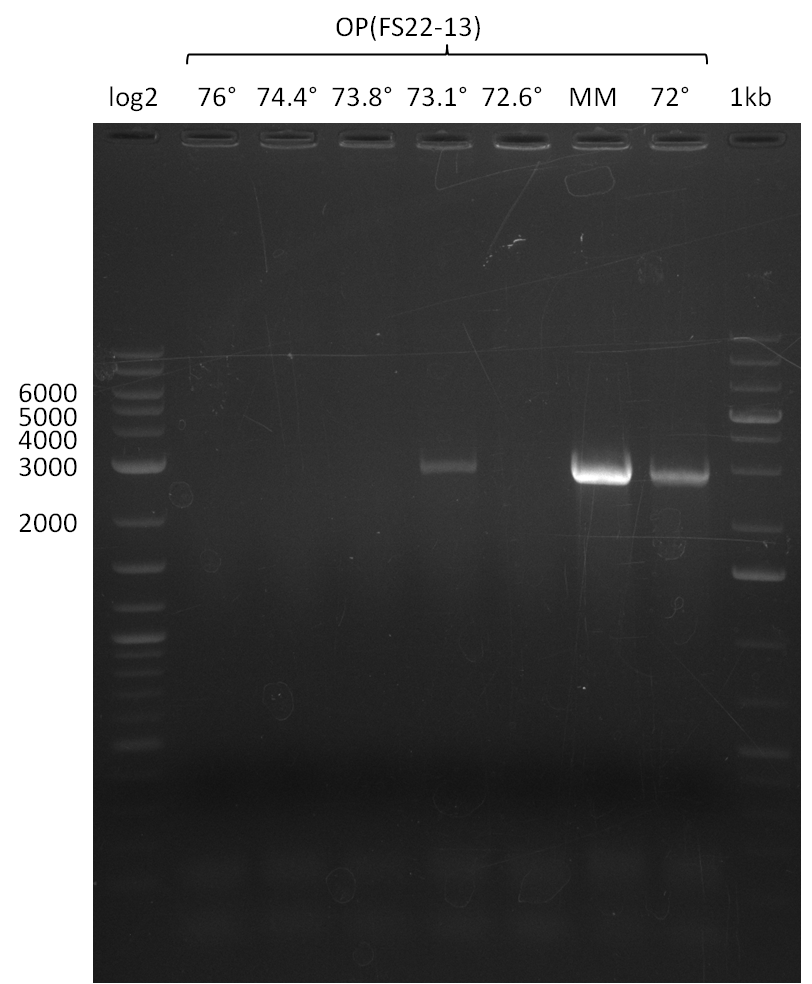
- Amplification of DelOP was successfull
- The gradient PCR shows that amplification worked best with an annealing temperature of 72.3°C, interestingly this annealing temperature was used for the remaining mastermix of the gradient PCR which included more DMSO than the other samples as mixing of DMSO often is not perfect and therefore the very last sample is of higher DMSO concentration
- Therefore PCR will be repeated with an annealing temperature of 72.3°C and 10% DMSO
13-08-2013
Restriction digest of fragment FS_22 to FS_13; 2.7 kb; 12-08-2013) with EcoRI-HF
Incubation at 37°C for 2 hours
| what | µl |
|---|---|
| FS_22 to FS_13 (12-08-2013) | 20 |
| EcoRI-HF | 1 |
| CutSmart Buffer | 2.5 |
| dd H2O | 1.5 |
| Expected fragment lengths [bp] | 1883, 960 |
Results:
- restriction digest shows the expected bands, therefore validation if PCR amplicon is sufficient for Gibson Assembly
Amplification from FS_22 to FS_13; 2.7 kb
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4 µl FS_22 |
| Primer rev | 4 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1/2 µl |
| dd H2O | 1/- µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 1:05 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successfull with 10% DMSO leading to one specific band of intended size
- PCR will be repeated with 10% DMSO to obtain the amount of product which is necessary for Gibson Assembly and test restriction digest
14-08-2013
Concentration measurement DelOP
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
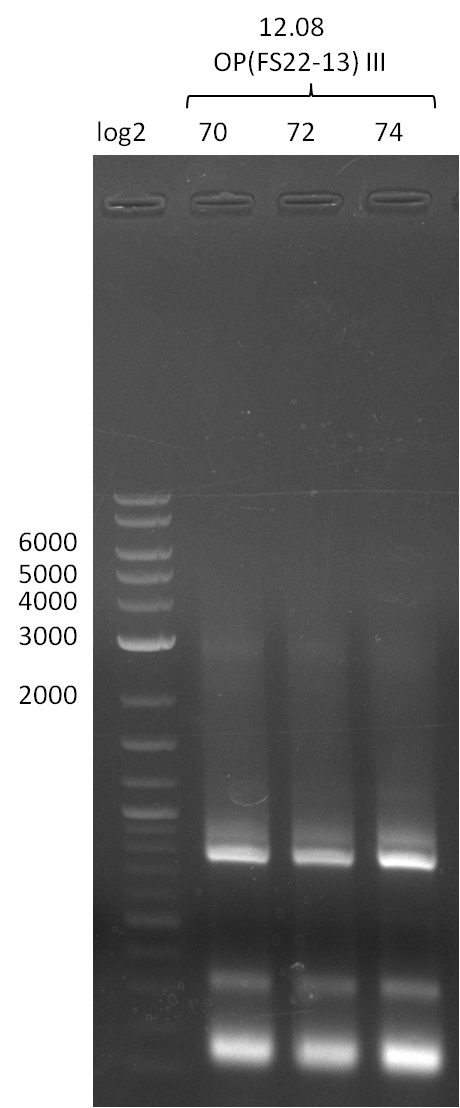
| DelOP | FS22-FS13 | 12-08-2013 | 11 ng/µl |
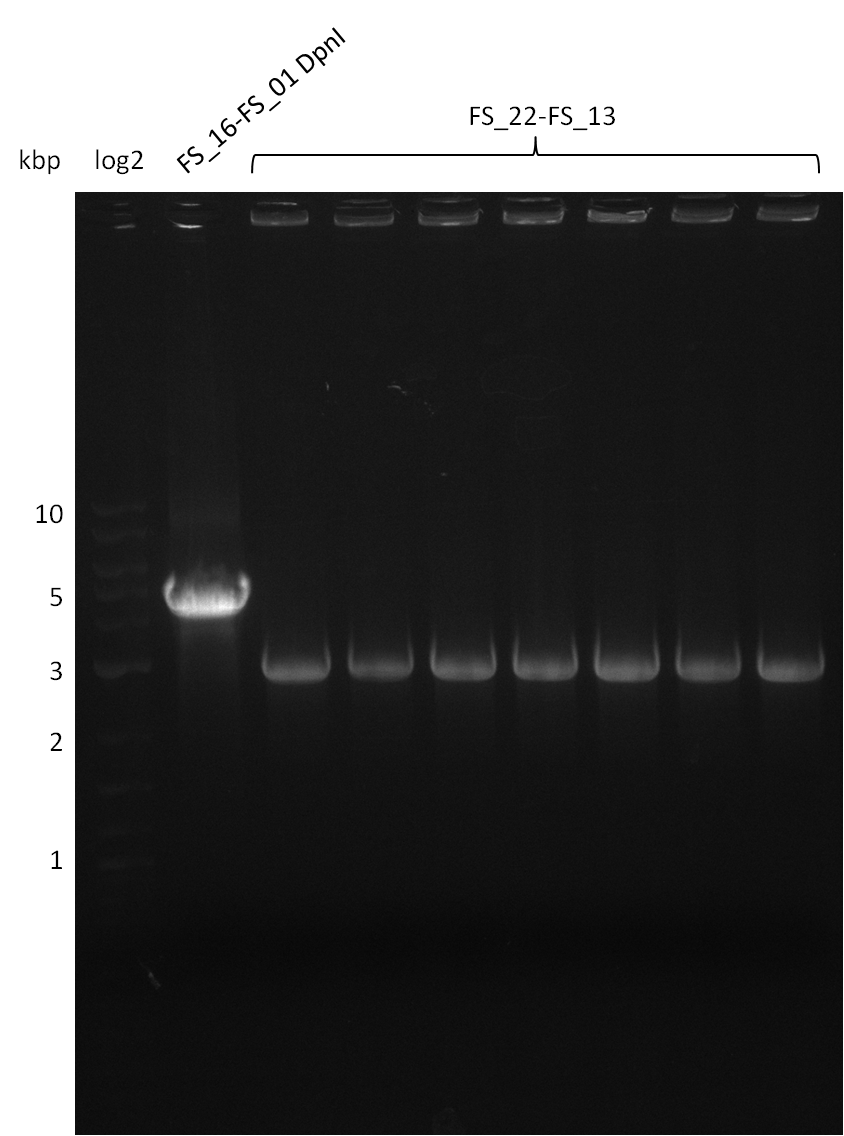
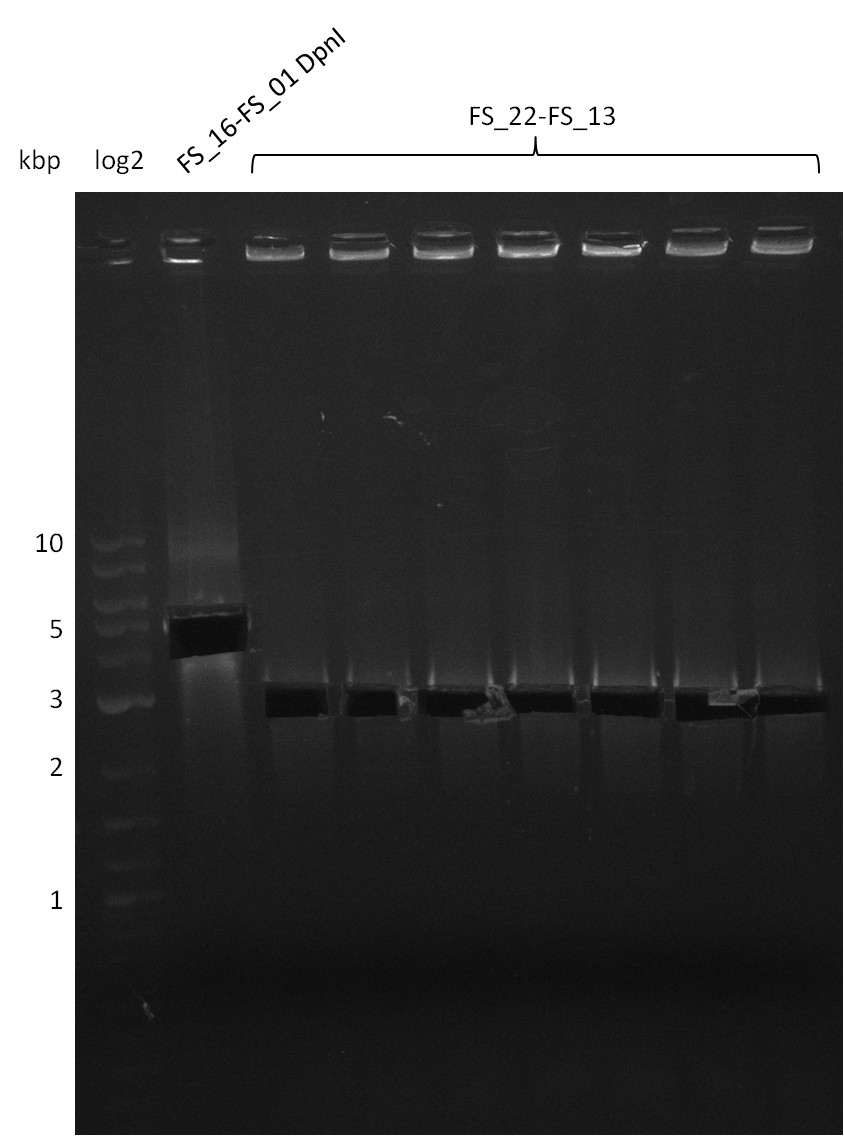
17-08-2013
Amplification from FS_22 to FS_13; 2.7 kb
6x20 µl
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 2.5 µl FS_22 |
| Primer rev | 2.5 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 2 µl |
| dd H2O | 3 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
18-08-2013
Amplification from FS_22 to FS_13; 2.7 kb
6x20 µl
- Reaction
| Reagent | DelOP |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 2.5 µl FS_22 |
| Primer rev | 2.5 µl FS_13 |
| Phusion Ready Mix | 10 µl |
| DMSO | 2 µl |
| dd H2O | 3 µl |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 1:00 | |
| 1 | 72 | 5 min |
| 1 | 10 | inf |
Results:
- Amplification of DelOP was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
15-08-2013
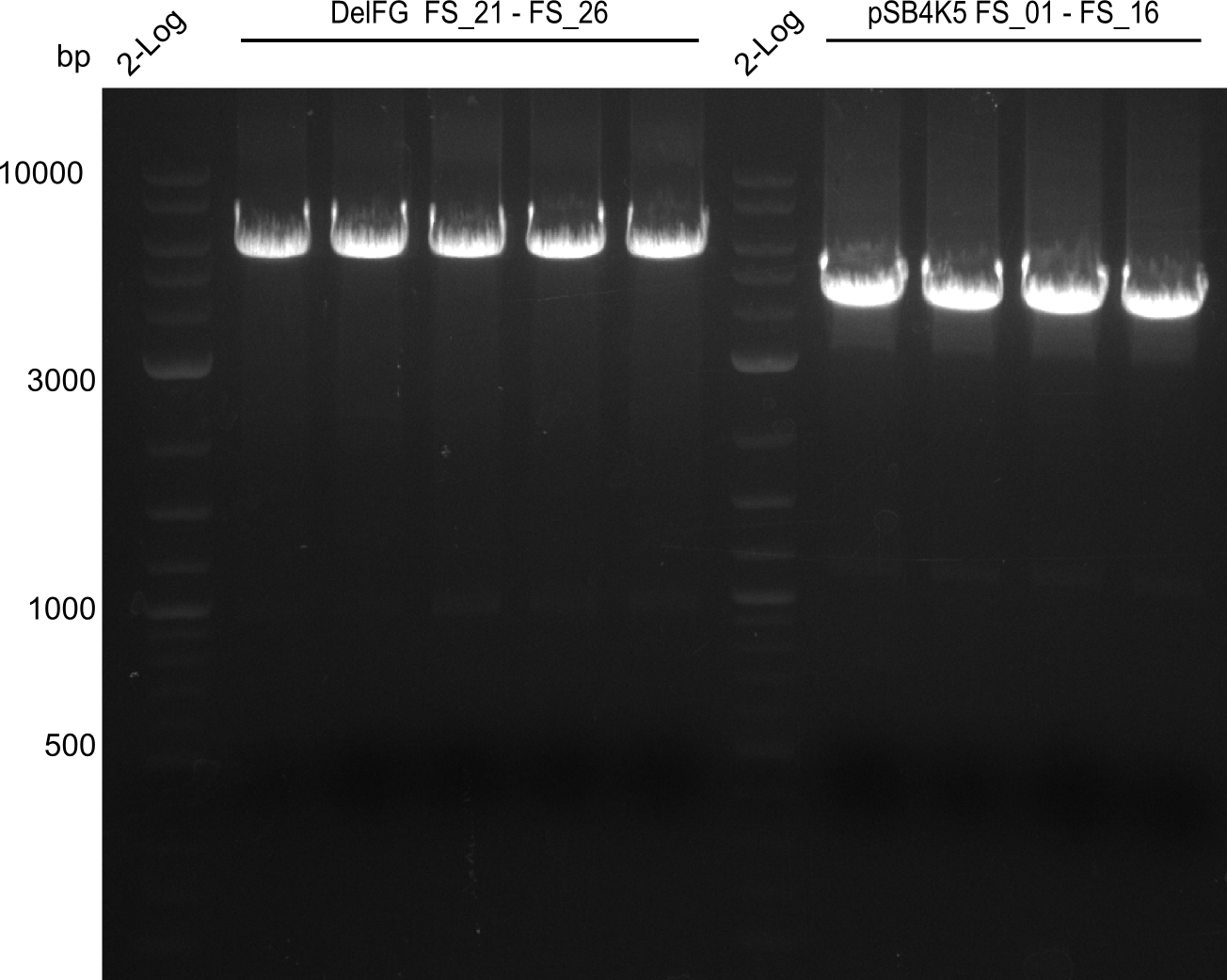
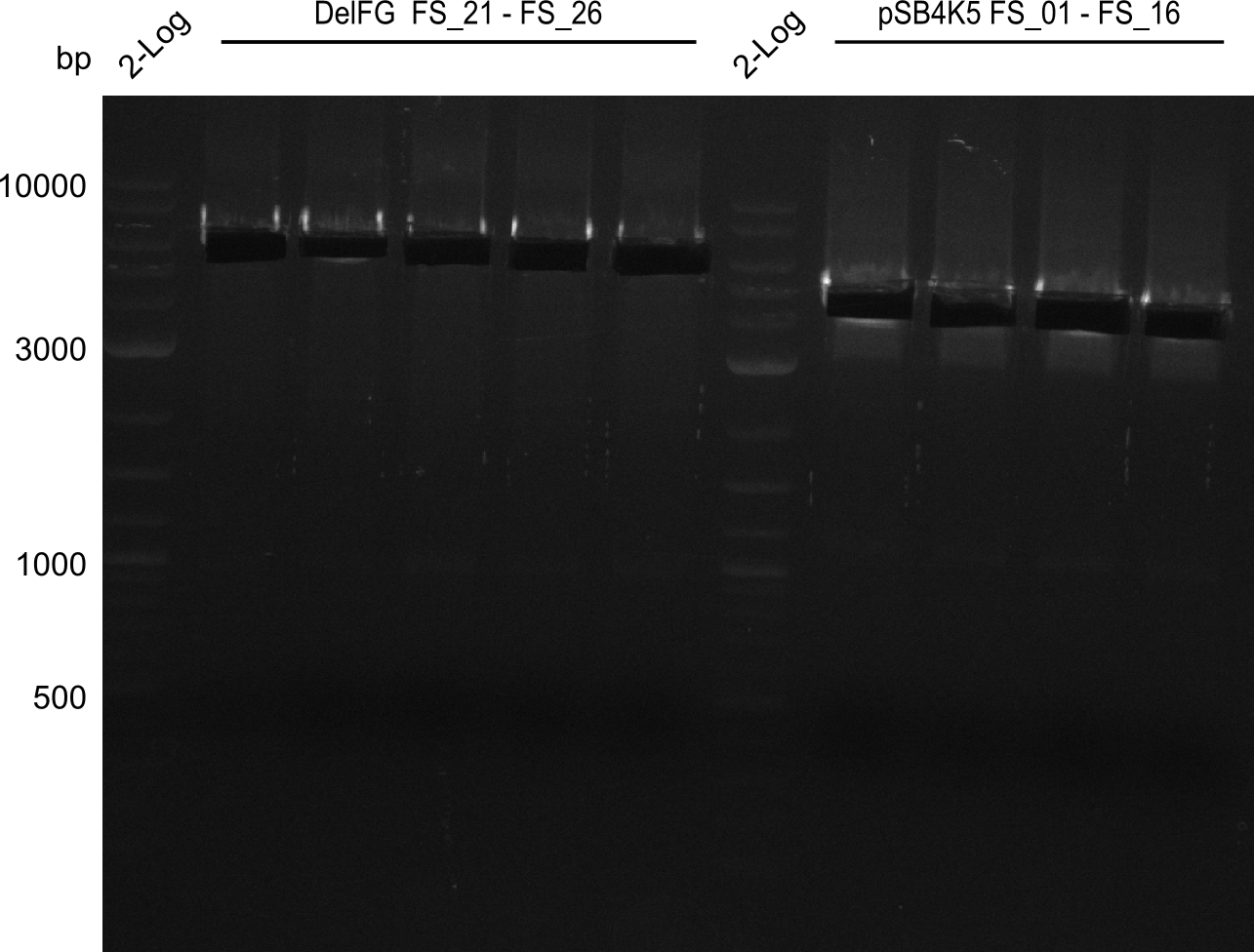
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(2x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
Restriction digest of pSB4K5 (FS_01 to FS_16; 4.2 kb; 15-08-2013) with DpnI
Incubation at 37°C for about 6 hours
| what | µl |
|---|---|
| FS_16 to FS_01 (15-08-2013) | 30 |
| DpnI | 2 |
| CutSmart Buffer | 4 |
| dd H2O | 4 |
Results:
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
16-08-2013
Concentration measurement
The concentration of the gel purified, and DpnI digested fragment was measured using a NanoDrop Instrument.
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| pSB4K5 DpnI digested | FS01-FS16 | 15-08-2013 | 30 ng/µl |
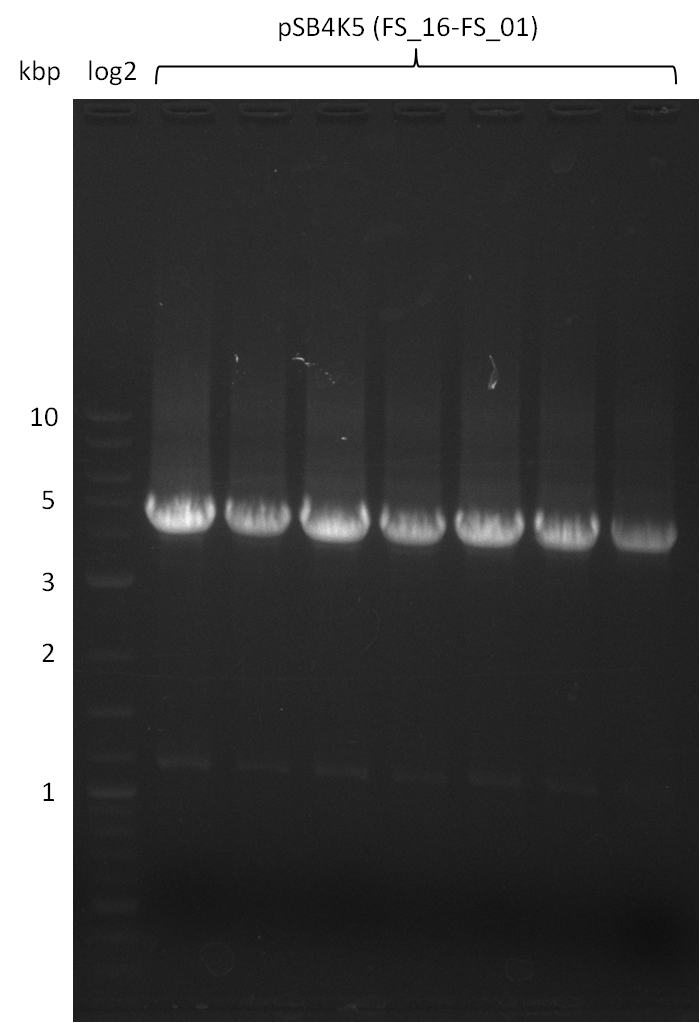
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(3x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
17-08-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(3x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Results:
- Amplification worked very well, we had bright bands on the right height.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
Restriction digest of pSB4K5 (FS_01 to FS_16; 4.2 kb; 17-08-2013) with DpnI
Incubation at 37°C for about 6 hours
| what | µl |
|---|---|
| FS_16 to FS_01 (17-08-2013) | 30 |
| DpnI | 2 |
| CutSmart Buffer | 4 |
| dd H2O | 4 |
Results:
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
18-08-2013
Amplification from FS_01 to FS_16; 4.2 kb
- Reaction
(3x50µl)
| what | µl |
|---|---|
| Template pSB4K5 | 1 |
| FS_01: (1/10) | 2.5 |
| FS_16: (1/10) | 2.5 |
| Phusion Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 16.5 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 62 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 5 min |
| 1 | 8 | inf |
Restriction digest of pSB4K5 (FS_01 to FS_16; 4.2 kb; 18-08-2013) with DpnI
Incubation at 37°C for about 6 hours
| what | µl |
|---|---|
| FS_16 to FS_01 (18-08-2013) | 30 |
| DpnI | 2 |
| CutSmart Buffer | 4 |
| dd H2O | 4 |
Results:
- Digested fragment was run on a gel.
- Bands were cut out and DNA purified using QIAquick Gel Extration Kit.
13-08-2013
Amplification from FS_21 to FS_07; 5.2 kb
- Reaction
| Reagent | µl |
|---|---|
| Template | D.acidovorans SPH-1 colony |
| Primer fw | 4 µl FS_21 |
| Primer rev | 4 µl FS_07 |
| Phusion Ready Mix | 10 µl |
| DMSO | 1 µl |
| dd H2O | 1 µl |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 2:00 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG worked, but amount of DNA was insufficient
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- PCR will be carried out with another cylcer which by experience of the last amplifications led to higher product yields.
15-08-2013
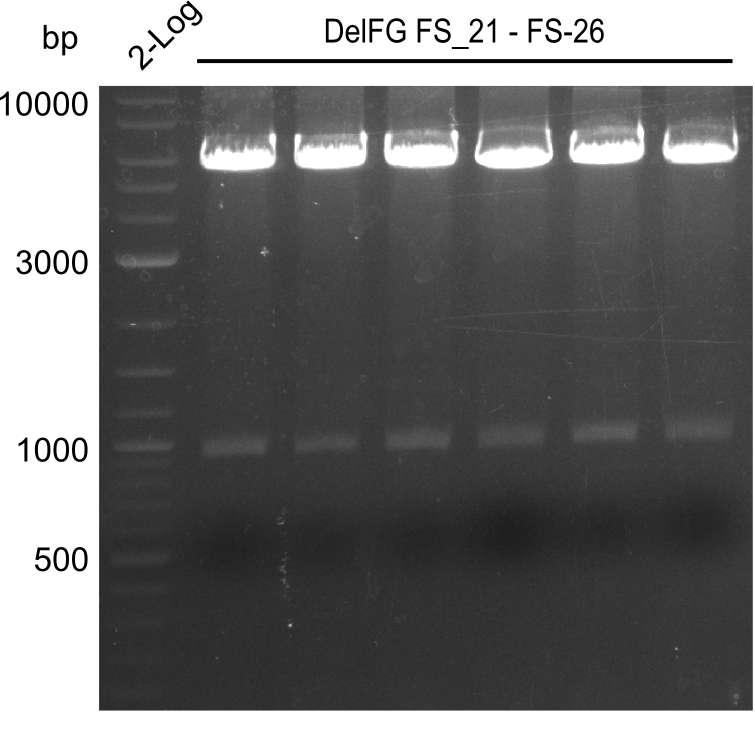
Amplification from FS_21 to FS_26 ; 5.5 kb
- Reaction
(5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- obtained DNA will consequently be used for Gibson Assembly
- PCR will be repeated to get higher amounts of DNA and increase concentration by purifying several gel slices using the same QIAquick spin column
16-08-2013
Concentration measurement DelFG
all fragments were gel purified, FG were additionally purified with the nucleotide removal kit
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| DelFG | FS21-FS26 | 29-07-2013 | 61.4 ng/µl |
| DelFG after nucleotide removal | FS21-FS26 | 29-07-2013 | 30 ng/µl |
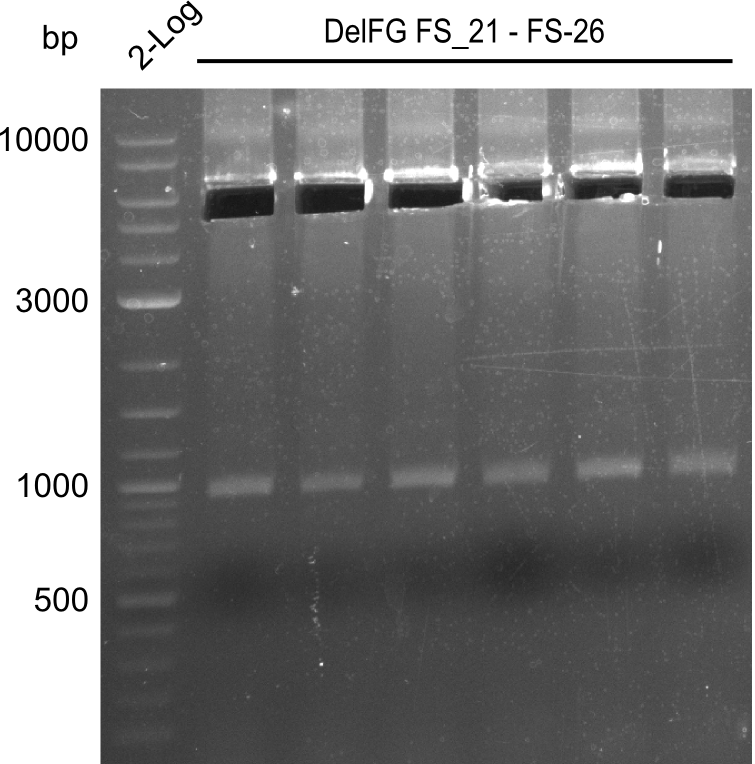
Amplification from FS_21 to FS_26 ; 5.5 kb
- Reaction
(5x20 µl)
| what | µl |
|---|---|
| D. acidovorans SPH-1 colony | 1 |
| FS_21: (1/10) | 2 |
| FS_26: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:15 | |
| 1 | 72 | 10 min |
| 1 | 10 | inf |
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- obtained DNA will consequently be used for Gibson Assembly
15-08-2013
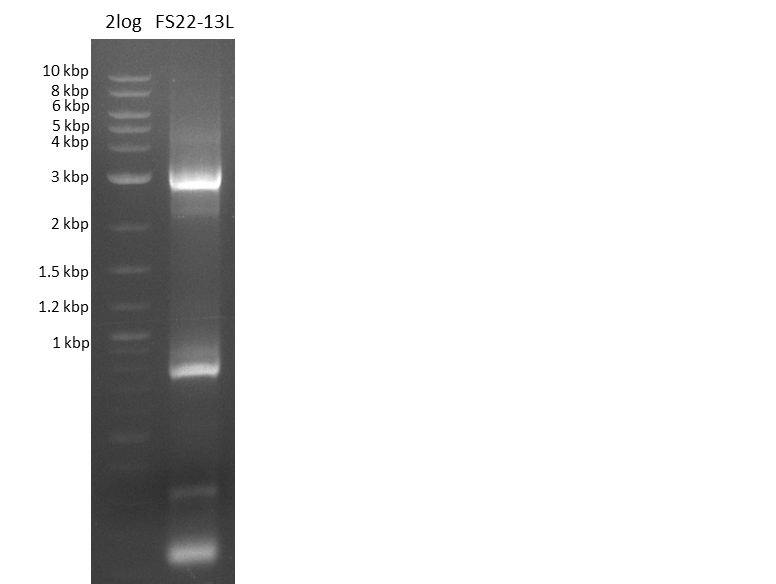
Gibson Assembly and electroporation
Assembly of our fragments
Those gel-purified fragments were added for the Gibson assembly of our construct pFSN.
| Fragment | Primer | Date | Concentration (ng/µl) | Volume (µl) |
|---|---|---|---|---|
| pSB4K5 | FS_01 - FS_16 | 14-08-13 | 13 | 1.35 |
| AF | FS_02 - FS_05 | 13-08-13 | 49.75 | 2.65 |
| FG | FS_21 - FS_26 | 07-08-13 | 24.5 | 2.69 |
| G | FS_35_SR_01 - FS_23 | 08-08-13 | 34.9 | 2.1 |
| OP | FS_22 - FS_13 | 13-08-13 | 36.3 | 0.89 |
| L | FS_14 - FS_15 | 07-08-13 | 51 | 0.31 |
| Gibson MasterMix NEB | 2x | 10 | ||
- Gibson Assembly according to protocols of NEB, 1h at 50°C
Gibson backbone (pSB4K5)
The backbone pSB4K5 was also incubated with the Gibson mastermix. We will use this as one negative control in the electroporation to test the efficiency.
| Fragment | Date | Concentration (ng/µl) | Volume (µl) |
|---|---|---|---|
| pSB4K5 | 14-08-13 | 13 | 1.35 |
| H2O | 8.65 | ||
| Gibson MasterMix NEB | 2x | 10 | |
- Gibson Assembly according to protocols of NEB, 1h at 50°C
Electroporation
The following samples were electroporated
| mixture | volume used | cells | |
|---|---|---|---|
| 5 µl Gibson Assembly of pFSN construct | 10 µl H2O | 1 µl | 50 µl DH10β old |
| 5 µl Gibson Assembly of pFSN construct | 10 µl H2O | 14 µl | 50 µl DH10β old |
| 5 µl Gibson Assembly of Backbone (control) | 10 µl H2O | 1 µl | 50 µl DH10β old |
| pSB4K5 14-08-13 | 1.35 µl | 50 µl DH10β old | |
| pSB6A1 Hanna [69.57 ng/µl] 1:10 | 1 µl | 50 µl DH10β Fanny new | |
- Cells were transdered into 400 µl SOC-medium (NEB) after electroporation
- incubation and growth of cells for 1 hour at 37°C
- cells were centrifuged for 3 min at 6000rpm, supernatant discarded and cells resuspended in remaining SOC-medium
- cells were plated on agar plates containing kanamycine, 10µl on one plate, remaining µl on another plate for electroporation of the construct pFSN and on agar plates containing ampicillin for electroporation of the control backbone pSB6A1
20130816ElectroporationA.JPG
1 µl of diluted Gibson electroporated; 10 µl of E.coli plated |
20130816ElectroporationB.jpg
1 µl of diluted Gibson electroporated; Rest of E.coli plated |
20130816ElectroporationC.jpg
14 µl of diluted Gibson electroporated; 10 µl of E.coli plated |
20130816ElectroporationD.jpg
14 µl of diluted Gibson electroporated; Rest of E.coli plated |
P1010689.JPG
1 µl of diluted Gibson (backbone control) electroporated; 10 µl of E.coli plated |
P1010690.JPG
1 µl of diluted Gibson (backbone control) electroporated; Rest of E.coli plated |
P1010692.JPG
1 µl of backbone control electroporated; 10 µl of E.coli plated |
P1010693.JPG
1 µl of backbone control electroporated; Rest of E.coli plated |
P1010704.JPG
Test of electrocompetence DH10β with 1 µl pSB6A1; 10 µl of E.coli plated |
P1010705.JPG
Test of electrocompetence DH10β with 1 µl pSB6A1; Rest of E.coli plated |
16-08-2013
Colony PCRs
As a lot of colonies grew we screened if DelL and pSB4K5 are present.
Different probes:
- A: 1 µl of diluted Gibson electroporated; 10 µl of E.coli plated
- B: 1 µl of diluted Gibson electroporated; Rest of E.coli plated
- C: 14 µl of diluted Gibson electroporated; 10 µl of E.coli plated
- D: 14 µl of diluted Gibson electroporated; Rest of E.coli plated
- Only red colonies were chosen for screening
- The expected amplicon size is 1.5 kb
- Reaction mixture
| Reagent | A1, A2 | B1-B5 | C1-C3 | D1-D40 |
|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | 2 red colonies A (A1, A2) | 5 red colonies B (B1-B5) | 3 red colonies C (C1-C3) | 40 red colonies D (D1-D40) |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl |
- PCR Conditions
--> T100
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 12 | 95 | 1:00 |
| 66 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 95 | 1:00 |
| 63 | 5 | |
| 72 | 1:30 min | |
| 1 | 12 | inf |
Result:
- None of the screened colonies led to the correct amplicon, therefore the correct assembly has not worked in any of the white colonies
- screening will be repeated, with white colonies as template, as bacteria including the correct 32 kbp backbone might not be capable of expressing mRFP anymore due to the large insert between lac promotor and mRFP
17-08-2013
Colony PCRs
Different probes:
- A 1 µl of diluted Gibson Assembly electroporated; 10 µl of E.coli plated
- B 1 µl of diluted Gibson Assembly electroporated; remaining E.coli plated
- C 14 µl of diluted Gibson Assembly electroporated; 10 µl of E.coli plated
- D 14 µl of diluted Gibson Assembly electroporated; remaining E.coli plated
- Only white colonies where chosen for screening
- The expected amplicon size is 1.5 kb
- Reaction mixture
| Reagent | B1w | C1w | D1w-D10w |
|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl |
| Colonies | 1 white colony B (B1w) | 1 white colony C (C1w) | 10 white colonies D (D1w-D10w) |
| DreamTaq | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl |
- PCR Conditions
--> T100
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 12 | 95 | 1:00 |
| 66 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 95 | 1:00 |
| 63 | 5 | |
| 72 | 1:30 min | |
| 1 | 12 | inf |
Result:
- Colony D8w shows expected amplicon of 1.5kbp
- The other colonies were screened negative.
- D8w will be transfered to liquid culture to repeat screening PCR as well as for further validation beeing screening for the other insert fragments as well as test restriction digest
- Furthermore another electorporation will be carried out to yield more clones positive for screening with the Primers VR and FS_14, therefore the remaining 10µl of the Gibson assembly will be purified using isopropanol precipitation
Electroporation
We electroporated the Gibson assembly (15-08) again to increase the chance of a positive clone. We purified it by isopropanol purification.
| mixture | volume used | Cells | |
|---|---|---|---|
| Assembled fragments (15-08) isopropanol precipitated | 15 µl | 50 µl DH10β | |
- Cells were transdered into 400 µl SOC-medium (NEB) after electroporation
- incubation and growth of cells for 1 hour at 37°C
- cells were centrifuged for 3 min at 6000rpm, supernatant discarded and cells resuspended in remaining SOC-medium
- cells were plated on agar plates containing kanamycine, 10µl on one plate, remaining µl on another plate for electroporation of the construct pFSN
P1010701.JPG
15 µl of Gibson 15-08 (isopropanol precipitated) electroporated; 10 µl of E.coli plated |
P1010702.JPG
15 µl of Gibson 15-08 (isopropanol precipitated) electroporated; Rest of E.coli plated |
18-08-2013
Colony PCRs
Different probes:
- D: 14 µl of diluted Gibson Assembly electroporated; remaining E.coli plated
- E: 1 µl isopropanol precipitated Gibson Assembly (15-08) ; 10 µl of E.coli plated
- F: 1 µl isopropanol precipitated Gibson Assembly (15-08); remaining E.coli plated
- The expected amplicon size is 1.5 kb
- 3 colonies were sceened in one PCR
- Reaction mixture
| Reagent | D8w | D11w-D24w | E1w-E19w | F1w-F20w | E1r-E10 | F1r-F20r |
|---|---|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | Colony D8w liquid culture | 24 white colonies D (D1w-D24w) | 19 white colonies E (E1w-E19w) | 30 white colonies F (F1w-F20w) | 10 red colonies E (E1r-E10r) | 20 red colonies F (F1r-F20r) |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
- PCR Conditions
--> T100
| Cycles-PCR | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 2:00 |
| 12 | 95 | 1:00 |
| 66 ↓ 0.5 | 5 | |
| 72 | 1:30 min | |
| 18 | 95 | 1:00 |
| 63 | 5 | |
| 72 | 1:30 min | |
| 1 | 12 | inf |
Result:
- colony D8w again showed the expected amplicon in the colony PCR and will therefore be further analyzed for the other fragments as well as restriction digested and partially sequenced as soon as screening primers arive
- Furthermore colonies E16w and F25w are screened positive for the fragment DelL
19-08-2013
Concentration measurement
The concentration of the gel purified, and DpnI digested fragments was measured using a NanoDrop Instrument.
| Fragment | Primer | Date PCR | Concentration |
|---|---|---|---|
| pSB4K5 DpnI digested | FS_01-FS_16 | 17-08-2013 | 101.2 ng/µl |
| pSB4K5 DpnI digested | FS_01-FS_16 | 18-08-2013 | 159.2 ng/µl |
19-08-2013
Colony PCRs for testing ligation of pSB4K5 and DelL
- Reaction mixture
| Reagent | E13w | E14w | E15w | F25w | F26w | F27w |
|---|---|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | Colony E13w liquid culture | Colony E14w liquid culture | Colony E15w liquid culture | Colony F25w liquid culture | Colony F26w liquid culture | Colony F27w liquid culture |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
Result:
- Colony F26w was screened positive, as it shows the expected band at 1.4kbp, the others were screened negative
- 8 ml of colony D8w and colony D26w (pFSN-1) were midi-preped according to protocol 'Preparation of large plasmids'.
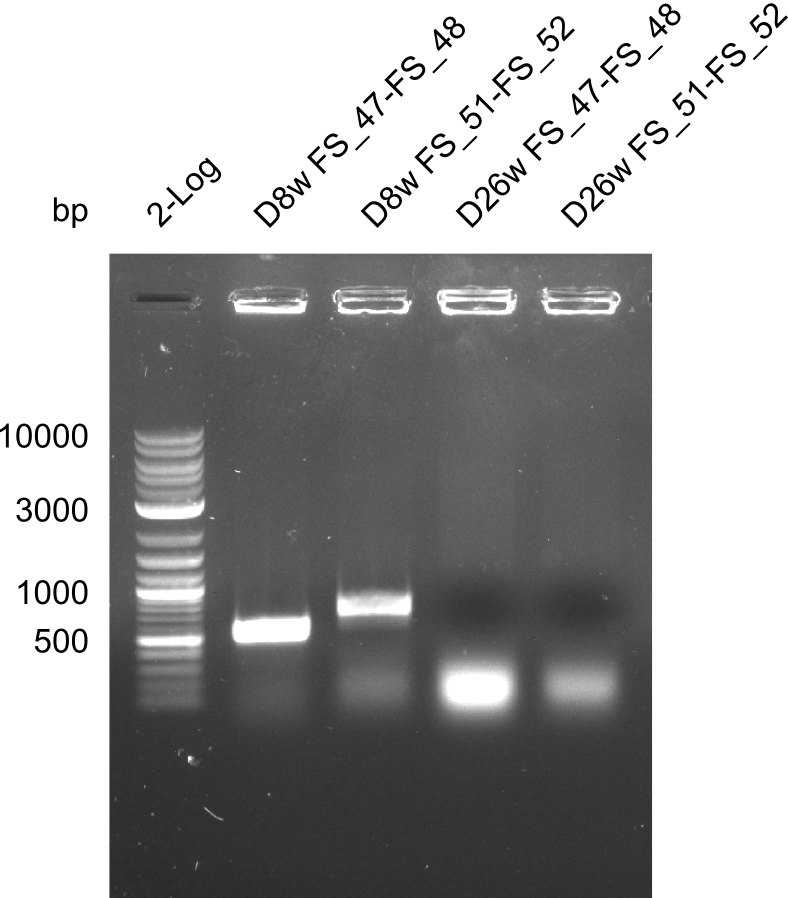
Colony PCRs for testing ligation of pSB4K5 and DelAF, as well as DelAF and DelFG
- Reaction mixture
| Reagent | D8w | D8w | F26w | F26w |
|---|---|---|---|---|
| Primer_fw (1/10) | 2 µl FS_47 | 2 µl FS_51 | 2 µl FS_47 | 2 µl FS_51 |
| Primer_rv (1/10) | 2 µl FS_48 | 2 µl FS_52 | 2 µl FS_48 | 2 µl FS_52 |
| Colonies | Colony D8w liquid culture | Colony D8w liquid culture | Colony F26w liquid culture | Colony F26w liquid culture |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl |
| Expected band | 547 bp | 724 bp | 547 bp | 724 bp |
Results:
- The screening was positive for clone D8w with both primer combinations.
- Thus this clone contains the backbone pSB4K5, the genes DelA, DelB, DelC, DelD, DelE, DelF, DelL and at least a part of the gene DelG.
- For colony F26w the screenings were negativ.
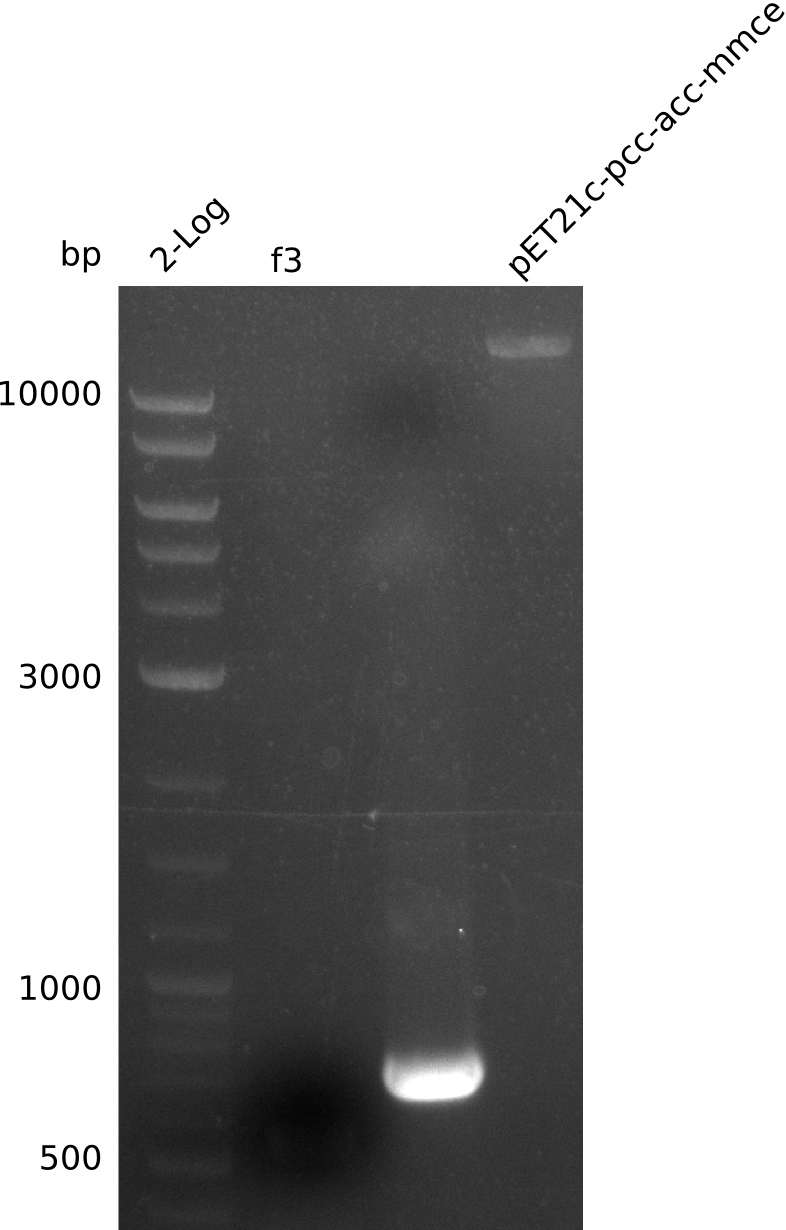
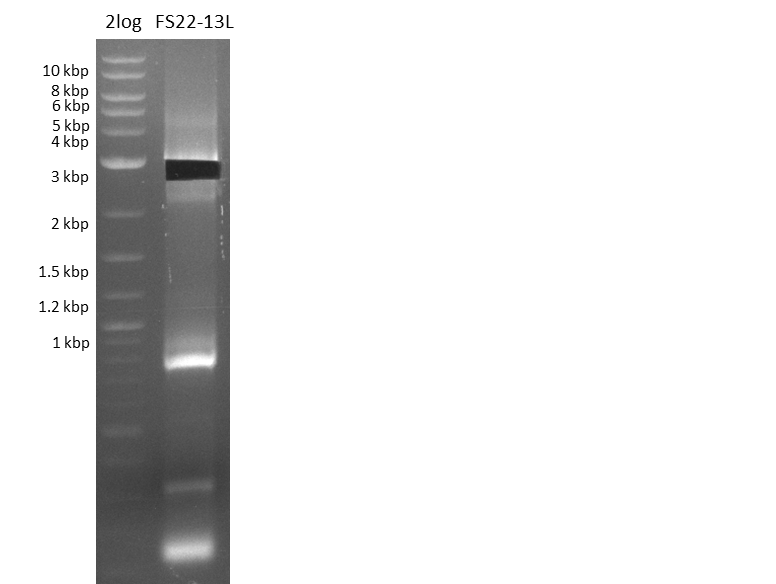
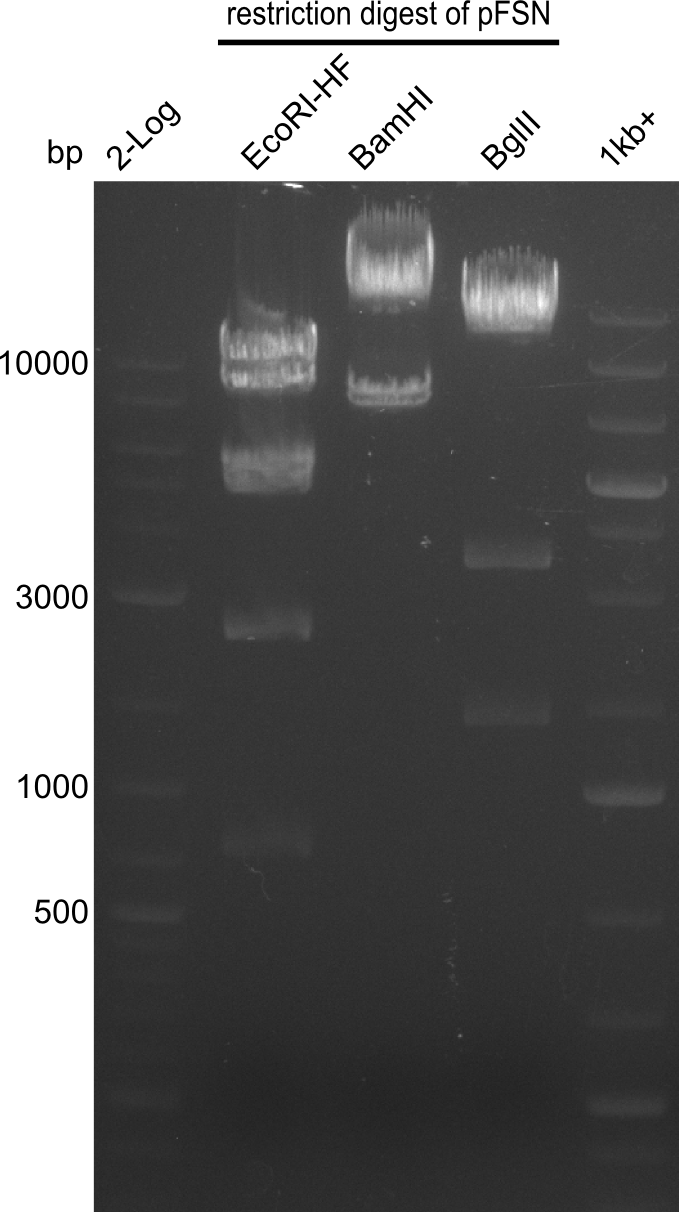
Restriction digest of pFSN (31.435 kbp)
Incubation at 37°C for about 6 hours
| Reagent | µl | ||
|---|---|---|---|
| pFSN | 1 | ||
| EcoRI | BamH1 | BglII | 2 |
| CutSmart Buffer | 0.5 | ||
| dd H2O | 1.5 | ||
| Expected fragment lengths [bp] EcoRI | 9798, 7856, 5276, 4624, 2529, 1212 | ||
| Expected fragment lengths [bp] BamHI | 23860, 7435 | ||
| Expected fragment lengths [bp] BglII | 14223, 11919, 3291, 1892 | ||
Results:
- All digests displayed the expected bands and thus were positive.
- Therefore the pFSN plasmid is very likely our desired construct.
20-08-2013
Sequencing
- The construct pFSN was isolated from overnight culture using the 'Preparation of large plasmids' protocol.
- Single read sequencing was carried out by GATC Biotech
- obtained .abi files were analyzed with UniPro Ugene, part of sequence showing convenient chromatogram was chosen for alignment using ClustalOmega
Sequence-Files from GATC Biotech
Alignments
Results:
- Sequencing of the final construct pFSN for all internal Gibson sites validated the intended sequence
- Sequence of mRFP shows mutations within the primer site, therefore it can be concluded that mRFP is not functional
- Colonies of D8w will be further analyzed by FACs to investigate whether mRFP is functional or not
- Additionaly, SDS-page of clone D8w will be conducted to check for expression of the inserted Del genes
- Concluding the results of colony PCRs, restriction digests and sequencing it can be concluded, that we successfully amplified 28 kbp of genomic DNA from our donator organism Delftia acidovorans SPH-1 distributed from the DSMZ, ligated the thereby obtained 6 fragments including the standard biobrick backbone pSB4K5 sucessfully in the intended order and consequently transformed a plasmid of 32 kbp in total into E. Coli using electroporation
21-08-2013
- Another 8 ml of colony D8w (pFSN-1) were midi-preped according to protocol 'Preparation of large plasmids'.
 "
"