Team:Heidelberg/NRPSDesigner
From 2013.igem.org


NRPS-Designer. Designing custom peptides.
This tool allows you to design any customized peptide you can build using the most common NRPS domains. This gives you the oppurtunity to include non-proteinogenic amino acids and secondary modifications without going through chemical synthesis or tRNA reprogramming. On top of providing you with a target pathway sequence the tool is also integrated with the parts registry to include any further BioBricks and with Gibthon to directly go from NRP design to Gibson cloning strategy within 10 minutes.
You should also read our documentation and our RFC100!
Week 12
In this week we implemented a first draft of the NRPS-Designer algorithm in C++ and designed a databse structure, as well as some scripts for entry making
Week 13
While revising the database structure and the input interface we integrated pfam for semi-automated domain prediction.
Week 14
Since we noticed, that writing the whole interface ourselves will be very nasty and time-consuming we put our whole concept into the framework Django. Meanwhile our database design evolved further an the pfam integration was improved.
Week 15
Besides the regular changes to the database design, we integrated the graphical user interface in Django and also included OpenBable for displaying chemical structures. Furthermore we visualised our concept this week.
Week 16
This week we mainly made changes to the NRP-Design interface and integrated ajax.
Week 17
Besides fixing the relations in our database and improving th interface, we integrated Gibthon - the tool for Gibson primer design developed by the 2011 Cambridge team.
Week 18
This week we started filling our database, using the NRPS-PKS database. Besides that we introduced session data and updated the designer algorithm.
Week 19
As we noticed, that different prediction tools give different domain borders, we further evaluated them and finally integrated antiSMASH
Week 20
This week we started the manual sequence annotation and database curation and integrated the multiple sequence alignment in the database input interface.
Week 21
During this week we integrated our tool with the parts regisrty, did more manual database curation, added more hidden Markov-Models to antiSMASH and improved Gibthon.
Week 22
In order to do request management we introduced celery to our system. Furthermore we optimised the multiple sequence alignment and did lots of manual database curation.
Week 23
In the final week we added several linkouts, including SBOL and GenBank files and fixed lots of bugs.
Methods:
Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet.Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. .
Initial database setup
First of all we began to design a database - which we modified, edited and changed all-over again numerous times - which ought to be capable of saving the data concerning NRPS origins, pathways, modules, specificity and lots of more information on them we needed for a later prediction of domains capable of producing a certain NRP. We focused on saving the data in a non-redundantly and easy to rerieve manner. Therefore we first set up a GitHub repository which contained the SQL-file of our first database-design. For it's implementation we used phpmyadmin, which spared us writing the whole thing in SQL. Creating forms to "easily" fill our newly implemented database we started with - what later turned out to be a quite naive idea - writing php scripts which connected to our MySQL-database using nasty index-files containing root-username and passwort of different localhosts.
Draft of command-line application
We thought of a common XML format for NRPs. Additionally a first command line application was written in C++ for extracting data about an NRP from those XML files.
Pathway finding implemented in C++
The basic C++ code needed for MySQL requests depending on a defined input was written. It fetches all the domains, that fit a certain substrate and calculates a pathway, which would theoretically produce the designed NRP.
Database redesign
The database was redesigned in order to better represent the natural composition of NRPS.
Extensions of user interface
The php-scripts for filling our database were adapted to the new database-structure and customized so that different tables in the database could be filled with them.
Nils finally uses a real operating system
After weeks of agony and trouble with his MacBook, Nils finally decided to follow the advice of master Ilia and made him install [http://aptosid.com/ aptosid] on his computer. Of course, because aptosid (based on Debian Sid) is not as user-friendly as Ubuntu, Nils and Ilia spent quite some time in order to change the background image from Fred the crab (the mascot of aptosid) to something else. Anyway, at least Nils has now a Linux distribution and maybe we will be able to convince our advisor Tim to also get rid of his Mac operating system.
Database redesign
The domains are now saved all in one table and they each have a domain type specified. Due to this setup the database can now also cover modification domains.
GUI extension
The php scripts were extended to allow for entries to the new database. Additionally an XML export funciton was added to the design interface.
Draft of PFAM querying
In the experimental parts, we determined a first rough estimate of NRPS domain boundaries based on Pfam. Thus we also wanted our software to use the Pfam predictions to make it easier to semi-automatically add new domains into our database. A first test Python script was written which queried the Pfam API using the Requests library according to the online documentation and then parsed the XML output. The Biopython library also proved to be of great help.
Concept transferred to django framework
Having distributed diverse tasks e.g. to write php scripts, we figured out that we would be constantly be reinventing the wheel and doing web-development like it was done a decade ago. Thus, we started looking at different web frameworks, which would make a lot of standardized tasks such as user login easy. We then decided to use Django, a Python framework, mainly because of the language. Some of us already knew Python, it is in general a language that is very easy to pick up and the Biopython project is pretty comprehensive. Additionally, a previously very successful iGEM Project (Gibthon) also used Django. Compared to most other choices (except Ruby on Rails, which has the disadvantage of being written in Ruby) it also seemed to be the most mature choice. We also thought of using a Perl or a Haskell framework, such as Yesod, but some team members disliked those languages (Haskellers disliked Perl and vice versa; the fact that the first Perl6 compiler was written in Haskell could not convince the Perl people either).
We then started reading the Django tutorials and the (great!) documentation and started porting our previous MySQL database designs to Django models. This was made a lot easier by an automated Django utility which converts a database into a draft models file, which we could then adapt to our needs. The rest of the team was impressed after seeing the admin site, which again was very easy to get up and running with Django. This week our official lab-youtube-song was this [http://www.youtube.com/watch?v=jMq5joFVxpQ one.]
Database redesign
Working with Django models rather than MySQL tables made it a lot easier to get a better grasp of the structure of our database and possible shortcomings. In particular, we noticed that our original idea of an "Origin" representing a large DNA sequence was problematic, as it would life a lot harder in regards to automated prediction: For example, coding sequences would have to be extracted out of the DNA sequence using a tool like Glimmer or by running through all 6 reading frames as Pfam does. Also it was not immediately obvious how big a DNA sequence in the database could or should be (for example, a biosynthetic cluster for NRPS might be split into two or more "Origins" depending on who added the entries).
Thus, we decided to change the models (which was very easy with Django) as follows: The "Origin" table now represented an actual source of DNA, out of which a domain could be amplified by PCR, such as a particular species or a biobrick. The DNA sequence were now saved in the new "Cds" (Coding sequence) table. The foreign key of the domain table entries also pointed at "Cds" entries rather than "Origins".
PFAM integration
Having learned a lot about one of the core concepts of Django, namely the models, we also started experimenting with the views and template systems, which basically are responsible for the rendering of the actual web pages. Thus, we tried to integrate the Pfam scripts we had previously written into Django views. Pfam actually includes a very nice help page (with gems such as: " IE, being IE, needs extra help [..] ")for its [http://pfam.sanger.ac.uk/help#tabview=tab9 domain graphics], which basically beautifully render JSON documents using Prototype.js and other JS libraries. The JSON representation of the different domains is also calculated by the Pfam server after the domain prediction. Therefore, we adapted the original Pfam script in order to actually extract this JSON file and then it was passed on to the Pfam domain graphics javascript. This had to be done asynchronously (user enters DNA sequence, presses button, then the sequence is passed to the server, the Django function which queries Pfam API is executed and finally the Pfam JSON File is returned to the user's browser). Here, the extensibility of Django via reusable apps came in handy, because the [http://www.dajaxproject.com/dajaxice/ Dajaxice] app made the AJAX requests a matter of a few lines of code.
Database entries
In order to fill the database with the standard monomers, the L- and D-conformation of the 20 proteinogenic amino acids, as well as those of ornithine were taken from pubchem and entered into the database via the standard admin page. To allow for displaying the structures in the user interface the 3D-SDF formatted text files were also copied to the database.
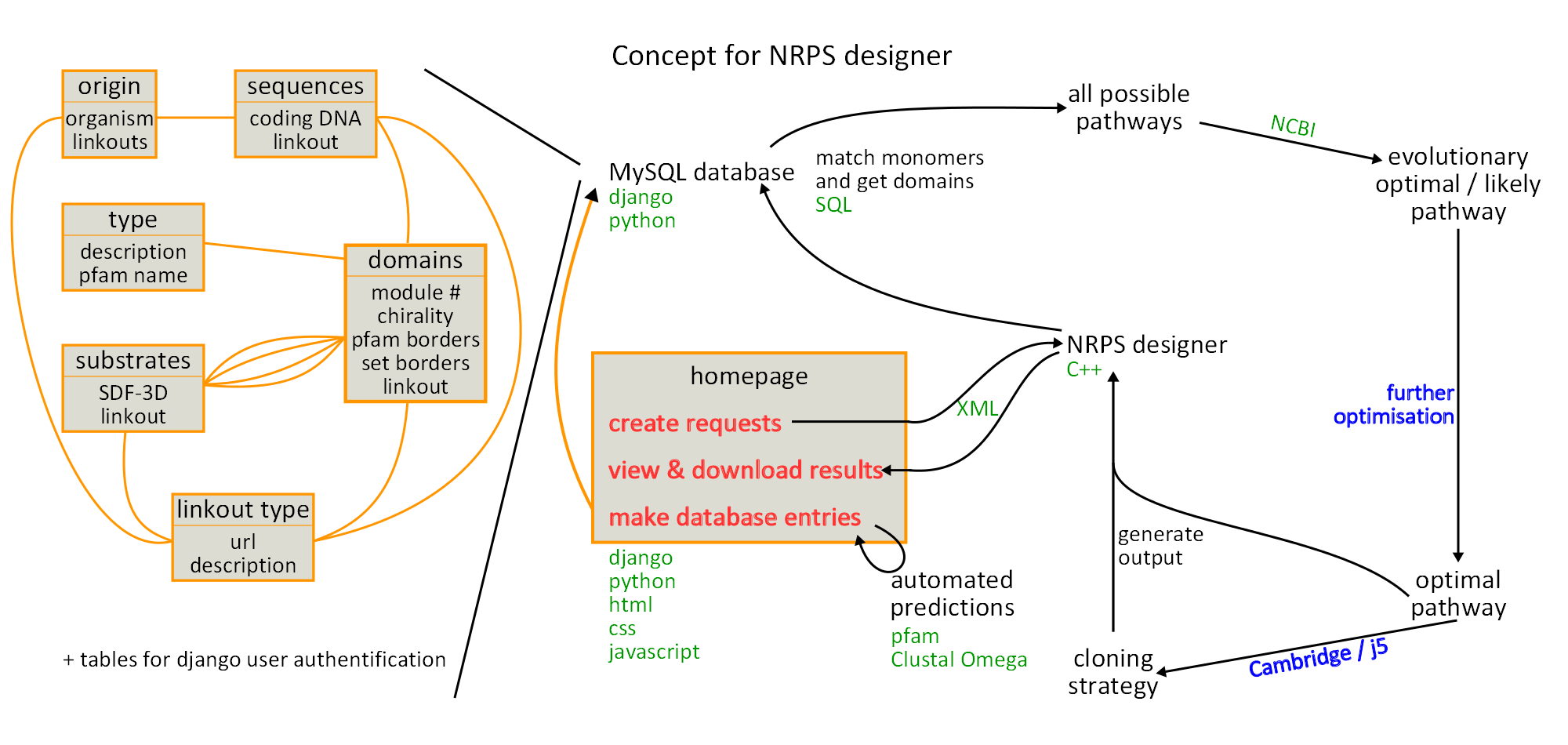
Concept visualisation
For presentation purposes and in order to have a visual representation of all the different aspects of our project we drew a huge graphic on the blackboard. After two days of revision we redrew the whole thing using inkscape. The result is displayed in figure 16.1.
Graphical user interface and Django-registration
This week we started trying to get the basics necessary for a decent user experience up and running. The pages for the Pfam input and the selection of NRP monomers were created. For this we used [http://jqueryui.com/ jQuery UI] and we very especially fond of its tabs. After playing around with some html and css, we soonly decided to integrate [http://getbootstrap.com/2.3.2/ bootstrap 2]for styling our interface. As our Django models and forms would deal a lot with many-to-many relationships and as the default multichoice html box is extremely user-unfriendly, we searched for JS libraries capable of improving this experience. We initially decided to use [http://harvesthq.github.io/chosen/ chosen.js], but then we quickly switched to [http://ivaynberg.github.io/select2/ select2], which provides many more functionalities.
Also in order to enable the smooth handling of user registration, login etc., the django-registration was roughly integrated to the rest of the software.
Use of OpenBable for structure display
OpenBable was included in the tool in order to display the currently designed NRP. The peptide bonds are formed automatically based on the SDF representations saved for each substrate in the database. One of the challenges faced was the correct conformation and the aesthetic display of the peptide chain.
Monomer selection page aesthetic improvements
This week we focused on improving the aesthetics of the NRP selection page. Most of the page layout was shuffled around, so that everything important (Monomer selection, buttons, peptide graphic) fit onto a laptop screen and no scrolling around is necessary. For this we also removed the help text, which added a lot of bloat to the page. We'll try to find a fancier way to document things and help the user later on. We also used a nice feature of s[http://ivaynberg.github.io/select2/ select2], which basically allows you to determine how the dropdown selection options are styled. More specifically, each monomer is displayed together with its 2d structure as generated by the OpenBabel script we had already written.
AJAX functionality
In order to facilitate the communication of the web interface with the C++ code, we started testing the AJAX functionality. One thing we noticed was that the (otherwise great) Dajaxice library, can actually cause problems due to relative URLs, as it does not correctly utilize the wsgi settings. This was especially problematic, as the development and production environments differed, so that a very ugly hack would have to be written for this to work. Instead, we decided to use a more barebones/standard AJAX implementation by actually using jQuery. A test was written with the monomer selection being passed via AJAX and then a popup alert (yay!) which produced an XML out of this selection. This XML could be passed into the C++ function.
Designer interface - modifications
A selection box for the modifications a substrate can have was added to the designer interface. This will also be adopted for the chemical structure and the domains will be included in the NRPS design.
Integration of Gibthon
The Gibthon software by Cambridge 2010 was integrated in our software to provide a cloning strategy for the user. Some features (such as the primer folding analysis) were excluded at first, in order to prevent possible bugs and to runtime when testing the funtionality. During this process of thesting, we also ran into bugs in Gibthon and fixed them in order to improve both tools.
== NRPS-Designer algorithm update ==
Database updates
As many organisms have several NRPS pathways and some NRPS pathways are present in different organisms (e.g. different bacterial strains) the database was changed in order to separate origin and product. The coding sequences now function as many to many connection between origin and products.
Integrating NRPS-PKS
We manually put all the organisms and pathways annotated in NRPS-PKS in our database. This basis was then used to write simple scraping script.
Session data
The user can now have his own personalised data kept in the database. This contains peptides designed and the results of the domains and primer predictions. An additional interface for the organisation of the NRPs was created.
Tool selection for domain prediciton
We tried to determine, which tool would be most appropriate for automated determination of domains. Thus we used the first CDS of the teicoplanin NRPS, as curated in the NRPS-PKS (SBSPKS) database. We then compared the automated prediction with the tool of Maryland, as well as antiSMASH2.
| A domain | T domain | C domain | A domain | T domain | E domain | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | |
| NRPS-PKS | 1 | 491 | 505 | 564 | 604 | 1021 | 1015 | 1511 | 1526 | 1584 | 1608 | 2053 |
| Maryland | 18 | 503 | 504 | 567 | 599 | 1012 | 1008 | 1524 | 1524 | 1587 | 1604 | 2063 |
| antiSMASH | 35 | 426 | 502 | 567 | 598 | 891 | 1056 | 1447 | 1522 | 1589 | 1602 | 1896 |
| pfam | 15 | 478 | 504 | 566 | 598 | 891 | 1036 | 1498 | 1524 | 1586 | 1620 (as C) | 1896 (as C) |
Note that Pfam can't differentiate between C and E domains. Pfam also does not return any predictions in regard to A-domain specificity. antiSMASH is able to predict the A-domain specificities as annotated in NRPS-PKS, while Maryland's tool can only predict the second A-domain (Tyrosine), while the first is incorrectly predicted to be specific for Leucine (curated: HpG, 4-hydroxyphenyl glycine).
The same analysis was also repeated for thaxtomin.
| A domain | NM domain | T domain | C domain | |||||
|---|---|---|---|---|---|---|---|---|
| Start | End | Start | End | Start | End | Start | End | |
| NRPS-PKS | 1 | 506 | 509 | 712 | 944 | 1005 | 1029 | 1458 |
| Maryland | 5 | 521 | 468 | 879 | 942 | 1006 | 1028 | 1455 |
| antiSMASH | 42 | 445 | 511 | 732 | 940 | 1007 | 1025 | 1325 |
| pfam | 22 | 486 | 538 | 638 | 942 | 1006 | 1023 | 1326 |
In this case, both tools correctly predict the L-Phenylalanine specificity of the A-domain.
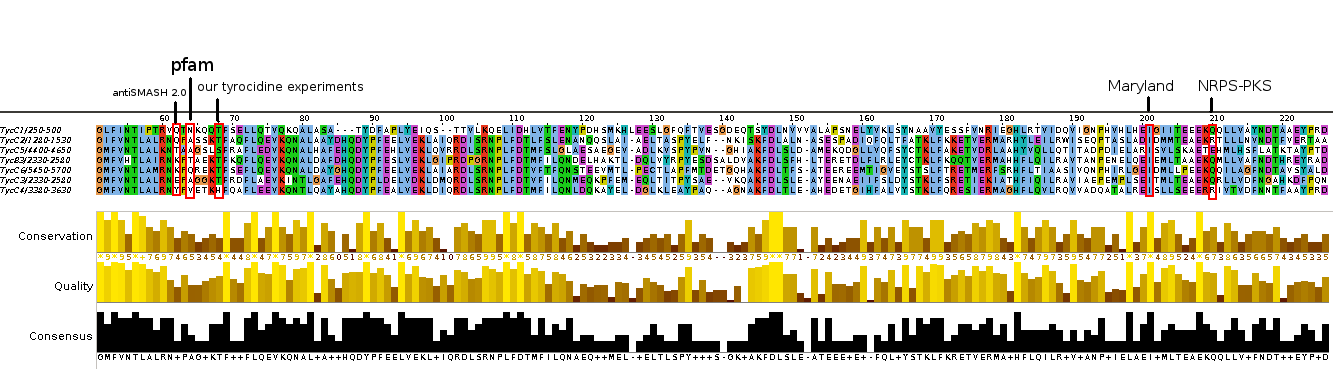
Analysis of tycC from the tyrocidine-cluster.
| C domain | A domain | T domain | C domain | A domain | T domain | C domain | A domain | T domain | C domain | A domain | T domain | C domain | A domain | T domain | C domain | A domain | T domain | TE domain | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | Start | End | ||
| Marahiel (C-T), Ilia (A-T), Philipp (C-A) | 953 | 963 | 1033 | 1050 | 1345 | 1990 | 2000 | 2070 | 2087 | 3028 | 3038 | 3108 | 3125 | 3571 | 4063 | 4073 | 4143 | 4160 | 5107 | 5117 | 5187 | 5204 | 6150 | 6160 | 6230 | 6247 | |||||||||||||
| NRPS-PKS | 13 | 447 | 442 | 957 | 972 | 1036 | 1058 | 1483 | 1478 | 1994 | 2009 | 2073 | 2095 | 2520 | 2515 | 3032 | 3048 | 3111 | 3133 | 3558 | 3553 | 4066 | 4082 | 4146 | 4168 | 4593 | 4588 | 5111 | 5126 | 5190 | 5212 | 5639 | 5636 | 6151 | 6169 | 6233 | 6256 | 6466 | |
| Maryland | 8 | 438 | 441 | 964 | 972 | 1036 | 1053 | 1474 | 1477 | 2001 | 2009 | 2073 | 2090 | 2511 | 2516 | 3039 | 3047 | 3111 | 3128 | 3549 | 3552 | 4074 | 4082 | 4146 | 4163 | 4584 | 4587 | 5118 | 5126 | 5190 | 5207 | 5630 | 5637 | 6160 | 6169 | 6233 | 6254 | 6482 | |
| antiSMASH | 8 | 305 | 490 | 887 | 970 | 1039 | 1054 | 1339 | 1526 | 1924 | 2007 | 2075 | 2090 | 2376 | 2563 | 2962 | 3045 | 3113 | 3128 | 3413 | 3601 | 3997 | 4080 | 4149 | 4165 | 4449 | 4636 | 5041 | 5124 | 5193 | 5208 | 5494 | 5682 | 6083 | 6167 | 6236 | 6253 | 6482 | |
| pfam | 7 | 306 | 470 | 944 | 972 | 1036 | 1052 | 1341 | 1506 | 1981 | 2009 | 2073 | 2089 | 2378 | 2543 | 3019 | 3047 | 3111 | 3127 | 3416 | 3581 | 4054 | 4082 | 4146 | 4162 | 4450 | 4616 | 5098 | 5126 | 5190 | 5206 | 5495 | 5662 | 6141 | 6169 | 6233 | 6254 | 6482 | |
In regards to prediction of A-domain specificity, antiSMASH predictions and the curated NRPS-PKS amino acids were the same. On the other hand, Maryland predicted did not get any hit for A6 (Leu) and for A1/A3 respectively it predicted to possible amino acids (Asn+Asp compared to Asn / Tyr + Trp compared to Tyr in antiSMASH and NRPS-PKS).
TycB3 C-A domain
| C domain | A domain | |||
|---|---|---|---|---|
| Start | End | Start | End | |
| paper primer position | 2520 (MLTAA..) | |||
| NRPS-PKS | 2100 | 2527 | 2540 | 3029 |
| Maryland | 2095 | 2518 | 2521 | 3039 |
| antiSMASH | 2096 | 2381 | 2570 | 2961 |
| pfam | 2094 | 2383 | 2550 | 3019 |
== Inegration of antiSMASH ==
Domain visualisation using pfam
We added the domain visualisation as it is in pfam to the domain types in the database in order to display the resulting NRPS structure.
Please see our documentation for our final results.
Please see our documentation for our final results.
SBOL and GenBank
Proper output of the designed NRPS as well as the primers, was created both for GenBank and for SBOL. Moreover the tool internal communication was shifted to SBOL.
Database Curation
The final state of the database after a major portion of SBSPKS was curated is displayed in the table below.
| Pathway | Gene | Module | SBSPKS domains | HMM domains | Specificity (Chirality C, TE) | Specificity (CA Substrate) | SBSPKS | SVM (0) | Stachelhaus (0) | Minowa (1) | Literature | Modifications | E-domain contained | Literature references | Comment |
| Actinomycin | acmA | 1 | A | A | - | 4-MHA | 4-MHA | hydrophobic/alphatic | PIP | 4-MHA | 4-MHA | - | http://www.ncbi.nlm.nih.gov/pubmed/9573200 | "T-domain after A is definitely missing C-starter can go with or without 4-MHA" | |
| acmB | 2 | C-A-T | CSt-A-T | -/4-MHA | Thr | Thr | Thr | Thr | Thr | Thr | - | ||||
| 3 | C-A-T-E | C-A-T-E-com | L | Val | D-Val | Val | Val | Val | Val | - | X | ||||
| acmC | 4 | C-A-T | com-C-A-T | D | Pro | Pro | Pro | Pro | Pro | Pro | - | ||||
| 5 | C-A-M-T | C-A-NM-T | L | Gly | Gly | Gly | Gly | Gly | Gly | N-Methylierung | |||||
| 6 | C-A-M-T-TE | C-A-NM-T-TE | L, L | Val | Val | Val | Val | Val | Val | N-Methylierung | |||||
| ACV | acvA | 1 | A-T | A-T | - | Aad | Aad | Aad | Aad | Aad | Aad | - | "A: http://www.ncbi.nlm.nih.gov/pubmed/9266851 E: http://www.ncbi.nlm.nih.gov/pubmed/21889568 TE: http://www.ncbi.nlm.nih.gov/pubmed/10715209 " | ||
| 2 | C-A-T | C-A-T | D (pred) / L (sbspks, lit) | Cys | Cys | Cys | Cys | Cys | Cys | - | |||||
| 3 | C-A-T-E-TE | C-A-T-E-TE | L, D | Val | Val-D | Val | Val | Val | Val | - | X | ||||
| A47934 | staA | 1 | A-T | A-T | HpG | DHpG | HpG | HpG | HpG | HpG | http://www.ncbi.nlm.nih.gov/pubmed/12060705 | "- staB E-domain is nonfunctional because of His-Pro mutation - CX in the end is either non-functional or L-selective" | |||
| 2 | C-A-T-E | Cglyc-A-T-E | D | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | C-glyc linking hydroxy | X | ||||
| staB | 3 | C-A-T-E | Cglyc-A-T-E*-com | D | DHpG | HpG | DHpG | DHpG | DHpG | DHpG | C-glyc linking hydroxy | X | |||
| staC | 4 | C-A-T-E | com-Cglyc-A-T-E | L | HpG | HpG2Cl | HpG | HpG | HpG | HpG | C-glyc linking hydroxy | X | |||
| 5 | C-A-T-E | Cglyc-A-T-E | D | HpG | HpG | HpG | HpG | HpG | HpG | C-glyc linking hydroxy | X | ||||
| 6 | C-A-T | Cglyc-A-T | D | bht | Tyrb-O | bht | bht | bht | bht | C-glyc linking hydroxy | |||||
| staD | 7 | C-A-T-E-TE | com-Cglyc-A-T-CX-TE | L, L | DHpG | DHpG | DHpG | DHpG | DHpG | DHpG | C-glyc linking hydroxy | ||||
| Arthrofactin | arfA | 1 | C-A-T | C-A-T | Leu | Leu | Leu | Leu | Leu | Leu | http://www.ncbi.nlm.nih.gov/pubmed/14522057 | Paper claims that there are epimerisation domains, but the dual domains sound more reasonable. | |||
| 2 | C-A-T | CDual-A-T | Asp | Asp | Asp | Asp | Asp | Asp | X | ||||||
| arfB | 3 | C-A-T | CDual-A-T | Thr | Thr | Thr | Thr | Thr | Thr | X | |||||
| 4 | C-A-T | CDual-A-T | Leu | Leu | Leu | Leu | Leu | Leu | X | ||||||
| 5 | C-A-T | CDual-A-T | Leu | Leu | Leu | Leu | Leu | Leu | X | ||||||
| 6 | C-A-T | CDual-A-T | Ser | Ser | Ser | Ser | Ser | Ser | X | ||||||
| arfC | 7 | C-A-T | CDual-A-T | Leu | Leu | Leu | Leu | Leu | Leu | X | |||||
| 8 | C-A-T | C-A-T | L | Ser | Ser | Ser | Ser | Ser | Ser | ||||||
| 9 | C-A-T | CDual-A-T | Ile | Ile | Ile | Ile | Ile | Ile | X | ||||||
| 10 | C-A-T | C-A-T | L | Ile | Ile | Ile | Ile | Ile | Ile | ||||||
| 11 | C-A-T-TE-TE | C-A-T-TE-TE | L,L | Asp | Asp | Asp | Asp | Asp | Asp | ||||||
| Bacitracin | bacA | 1 | A-T | A-T | Ile | Ile | Ile | Ile | Ile | Ile | http://www.ncbi.nlm.nih.gov/pubmed/9427658 | ||||
| 2 | C-A-T | Cy-A-T | L | Cys | Cys | Cys | Cys | Cys | Cys | ||||||
| 3 | C-A-T | C-A-T | L | Leu | Leu | Leu | Leu | Leu | Leu | ||||||
| 4 | C-A-T-E | C-A-T-E | L | Glu | Glu | Glu | Glu | Glu | D-Glu | ||||||
| 5 | C-A-T | C-A-T | D | Ile | Ile | Ile | Ile | Ile | Ile | ||||||
| bacB | 6 | C-A-T | C-A-T | L | Lys | Lys | Lys | Lys | Lys | Lys | |||||
| 7 | C-A-T-E | C-A-T-E | L | Orn | Orn | Orn | Orn | Orn | D-Orn | ||||||
| bacC | 8 | C-A-T | com-C-A-T | D | Ile | Ile | Ile | Ile | Ile | Ile | |||||
| 9 | C-A-T-E | C-A-T-E | L | Phe | Phe | Phe | Phe | Phe | D-Phe | ||||||
| 10 | C-A-T | C-A-T | D | His | His | Tyr | His | His | His | ||||||
| 11 | C-A-T-E | C-A-T-E | L | Asp | Asp | Asp | Asp | Asp | D-Asp | ||||||
| 12 | C-A-T-TE | C-A-T-TE | D, L | Asn | Asn | Asn | Asn | Asn | Asn | ||||||
| CDA | cdaPSI | 1 | C-A-T | CSt-A-T | Ser | Ser | Ser | Ser | Ser | Ser | http://www.ncbi.nlm.nih.gov/pubmed/12445768 | Not sure why Cglyc instead of C since there is no modification here. | |||
| 2 | C-A-T | C-A-T | L | Thr | Thr | Thr | Thr | Thr | Thr | ||||||
| 3 | C-A-E | C-A-T-E | L | Trp | Trp | Trp | Trp | Trp | Trp | X | |||||
| 4 | C-A-T | C-A-T | D | Asp | Asp | Asp | Asp | Asp | Asp | ||||||
| 5 | C-A-T | C-A-T | L | Asp | Asp | Asp | Asp | Asp | Asp | ||||||
| 6 | C-A-E | C-A-T-E | L | HpG | HpG | HpG | HpG | HpG | HpG | X | |||||
| cdaPS2 | 7 | C-A-T | Cglyc-A-T | D | Asp | Asp | Asp | Asp | Asp | Asp | no glycosylation here | ||||
| 8 | C-A-T | C-A-T | L | Gly | Gly | Gly | Gly | Gly | Gly | ||||||
| 9 | C-A-E | C-A-T-E | L | Asn | Asn | Asn | Asn | Asn | Asn | X | |||||
| cdaPS3 | 10 | C-A-T | com-C-A-T | D | Glu/Glu3Me | GluMe3 | Asp/Asn | Glu | Asn | GluMe3 | |||||
| 11 | C-A-T-TE | C-A-T-TE | L, L | Trp | Trp | Trp | Trp | Trp | Trp | ||||||
| Cyclosporin | simA | 1 | C-A-T | C-A-T | ??? | D-Ala | D-Ala | Pro | D-Ala | Ala | D-Ala | "http://www.ncbi.nlm.nih.gov/pubmed/8376400 domain order: http://www.ncbi.nlm.nih.gov/pubmed/16895337 D-Ala:http://www.ncbi.nlm.nih.gov/pubmed/8175682" | final C-domain is never metioned anywhere, but since the product is cyclic it might have similar function as a thioesterase. Thus it is uncertain, how the stereochemistry of the first alanine is achieved. | ||
| 2 | C-A-T | C-A-NM-T | L | Leu | Leu | val,leu,ile,abu,iva | Leu | Leu | Leu | N-Methylierung | |||||
| 3 | C-A-T | C-A-NM-T | L | Leu | Leu | val,leu,ile,abu,iva | Leu | Leu | Leu | N-Methylierung | |||||
| 4 | C-A-T | C-A-NM-T | L | Val | Val | val,leu,ile,abu,iva | Val | Val | Val | N-Methylierung | |||||
| 5 | C-A-T | C-A-NM-T | L | Bmt | Bmt | phe,trp,phg,tyr,bht | Bmt | Bmt | Bmt | N-Methylierung | |||||
| 6 | C-A-T | C-A-T | L | Abu | Abu | val,leu,ile,abu,iva | Abu | Abu | |||||||
| 7 | C-A-T | C-A-NM-T | L | Gly (NM > Sar) | Glu | Pro | Gly | Sar | sarcosine | N-Methylierung | |||||
| 8 | C-A-T | C-A-NM-T | L | Leu | Leu | val,leu,ile,abu,iva | Leu | Leu | Leu | N-Methylierung | |||||
| 9 | C-A-T | C-A-T | L | Val | Val | Pro | Val | Val | Val | ||||||
| 10 | C-A-T | C-A-NM-T | L | Leu | Leu | val,leu,ile,abu,iva | Leu | Leu | Leu | N-Methylierung | |||||
| 11 | C-A-T | C-A-T | L | Ala | Ala | Pro | Ala | Ala | Ala | ||||||
| 12 | C | C | ??? | ||||||||||||
| Gramicidin | gramicidin synthetase 1 | 1 | A-T-E | A-T-E | D | Phe | D-Phe | Phe | Phe | Phe | Phe | X | http://www.ncbi.nlm.nih.gov/pubmed/1560782 | "On the genomic sequence for first synthetase are an additional TE and an additional Com-C domain on separate gene products. Two primary peptides can be fused to one ring." | |
| grsB | 2 | C-A-T | com-C-A-T | L | Pro | Pro | Pro | Pro | Pro | Pro | |||||
| 3 | C-A-T | C-A-T | L | Val | Val | Val | Val | Val | Val | ||||||
| 4 | C-A-T | C-A-T | L | Orn | Orn | Orn | Orn | Orn | Orn | ||||||
| 5 | C-A-T-TE | C-A-T-TE | L, L | Leu | Leu | Leu | Leu | Leu | Leu | ||||||
| Lichenycin | licA | 1 | C-A-T | CSt-A-T | Gln | Gln | Gln | Gln | Gln | Gln | http://www.ncbi.nlm.nih.gov/pubmed/9864322 | Separate TE domain has unknown function and can't be predicted. The first C-Domain and the Thioesterase put a beta-hydroxy fatty acid in the circular peptide | |||
| 2 | C-A-T | C-A-T | L | Leu | Leu | Leu | Leu | Leu | Leu | ||||||
| 3 | C-A-T-E | C-A-T-E | L | Leu | D-Leu | Leu | Leu | Leu | Leu | X | |||||
| licB | 4 | C-A-T | com-C-A-T | D | Val | Val | Val | Val | Val | Val | |||||
| 5 | C-A-T | C-A-T | L | Asp | Asp | Asp | Asp | Asp | Asp | ||||||
| 6 | C-A-T-E | C-A-T-E | L | Leu | D-Leu | Leu | Leu | Leu | Leu | X | |||||
| licC | 7 | C-A-T-TE | com-C-A-T-TE | D, L | Ile | Ile | Ile | Ile | Ile | Ile/Val/Leu | |||||
| licTE (deleted) | TE | TE | |||||||||||||
| Syringomycin | syrE | 1 | C-A-T | Cst-A-T | Ser | Ser | Ser | Ser | Ser | Ser | http://www.ncbi.nlm.nih.gov/pubmed/9830033 | syrB1 is 9th module somehow attaching to C-TE after 8th module. | |||
| 2 | C-A-T | Cdual-A-T | L | Ser | D-Ser | Ser | Ser | Ser | Ser | X | |||||
| 3 | C-A-T | Cdual-A-T | D | Dab | D-Dab | gly,ala,val,leu,ile,abu,iva | Dab | Dab | Dab | X | |||||
| 4 | C-A-T | Cdual-A-T | D | Dab | Dab | gly,ala,val,leu,ile,abu,iva | Dab | Dab | Dab | X | |||||
| 5 | C-A-T | C-A-T | L | Arg | Arg | Arg | Arg | Arg | Arg | ||||||
| 6 | C-A-T | C-A-T | L | Phe | Phe | Phe | Phe | Phe | Phe | ||||||
| 7 (not curated) | C-A-T | C-A-T | L | DhBu-3OH | Thr | Thr | Thr | maybe Thr is recognised and Cdual is dehydrating instead of racemising | |||||||
| 8 | C-A-T | Cdual-A-T | X | Asp | Asp-3OH | Asp | Asp | Ala | Asp | X | |||||
| C-T-TE | C-T-TE | L, L | only TE kept | ||||||||||||
| syrB1 (del) | 9 | A-T | A-T | Thr | Thr-4Cl | Thr |
 "
"