Team:Heidelberg/NRPS
From 2013.igem.org


NRPS. Get to know the theory.
Overview
Everybody knows the dogma of molecular biology - DNA makes RNA makes protein, but it doesn't cover all nature's capabilities, as for example non-ribosomal peptide synthetases, or short NRPS. This alternative pathway for peptide formation is mainly found in bacteria and fungi, but can at also be functionally transferred to mamallian cells (pmid:22543310). Natural NRPs can have various functions from simple dyes up to metal chelators or antibiotics. NRPSs are large protein complexes adding amino or arylic acids, taken from a pool of more than 500 monomers, to a growing peptide chain without the need for a template. The molecular structure of NRPS is based on modules, each incorporating exactly one monomer and comprising several domains of different functions. The most important domains are the adenylation (specific substrate activation), the thiolation (peptide / monomer carrier) and the condensation domain (peptide bond formation), which are needed to form basic peptide chains. Besides these various other domains are known to introduce secondary modifications. Thanks to their remarkable modularity and the wide range of substrates, NRPSs bear the impressive biosynthetic potential to create novel non-ribosomal peptides of various functions in vivo or in vitro. The range of possible applications exceeding natural functions remains unforseen.
Modular Structure
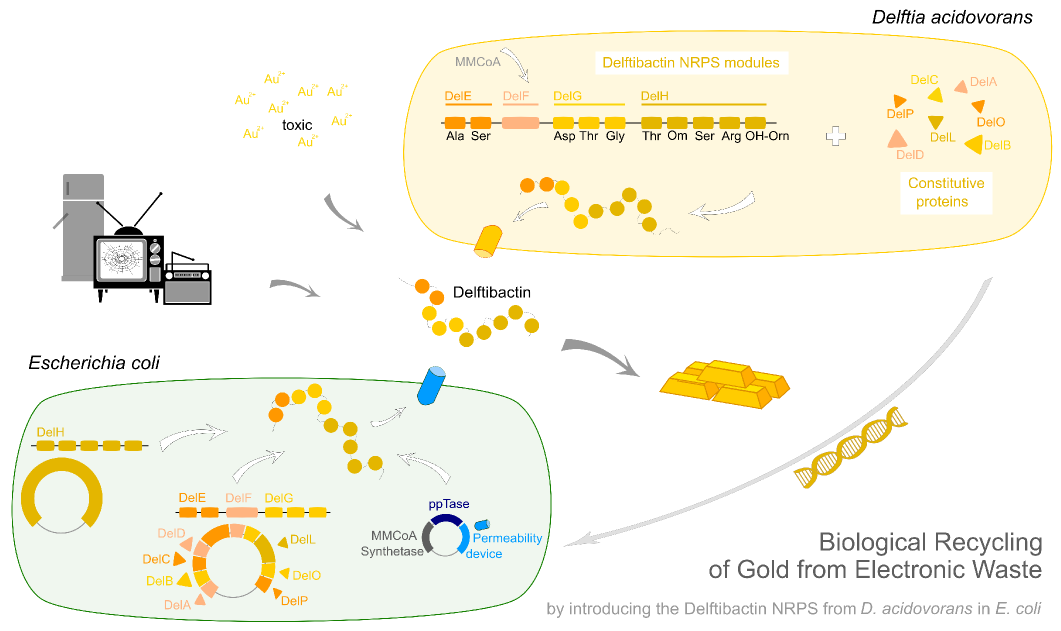
The biggest benefit of NRPS for synthetic biology is it's very modular structure. This starts at the gene level and goes as deep as domain or even single residue level. This modular organisation can be exploited by reorganisation of the modules in order to achieve different products.Biosynthetic gene cluster The genes necessary for the biosynthesis are normally organised in a gene cluster. This contains the genes coding for the non-ribosomal peptide synthetases, which can reach a size of up to 1.5 MDa (cyclosporin Fischbach), genes necessary for the monomers' biosynthesis, as well as tailoring enzymes introducing further modifications in the peptide. There is normally more than just one peptide synthetase. The proteins are often connected by communication domains in order to keep the structure of the assembly line. If one wants to transfer a whole pathway to a different host organisms one crucial apect, besides the successful cloning of the synthetases, is the monomer supply. Depending on the hosts's endogenous machinery one can leave out certain genes or has to include other pathways in order to keep up the supply.
One module - one amino acid Each NRP synthetase is organised in so called modules, where every single module is responsible for incorporating one amino acid in the growing peptide chain. Since all modules have similar minimal structure components one can reorder them easily to achive a different product. These minimal structure components are NRPS specific protein domains each providing the assembly line with a different function. The three levels of modularity - proteins, modules and domains are shown in figure 1.(Fischbach)
Chain elongation
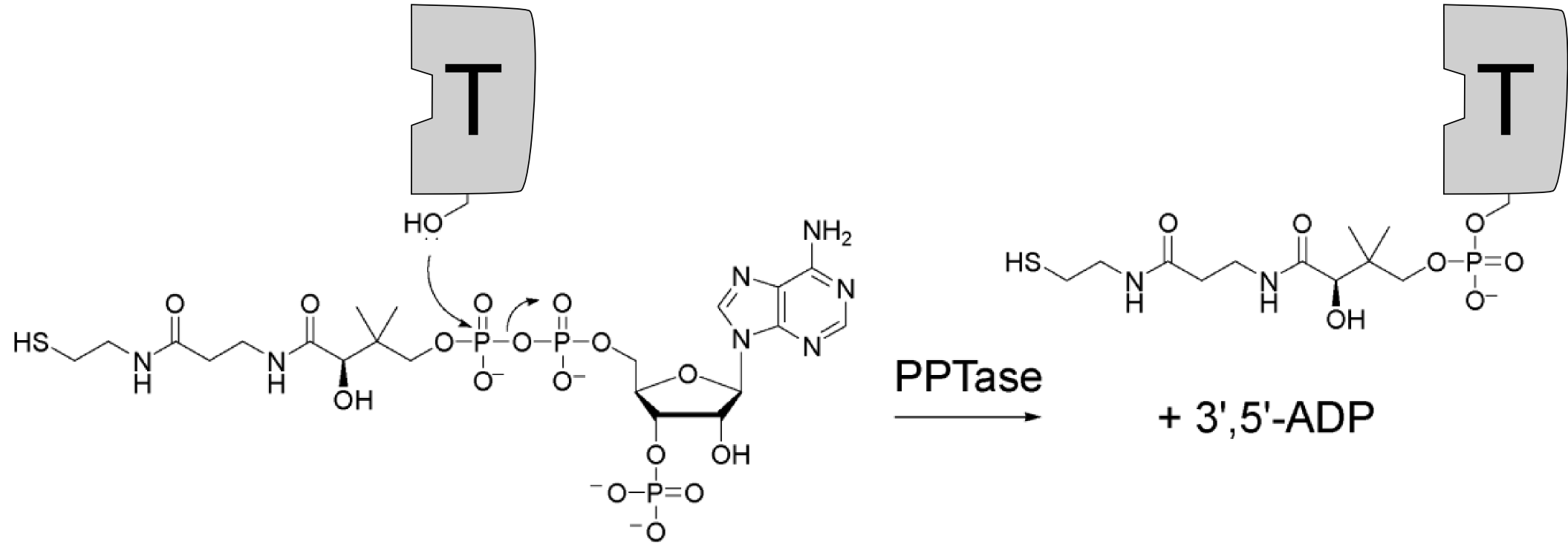
The mechansim of the peptide bond formation in NRPS is partially different from that in the ribosome. The biggest differences are the attachment of the growing peptide chain and the number of catalytic domains. In ribosomal synthesis one ribosome can add many amino acids, but in non-ribosomal synthesis the number of catalytic domains rises linearly with the number of amino acids incorporated. Thus the latter is only suitable for oligopeptide synthesis.Adenylation domain The actual attachement of the amino acid to the already modified T-domain is carried out by the adenlytation domain (A-domain). Condensation domain TErmination - ThioEsterase
Monomer range
Epimerisation and stereoselectivity Non-proteinogenic amino acidsModifying domains
N-Acylation O-Acylation Heterocyclisation1. Eisler R, Wiemeyer SN (2004) Cyanide hazards to plants and animals from gold mining and related water issues. Reviews of environmental contamination and toxicology: 21–54.
2. Eisler R (2004) Arsenic hazards to humans, plants, and animals from gold mining. In:. Reviews of environmental contamination and toxicology. Springer. pp. 133–165.
3. Donato DB, Nichols O, Possingham H, Moore M, Ricci PF, et al. (2007) A critical review of the effects of gold cyanide-bearing tailings solutions on wildlife. Environment international 33: 974–984.
4. Tay SB, Natarajan G, bin Abdul Rahim MN, Tan HT, Chung MCM, et al. (2013) Enhancing gold recovery from electronic waste via lixiviant metabolic engineering in Chromobacterium violaceum. Scientific reports 3.
5. Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, et al. (2013) Gold biomineralization by a metallophore from a gold-associated microbe. Nature chemical biology 9: 241–243.
6. Strieker M, Tanović A, Marahiel MA (2010) Nonribosomal peptide synthetases: structures and dynamics. Current opinion in structural biology 20: 234–240.
7. Caboche S, Leclère V, Pupin M, Kucherov G, Jacques P (2010) Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. Journal of bacteriology 192: 5143–5150.
8. Baltz RH (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. Journal of industrial microbiology & biotechnology 38: 1747–1760.
9. Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, et al. (1998) Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37: 1585–1595.
10. de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J (2012) PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC genomics 13: 299.
11. Reith F, Etschmann B, Grosse C, Moors H, Benotmane MA, et al. (2009) Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proceedings of the National Academy of Sciences 106: 17757–17762.
12. Cárcamo J, Ravera MW, Brissette R, Dedova O, Beasley JR, et al. (1998) Unexpected frameshifts from gene to expressed protein in a phage-displayed peptide library. Proceedings of the National Academy of Sciences 95: 11146–11151.
 "
"