|
|
| Line 135: |
Line 135: |
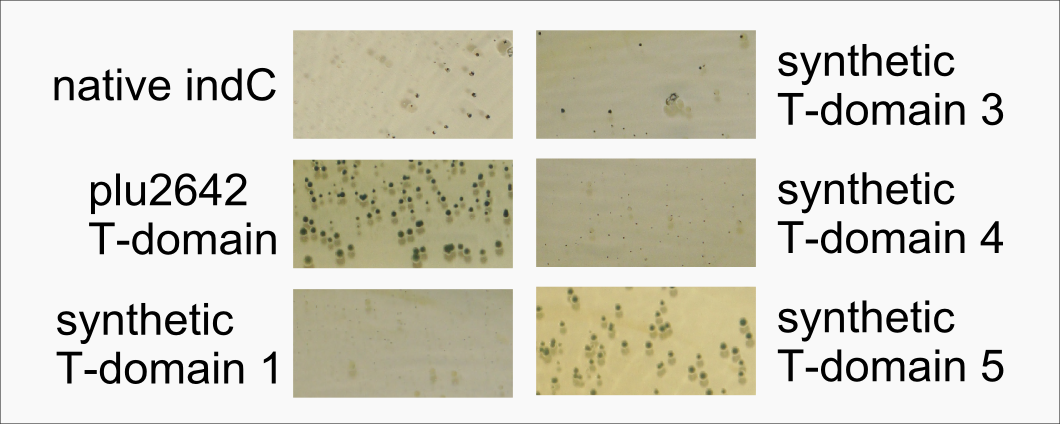
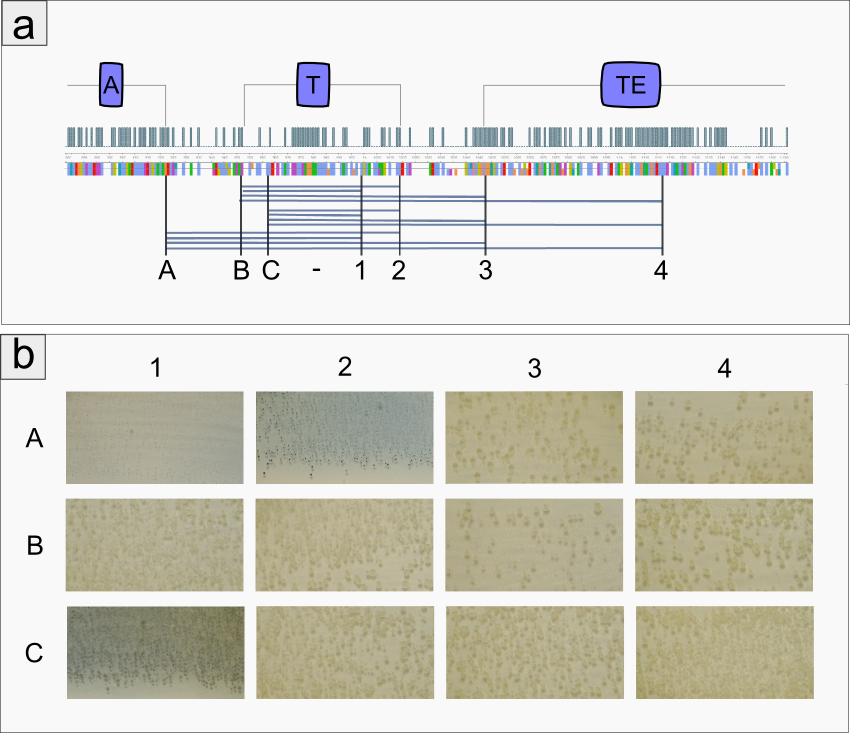
| | However, our data shows that the T-domain of indC can be replaced by the T-domain of bpsA only if specific T-domain border combinations are used (see Fig. x) to define the replaced fragment (Figure xa). We proved that indC retained its functionality when the T-domain of the native indC gene was replaced with T-domains of other NRPS modules (see Figure X), applying the T-domain border combination "A2" (see Fig. x). We also tried the T-domain border combination "B2" with the same T-domains, as these T-domain borders were proposed by the Pfam domain prediction tool (pfam.sanger.ac.uk). In this approach less transformants were capable of producing the plue pigment (data not shown), suggesting that the T-domain border combination "A2" can be seen as an improved variant compared to the Pfam prediction. | | However, our data shows that the T-domain of indC can be replaced by the T-domain of bpsA only if specific T-domain border combinations are used (see Fig. x) to define the replaced fragment (Figure xa). We proved that indC retained its functionality when the T-domain of the native indC gene was replaced with T-domains of other NRPS modules (see Figure X), applying the T-domain border combination "A2" (see Fig. x). We also tried the T-domain border combination "B2" with the same T-domains, as these T-domain borders were proposed by the Pfam domain prediction tool (pfam.sanger.ac.uk). In this approach less transformants were capable of producing the plue pigment (data not shown), suggesting that the T-domain border combination "A2" can be seen as an improved variant compared to the Pfam prediction. |
| | | | |
| - | <a class="fancybox fancyFigure" title="Exchange of the indC T-domain on a pSB1C3 derived expression plasmid" href="https://static.igem.org/mediawiki/2013/c/ca/Heidelberg_IndPD_Fig2.png" rel="gallery1">
| |
| | | | |
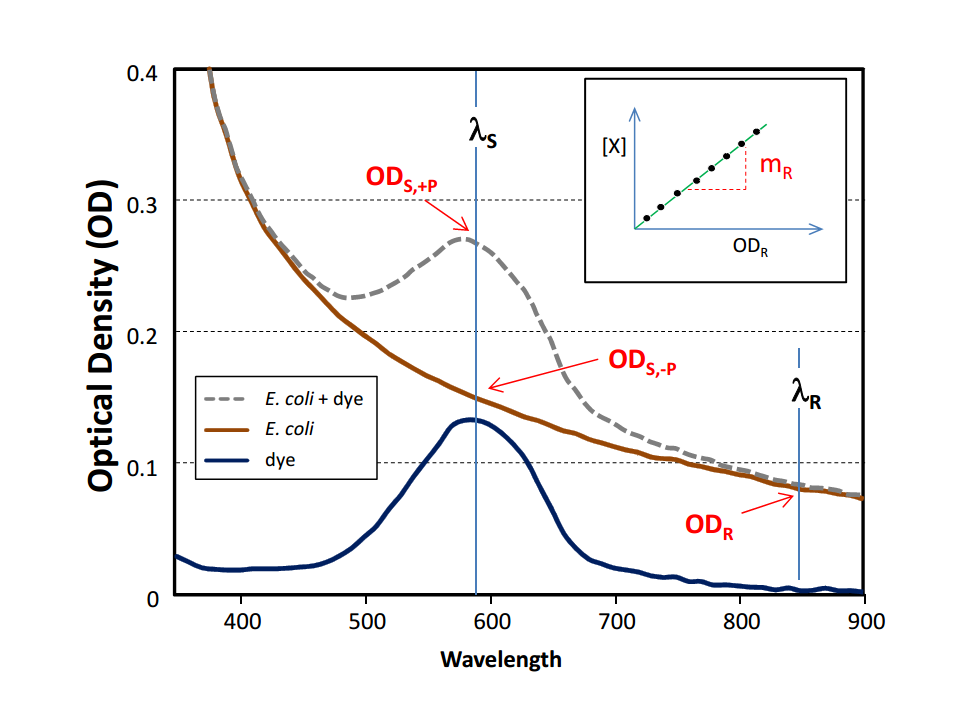
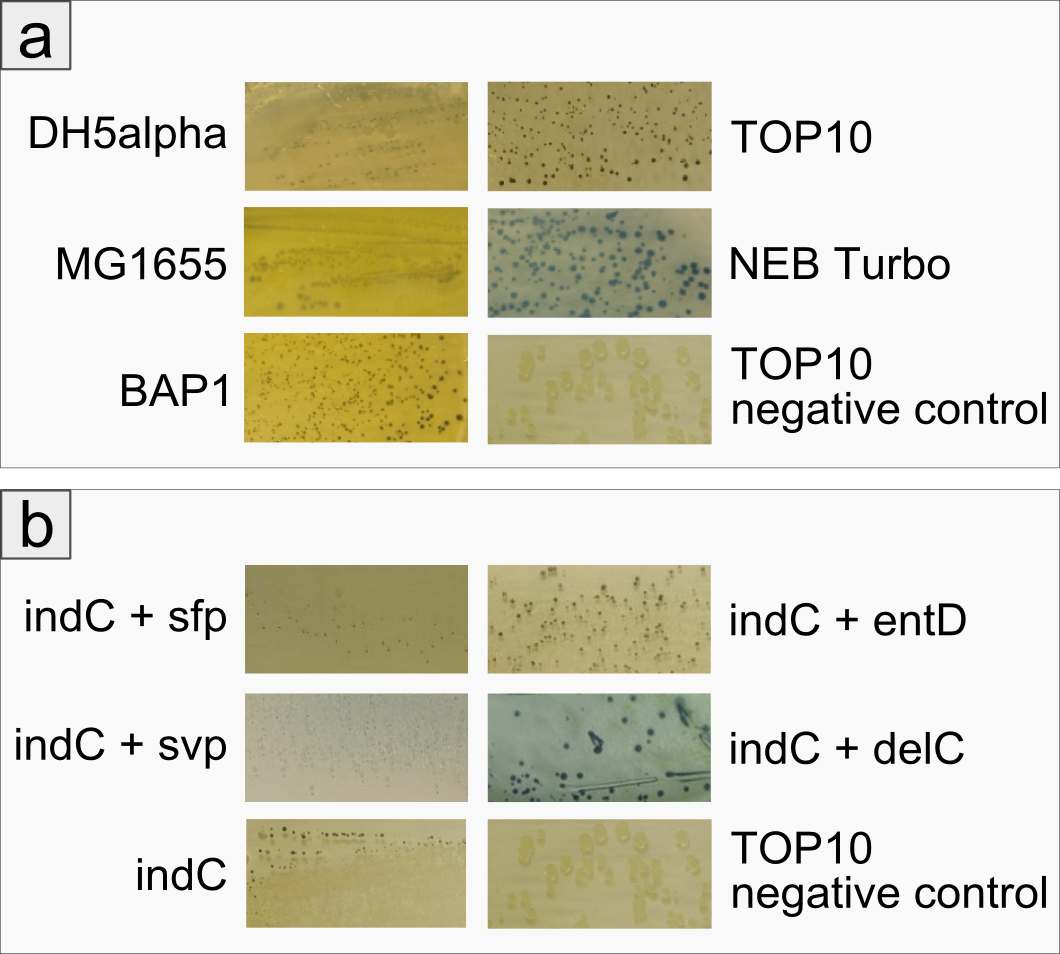
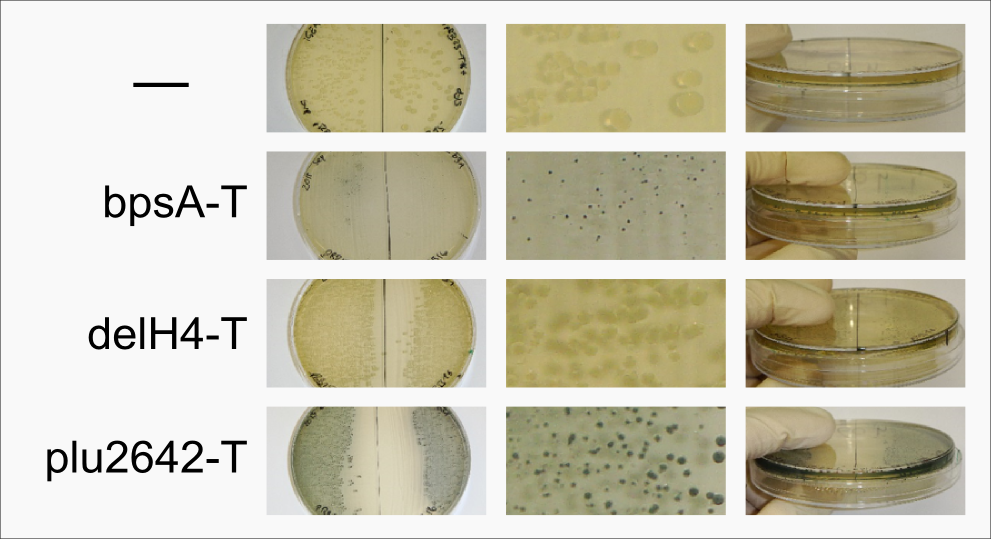
| - | <a class="fancybox fancyFigure" title="Comparison between different <em>E. coli</em> strains and PPTases." href="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png" rel="gallery1"> | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/c/ca/Heidelberg_IndPD_Fig2.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/c/ca/Heidelberg_IndPD_Fig2.png"></img> |
| | + | <figcaption><b>Figure 2</b>: Exchange of the indC T-domain on a pSB1C3 derived expression plasmid |
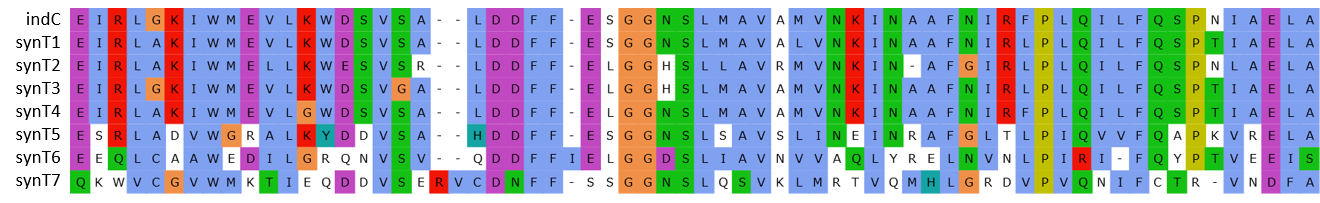
| | + | The indigoidine synthetase indC from <em>P. luminescens</em> consists of an adenylation domain with an internal oxidation domain, a thiolation domain and a thioesterase domain. We replaced the indC T-domain with T-domains of other NRPS modules (one of which is the indigoidine synthetase bpsA from <em>S. lavendulae</em>) and seven synthetic T-domains, which were customized to be introduced to indC, respectively. |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/6/6c/Heidelberg_IndPD_Fig3.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/6/6c/Heidelberg_IndPD_Fig3.png"></img> |
| | + | <figcaption><b>Figure 3</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <h2 id="results">Discussion</h2> |
| | + | We showed that it is possible to replace the T-domain of an indigoidine synthetase with T-domains of from other NRPS modules to proof that one can in principle also replace the T-domain of a given NRPS module, preserving its function. We propose a method to design synthetic NRPS domains and proof their functionality by introducing them into the indigoidine synthetase indC, thus resulting in a blue phenotype of positive transformants. Moreover, we submitted the RFC [99], describing "high throughput protocols for circular polymerase extension cloning and transformation" (Hi-CT). To further establish the indigoidine synthetase indC as a model for NRPS research we created a library of 58 engineered indC variants – giving insights in how synthetic buiding blocks should be designed, where the domain borders should be set, which domains from respective NRPS pathways and bacterial strains can be used, when creating novel engineered NRPS pathways. Together with our software "NRPS-Designer" and the labelling of engineered NRPS products, we pioneered the research on high-throughput methods for creation of novel NRPS systems, which could lead to a faster development in drug research, recycling, cell signaling and other applications one could think of when creating customized peptides with hundreds of possible monomers. |
| | | | |
| - | <img style="width:100%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png">
| |
| - | </a></p><figcaption><a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png" style="margin: 10px;"><b>Fig. 1</b> Overview of the Tyrocidine Cluster. Adapted from <span class="citation">[6]</span>. </a></figcaption><a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png" style="margin: 10px;">Fig. 2 Exchange of the indC T-domain on a pSB1C3 derived expression plasmid.
| |
| - | The indigoidine synthetase indC from <em>P. luminescens</em> consists of an adenylation domain with an internal oxidation domain, a thiolation domain and a thioesterase domain. We replaced the indC T-domain with T-domains of other NRPS modules (one of which is the indigoidine synthetase bpsA from <em>S. lavendulae</em>) and seven synthetic T-domains, which were customized to be introduced to indC, respectively.
| |
| - | </a>
| |
| | | | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/4/4b/Heidelberg_IndPD_Fig4.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/4/4b/Heidelberg_IndPD_Fig4.png"></img> |
| | + | <figcaption><b>Figure 4</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/6/64/Heidelberg_IndPD_Fig5.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/6/64/Heidelberg_IndPD_Fig5.png"></img> |
| | + | <figcaption><b>Figure 5</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/c/cf/Heidelberg_IndPD_Fig6.png"></img> |
| | + | <figcaption><b>Figure 6</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/f/f1/Heidelberg_IndPD_Fig7.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/f/f1/Heidelberg_IndPD_Fig7.png"></img> |
| | + | <figcaption><b>Figure 7</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/e/eb/Heidelberg_IndPD_Fig8.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/e/eb/Heidelberg_IndPD_Fig8.png"></img> |
| | + | <figcaption><b>Figure 8</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/6/6e/Heidelberg_IndPD_Fig9.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/6/6e/Heidelberg_IndPD_Fig9.png"></img> |
| | + | <figcaption><b>Figure 9</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/4/4d/Heidelberg_IndPD_Fig10.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/4/4d/Heidelberg_IndPD_Fig10.png"></img> |
| | + | <figcaption><b>Figure 10</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/3/38/Heidelberg_IndPD_Fig11.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/3/38/Heidelberg_IndPD_Fig11.png"></img> |
| | + | <figcaption><b>Figure 11</b>: |
| | + | </figcaption> |
| | + | </a> |
| | + | |
| | + | <a class="fancybox fancyGraphical" href="https://static.igem.org/mediawiki/2013/7/75/Heidelberg_IndPD_Fig_multiplot.png"> |
| | + | <img style="width:60%; margin-bottom:10px; padding:1%;border-style:solid;border-width:1px;border-radius: 5px;" src="https://static.igem.org/mediawiki/2013/7/75/Heidelberg_IndPD_Fig_multiplot.png"></img> |
| | + | <figcaption><b>Figure 12</b>: |
| | + | </figcaption> |
| | + | </a> |
| | | | |
| - |
| |
| - | <h4>Assembly of a synthetic NRPS </h4>
| |
| - | <p>We started the amplification of the desired single modules for the assembly of various constructs required for proving the aforementioned criteria. Primers were annealed to the unconserved linker regions between the NRPS domains that were predicted by <a href="http://pfam.sanger.ac.uk/">Pfam</a>. We successfully validated the correct amplification of 12 single DNA fragments and corresponding <a href="http://parts.igem.org/Part:pSB1C3">pSB1C3</a> backbones by electrophoresis (<strong>Fig. 4</strong>) <img src="XXXX" title="fig:Fig. 4 Gel Electrophoresis of our amplified fragments" alt="Fig. 4 Gel Electrophoresis of our amplified fragments" /><br /><br />Their functionality as isolated modules cannot be shown, as they simply take up single amino acids without linking them to another monomer.<br />To show the compatibility of the Tyrocidine modules with one another, we put module genes into non-native order via <a href="XX" title="wikilink">Gibson Assembly</a>. These constructs led to synthetic NRPSs and the production of five new peptides, i.e. one dipeptide, two tripeptides and two tetrapeptides.<br />We were able to successfully assemble all of our plasmids and continued our work with our synthetic Dipeptide NRPS and Tripeptide-I-NRPS (<strong>Fig. 5</strong>) by transforming them into the <em>E. coli</em> strain BAPI. <img src="Results%20Shuffling%20scheme%20di%20tri%2001.png" title="fig:Fig. 5: Shuffling genes encoding for modules of the Tyrocidine synthetase leads to novel NRPSs and referring peptides, e.g. the Dipeptide Proline-Leucine (left) or the Tripeptide Phenylalanine-Ornithine-Leucine (right)." alt="Fig. 5: Shuffling genes encoding for modules of the Tyrocidine synthetase leads to novel NRPSs and referring peptides, e.g. the Dipeptide Proline-Leucine (left) or the Tripeptide Phenylalanine-Ornithine-Leucine (right)." /><br /><br /> </p>
| |
| - | <h4>Expression and detection of the NRPS and its products</h4>
| |
| - | <p>The expression of the 212 kDa Dipeptide synthetase and the 380 kDa Tripeptide synthetase was shown by <a href="XXX" title="wikilink">SDS-PAGE</a> (<strong>Fig. 6</strong>).<br /><br /><img src="SDS%202nd%2020130830.png" title="fig:Fig. 6: SDS-PAGE confirming the expression of our engineered NRPSs. The 380 kDa tripeptide synthetase (Tri I) can be seen after 3 hours while 212 kDa Dipeptide synthetase (in the samples Di A and Di B) can be seen 12 hours after induction. Respective bands are indicated by blue arrows." alt="Fig. 6: SDS-PAGE confirming the expression of our engineered NRPSs. The 380 kDa tripeptide synthetase (Tri I) can be seen after 3 hours while 212 kDa Dipeptide synthetase (in the samples Di A and Di B) can be seen 12 hours after induction. Respective bands are indicated by blue arrows." /> Since SDS-PAGE is not sensitive enough to detect small peptides, we wanted to assess the presence of the newly synthesized peptides at different time points by the use of <a href="XX" title="wikilink">mass spectrometry</a>. Expression of the NRPSs was induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG). Samples were taken at different time points post induction. Since residual salts could potentially disturb the acquisition of small peptide abundances, we washed our LB-Cm culture in <a href="XX" title="wikilink">M9 minimal medium</a> to minimize this effect and improve the detection of our short NRPs. Supernatant and the bacterial pellet were processed separately. Final sample preparation for tandem mass-spectrometry was conducted at the <a href="XX" title="wikilink">neonate screening facility of the university medical center</a>.<br /><br />There the peptides were hydrolyzed during the butylation reaction. Finally the highly specific m/z profile allowed the identification of different amino acid abundances. Since Ornithine is a non-proteinogenic amino acid that is incorporated in our Tripeptide, we mainly focused on detecting Ornithine levels. The abundance of Ornithine in the Tripeptide samples was strongly elevated with time compared to our Dipeptide (<strong>Fig. 7</strong>).<br /><br />Due to variation of the general amino acid concentration in both used media and samples, we normalized the Ornithine values by the amount of all amino acids present in the respective medium (<strong>Fig. 8</strong>). The Ornithine level in the supernatant of the Tripeptide samples peaked 21 hours upon induction. Afterwards concentrations returned to basal levels. Samples prepared from bacteria pellets showed minor increases in Ornithine levels in comparison to the pure medium and our negative control (untransformed BAPI). To aquire additional data on the existance of our short peptides we sent two samples to analysis via high-resolution electrospray ionization (HR-ESI) mass spectrometry at the <a href="XX" title="wikilink">mass spectrometry facility of the Institue for Chemistry</a>, based on preliminary results from the Ornithine screening. However, we could not obtain conclusive data for our samples, because the background was too high (MS Results: <a title="File:Heidelberg MS Pro Leu.pdf" href="/File:Heidelberg_MS_Pro_Leu.pdf">
| |
| - |
| |
| - | Dipeptide
| |
| - |
| |
| - | </a>
| |
| - | and
| |
| - | <a title="File:Heidelberg MS Phe Orn Leu.pdf" href="/File:Heidelberg_MS_Phe_Orn_Leu.pdf">
| |
| - |
| |
| - | Tripeptide
| |
| - |
| |
| - | </a> ).</p>
| |
| - | <p>File: Ornithine_levels_MS.png| <strong>Fig. 7</strong>: Levels of Ornithine measured at neonate screening via mass spectrometry. Samples were bacteria cultures harbouring engineered NRPSs. Culture medium of Tripeptide-I showed highly elevated ornithine levels. File: Ornithine_levels_normalized_MS.png |<strong>Fig. 8</strong>: Normalized levels of Ornithine measured at neonate screening via mass spectrometry. Culture medium of Tripeptide-I showed highly elevated ornithine levels. Acquired intensities were divided by the signal of all amino acids detected in the referring sample.<br /> In summary, we were able to amplify single modules from the Tyrocidine NRPS cluster, and we shuffled them via Gibson Assembly. Two constructs coding for two entirely new NRPSs were successfully transformed. The synthetases are both well expressed on pSB1C3 in BAPI and their products, short NRPs, can be detected through mass spectrometry finally confirming the functionality of the engineered NRPSs.<br /><br /> </p>
| |
| - | <h3>Showing inter-species module compatibility by fusion of Tyrocidine modules to the Indigoidine synthetase</h3>
| |
| - | <p>Our module-shuffling approach was confirmed by mass spectrometry. Access to such special technical devices and the referring expertise is demanded for detection of small peptides but often limited. That is why we were thinking of a potential alternative to test for the synthesis of NRPs. We wanted to establish an assay accessible to the majority of the community for validation of the presence of custom peptides. Since the synthetase for indigoidine consists of only a single module, it could serve as a paradigm for the fusion to modules of other NRPSs. The pigment Indigoidine could potentially ease detection of peptides when they are fused to the dye that is visible by eye (see <a href="XX" title="wikilink">project on Indigoidine Domain Shuffling</a>).<br />Thus we wanted to investigate whether it is possible to fuse Indigoidine as a tag to NRPs of other pathways even originating from other species.<br /><br />In the following, we will showcase the extent of NRPSs’ modular compatibility by creating inter-species module-fusions between the Indigoidine synthetase from <em>P. luminescens</em> and modules of the Tyrocidine synthetase from <em>B. parabrevis</em>. In those fusion NRPs, Indigoidine serves as a tag that eases identification of <em>E. coli</em> clones synthesizing the customized peptide.<br /><br /></p>
| |
| - | <h4> Fusing single amino acids to Indigoidine</h4>
| |
| - | <p>First, we combined single modules of the Tyrocidine synthetase with the indC synthetase resulting in three distinct NRPSs producing Asparagin, Valine or Phenylalanine respectively, all tagged with Indigoidine (<strong>Fig. 9</strong>).<br />To assure compatibility the constructs were designed in such a way that the C domain of the C2 module was always used, given its specificity for Glutamine required for the Indigoidine production. SDS-PAGE showed the expected bands for the expression of the NRP synthetases in the transformed BAPI.<br /><br /><img src="Results%20Fusion%20ind%20tyc%20part1.png" title="fig:Fig. 9: Composition of three fusion constructs originating from Tyrocidine (tyc) synthetase modules and the Indigoidine synthetase (indC). First row indicates domains with referring modules. Coloured modules in the three rows below were fused together to create plasmids encoding novel NRPSs (rectangle)." alt="Fig. 9: Composition of three fusion constructs originating from Tyrocidine (tyc) synthetase modules and the Indigoidine synthetase (indC). First row indicates domains with referring modules. Coloured modules in the three rows below were fused together to create plasmids encoding novel NRPSs (rectangle)." /> The <em>E. coli</em> strain BAPI was used for expression of the NRPS fusions because it carries the required PPTase endogenously. All three of the fusion variants turned the colonies blue even before expression induction with IPTG. The blue pigment thus served a first indicator that peptide synthesis was successful. To further verify the existence of the fusion peptide, we ran comparative thin-layer chromatography (TLC). The native, purified Indigoidine ran further than our purified dipeptides suggesting that the amino acids were indeed fused to the pigment (<strong>Fig. 10</strong>). The peptides were detected under visible and UV light due to Indigoidine’s properties as a dye.<br /><br /><img src="XXX" title="fig:Fig. 10 XXXXXXX" alt="Fig. 10 XXXXXXX" /><br /><br />We sent a sample of our purified Val-Ind NRP and purified Indigoidine to the mass spectrometry facility at <a href="XX" title="wikilink">the Institute for Chemistry</a> handling these samples the same way as the samples sent from the Module Shuffling experiments. <strong>RESULTS</strong> (<a href="Media:MS%20Val%20Ind.pdf" title="wikilink">Val-Ind MS</a> and <a href="Media:MS%20Ind%20negativecontrol.pdf" title="wikilink">Negative Control Ind</a>)</p>
| |
| - | <h4 id="using-indigoidine-as-tag-for-non-ribosomal-peptides">Using Indigoidine as tag for non-ribosomal peptides</h4>
| |
| - | <p>To gather additional evidence for our functional Indigoidine tag, we assembled seven variants with up to four modules in front of the Indigoidine synthetase (Fig. 11) following the same approach as described above. <img src="Results_Fusion_ind_tyc_part2.png" title="fig:Fig. 11 Composition of seven fusion constructs originating from Tyrocidine (tyc) synthetase modules and the Indigoidine synthetase (indC). First row indicates domains with referring modules. Coloured regions in the second row were fused together to create plasmids encoding novel NRPSs (rectangle)" alt="Fig. 11 Composition of seven fusion constructs originating from Tyrocidine (tyc) synthetase modules and the Indigoidine synthetase (indC). First row indicates domains with referring modules. Coloured regions in the second row were fused together to create plasmids encoding novel NRPSs (rectangle)" /><br />Again the constructs pPW06, pPW09, pPW10, pPW11 and pPW12 turned BAPI colonies blue upon transformation. We tested the fusion of those peptides by comparative TLC with native Indigoidine. Even with increasing peptide length, synthesis did not seem to be affected and the dye-properties of the Indigoidine were still preserved (<strong>Fig. 12</strong>). <img src="XXX" title="fig:Fig. 12 XXXXXXX" alt="Fig. 12 XXXXXXX" /><br /><br />From this, we deduced that Indigoidine possesses characteristics required for a proper peptide tag that we would like to propose for use to the community (<a href="RFC%20100" title="wikilink">RFC 100</a>). For this purpose we have created a ccdB-dependent vector to ease the tagging of NRPs, accessible through the parts registry. Design of such constructs is enabled with our software, the <a href="XX" title="wikilink">NRPS-Designer</a> .<br /><br /></p>
| |
| - | <h4>Experimental validation of software predictions</h4>
| |
| - | <p>The NRPS-Designer predicts module boundaries and linker regions based on <a href="http://pfam.sanger.ac.uk/">Pfam</a>. We used these predictions for our module shuffling experiments, which all worked well in our constructs. However we feel that the borders and linker regions predicted are rather vague and want to evaluate the predictions to contribute more data to the NRPS Designer. Therefore we systematically varied the module/ boundaries of the A domain and C domain of C2 within the Val-Ind-construct in relation to the <a href="http://pfam.sanger.ac.uk/">Pfam</a> prediction. Based on this altered domain positioning, eight constructs were designed combining narrow, broad and original <a href="http://pfam.sanger.ac.uk/">Pfam</a> module regions (<strong>Fig. 13</strong>). Their functionality was always compared to the original construct based on the boundaries obtained from <a href="http://pfam.sanger.ac.uk/">Pfam</a><strong>RESULTS?!</strong>.<br /><img src="Results%20LV%20overview.png" title="fig:Fig. 13 Overview over our constructs investigating the different domain borders in relation to Pfam. dDrk colors are variations in the start-region of the A domain borders and light colors are variations in the end-region of the C domain." alt="Fig. 13 Overview over our constructs investigating the different domain borders in relation to Pfam. dDrk colors are variations in the start-region of the A domain borders and light colors are variations in the end-region of the C domain." /> To summarize, with our inner- and interspecies module shuffling, we successfully validated the concept of modularity. We have integrated our experimental data and fuelled it into our software to improve its prediction accuracy. To make NRPSs and their custom design more accessible to the community, we have submitted standardized versions of the modules we used for our shuffling experiments, to the parts registry (see the <a href="parts%20registry" title="wikilink">parts registry</a> entries).</p>
| |
| - | <h2 id="conclusion">Conclusion</h2>
| |
| - | <p>We can prove all mentioned claims since we have:</p>
| |
| - | <ol style="list-style-type: decimal">
| |
| - | <li>Exchanged modules of of the Tyrocidine synthetase,</li>
| |
| - | <li>Constructed a custom dipeptide- and a custom tripeptide-NRPS,</li>
| |
| - | <li>Detected both the engineered NRPSs and the synthetic peptides via SDS-PAGE and mass spectrometry,</li>
| |
| - | <li>Demonstrated compatibility of modules from the Tyrocidine synthetase of <em>B. parabrevis</em> with modules from the Indigoidine synthetase of <em>P. luminescens</em> and</li>
| |
| - | <li>Established Indigoidine as functional tag that allows for cost-efficient detection of newly synthesized NRPs</li>
| |
| - | </ol>
| |
| - | <p>The achievements were enabled through and fed back to the NRPS-Designer. This software tool incorporates a database utilizing our experimental on NRPS modules for guiding through user-oriented design of NRPSs. <strong>conclusion</strong></p>
| |
| - | <ul>
| |
| - | <li>proof of interchangeability, modularity and felxability of NRPS within a NRPS strain system and also between different species</li>
| |
| - | <li>new NRP tagging system with Indigoidine fusion experimentally prooved with up to four modules( amino acids). This highly efficient system was contributed to the community (ccdB-IndC).</li>
| |
| - | <li>supportive data for the NRPS designer, regarding pfam linker/domain border predictions</li>
| |
| - | </ul>
| |
| - | <h2 id="discussion">Discussion</h2>
| |
| - | <p>Module compatibility is the vital basis for any standardized work with NRPSs. Hence, the major objective of this project was to investigate flexible interchangeability of modules, which allows for customized synthesis of short peptides via NRPSs, as we propose in our standard (<a href="RFC%20100" title="wikilink">RFC 100</a>). Tyrocidine served as a paradigm for semi-rational rearrangements in the modular structure of NRPSs, a process, which we refer to as shuffling.
| |
| - | <br/>
| |
| - | Here, we present a clear line of evidence stating that it is possible to shuffle modules to produce functional NRPs. Modules of NRPSs can be interchanged by creating two different novel peptides through rearrangement of the respective modules that were amplified from <i>B. parabrevis</i>. The detection of these peptides was eased by the use of Ornithine, which is a non-proteinogenic amino acid and thus a proper marker for the synthesis of the desired peptide. Comparing the normalized levels of Ornithine in the different samples, we could conclude that the synthetic NRPS is in fact functional enabling the creation of customized peptides that can be detected via mass spectrometry.
| |
| - | <br/>
| |
| - | (<strong>THIS IS SO FAR ONLY RECAPITULATION OF RESULTS, BETTER REFLECT ACCHIEVEMENTS IN THE CONTEXT OF LITERATURE</strong>)
| |
| - | Other approaches <span class="citation">[16]</span> describe an extensive purification protocol, which requires large amounts of product, as high salt concentrations are an important interference factor. Small peptides are hardly specifically separable from salt-ions, hence high amounts could have bad influence on the signal at mass spectrometry. The method we describe above offers an useful and efficient way of detecting synthesis of the NRP.
| |
| - | <br/><br/>
| |
| - | Before our experiments, there was no evidence whether the synthetic peptides would be released to the medium or remained in the pellet. Showing that the resuspension of lyophilized supernatant in ethanol obtains a higher yield in ornithine content compared to the pellet samples, we can conclude that the small peptides are emitted into the medium. As salt concentrations did not seem to interfere with mass spectrometry measurements, the whole work up process has been successful. The final output of the tandem mass spectrometry demonstrated a highly elevated concentration of synthetic peptides in the medium compared to the cell interior, which leads to an improved protocol for standardized purification of short non-ribosomal peptides (<a href="RFC%20100" title="wikilink">RFC 100</a>).
| |
| - | <br/><br/>
| |
| - | Furthermore, we found that there was no need to provide additional Ornithine in the media since it was incorporated from <em>E. coli</em> as an intermediate product from L-glutamate. Moreover, this assay accounted for the functional incorporation of non-proteinogenic amino acids into artificial non-ribosomal peptides. A high variety of non-proteinogenic amino acids as constituents has already been described in literature <span class="citation">[17]</span>.
| |
| - | <br/><br/>
| |
| - | Interestingly, the Ornithine concentration in the samples peaked at the first day upon induction ([[Media: Ornithine levels normalized MS.png| Fig. 7) but dropped rapidly during the second day to basal level. The transient enrichment of Ornithine could reflect the stability of the NRPS or the synthesized Tripeptide. Most likely Ornithine is cleaved by the endogenous enzyme ornithine decarboxylase encoded by the speC gene to form CO2 and Putrescine <span class="citation">[18]</span>. As ornithine abundance and acidic conditions serve as activators for the expression of the Ornithine decarboxylase respectively <bib id="pmid
| |
| - | 1939141"/>, this would explain the time-shift due to increased expression. Although this decarboxylase cannot cleave Ornithine incorporated in peptides, it could contribute to the decay process of free ornithine. A standardized test (MIO test) for the presence of Ornithine decarboxylase could help to determine the decay mechanism. In addition, the products could be toxic for the cells and are therefore degraded faster (<strong>HAVE WE NORMALIZED FOR CELL AMOUNT BEFORE PROCESSING THE SAMPLE?- We just took 7.5ml of each sample at ~OD600=0.6</strong>). The kinetics of Ornithine as a proxy for the production of the Tripeptide synthesis would thus imply that harvest of the peptide after one day would give optimal yield. Computational metabolic analysis could provide more insights into processes leading to peptide decay and instability. Metabolic reconstruction can give new insights to correlations at gene regulation and therefore provide new insights integrated in a biological context <span class="citation">[19]</span>.
| |
| - | <br/><br/>
| |
| - | In the future, synthesized non-ribosomal peptides could be elongated even further. Therefore, a stepwise-assembly strategy should be considered since the complexity of cloning will increase with the number and size of DNA fragments as well. Of course, additional modules for shuffling will increase the amount of opportunities to create comprehensive constructs. Such an approach is guided by the NRPS-Designer, which frames our proposed standard (<a href="RFC%20100" title="wikilink">RFC 100</a>).
| |
| - | <br/>
| |
| - | As we have seen, different modules of the Tyrocidine cluster are interchangeable with one another from the same synthetase. However, these findings only prove the compatibility of modules within a single pathway and a single species. To offer a general standard for short oligopeptide synthesis, we needed to establish synthesis of oligopeptides by NRPSs that were composed of modules originating from various pathways and organisms.
| |
| - | <br/><br>
| |
| - | The bottleneck to test libraries of combinatorial NRPSs with an array of different modules is in fact the screening for functional enzymes <span class="citation">[20]</span> <span class="citation">[21]</span>. How to identify novel NRPSs consisting of compatible modules? Our experimental results (Fig. 10) strongly support the hypothesis that the synthesis of short peptides can be easily monitored when fused to Indigoidine. here obtained by module shuffling within the Tyrocidine cluster, could not be detected in a high-throughput manner yielded in the design and synthesis of fusion peptides that are composed of. The approach of combining one or more modules from the Tyrocidine cluster and the Indigoidine module, encoded by indC from <em>P. luminescens</em> represents an entirely new finding. NRPSs seem to offer a framework that does not only go beyond species borders, as already shown by Marahiel <span class="citation">[22]</span>, but the resulting fusion NRP is synthesized and even detectable by eye.
| |
| - | <br/><br/>
| |
| - | With this, we offer a novel and very efficient way of tagging NRPs with Indigoidine. The dye can be easily measured and quantified (<strong>ELABORATE ON THIS, CITE OTHER ASSAYS</strong>). This aspect furthermore appends a valuable feature of the NRPS-Designer. Users now have the opportunity to easily add an Indigoidine tag when they design their constructs, which is also described in our proposed RFC (<a href="RFC%20100" title="wikilink">RFC 100</a>). Noteworthy, our experiments we could show that the Indigoidine tag works fine when put behind modules for different amino acids, but the tagging clearly works best, when an apolar, small spacer amino-acid such as Glycine, Alanine or, as in our experiments, Valine is used. Since these amino acids adjacent to the pi electron system of the Indigoidine interfere least with the delocalized electron cloud of the mesomeric benzene ring systeme, electron excitation by electromagnetic radiation can occur more easily. Since the difference of energy betweeen HOMO and LUMO states is minimized a longer wavelength is observed (Planck’s constant).
| |
| - | <br/><br/>
| |
| - | Compared with other potential methods for <em>in-vivo</em> tagging of NRPs (<strong>HAS THIS BEEN DONE WITH NRPs BEFORE? - yes: <span class="citation">[23]</span> and <span class="citation">[24]</span></strong>), the Indigoidine tag has the apparent advantage that it is relatively small compared to e.g. a Haemagglutinine tag. Similarly, using fluorescent proteins (FPs) as tag is hardly feasible as the peptides that should be tagged are often smaller than the chromophore of those FPs. Imagining e.g. GFP synthesized by an NRPS is practically not feasible. Something similar accounts for other tags, such as the His tag, for which four to nine Histidines are required. Synthesizing a short peptide with several modules for Histidine is imaginable, but would double or triple the size of the required NRPS, and hence of the vector, which is highly ineffective (<strong>MORE SYSTEMATICALLY REVIEW TAGS</strong>).
| |
| - | <br/><br/>
| |
| - | As far as <em>in-vitro</em> approaches are concerned, there are, in principle, two ways. I) Either one could add a tagging agent to the cells or the medium before purification – which would be the case for e.g. Click-Chemistry. II) Or one could add a certain tag such as a His tag to the NRPS and perform the entire synthesis of the NRP <em>in vitro</em>. The latter approach has been widely used <span class="citation">[25]</span> <span class="citation">[26]</span>, is, however in vitro and hence less effective for a high-throughput advance as a functioning in-vivo-method (<strong>WHAT DO YOU WANT TO SAY HERE, EXCEPT THAT IN VITRO IS INFERIOR TO IN VIVO?</strong>). We ourselves thought of using methods such as Click-Chemistry for the purification of the short, synthetic peptides. Increased masses and easy cleavage enable high purities and can be determined via HPLC with UV and ESI-MS detection <span class="citation">[27]</span>. This approach, however, does not offer any opportunity to evaluate expression <em>in vivo</em>’ <span class="citation">[28]</span>.
| |
| - | <br/><br/>
| |
| - | Hence, using and Indigoidine-tag, which can be added to the nascent NRP in the process of its formation by adding a 4kbp Indigoidine-module to the NRPS is a novel and effective approach for labeling NRPs for quantitative expression analysis.<br/><br/><br/></p>
| |
| - | </div>
| |
| - | <div class="col-sm-12 jumbotron">
| |
| - | <div class="references">
| |
| - | <p>1. Mootz HD, Marahiel MA (1997) The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol 179: 6843–6850.</p>
| |
| - | <p>2. Linne U, Marahiel MA (2000) Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry 39: 10439–10447.</p>
| |
| - | <p>3. Marahiel MA, Stachelhaus T, Mootz HD (1997) Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev 97: 2651–2674.</p>
| |
| - | <p>4. Finking R, Marahiel MA (2004) Biosynthesis of nonribosomal peptides. Annu Rev Microbiol 58: 453–488.</p>
| |
| - | <p>5. Marahiel MA (2009) Working outside the protein-synthesis rules: insights into non-ribosomal peptide synthesis. J Pept Sci 15: 799–807.</p>
| |
| - | <p>6. Linne U, Stein DB, Mootz HD, Marahiel MA (2003) Systematic and quantitative analysis of protein-protein recognition between nonribosomal peptide synthetases investigated in the tyrocidine biosynthetic template. Biochemistry 42: 5114–5124.</p>
| |
| - | <p>7. Weber T, Marahiel MA (2001) Exploring the domain structure of modular nonribosomal peptide synthetases. Structure 9: R3–R9.</p>
| |
| - | <p>8. Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA (1998) Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J Biol Chem 273: 22773–22781.</p>
| |
| - | <p>9. Owen JG, Copp JN, Ackerley DF (2011) Rapid and flexible biochemical assays for evaluating 4’-phosphopantetheinyl transferase activity. Biochem J 436: 709–717.</p>
| |
| - | <p>10. Schwarzer D, Mootz HD, Marahiel MA (2001) Exploring the impact of different thioesterase domains for the design of hybrid peptide synthetases. Chem Biol 8: 997–1010.</p>
| |
| - | <p>11. Stein DB, Linne U, Hahn M, Marahiel MA (2006) Impact of epimerization domains on the intermodular transfer of enzyme-bound intermediates in nonribosomal peptide synthesis. Chembiochem 7: 1807–1814.</p>
| |
| - | <p>12. Stein DB, Linne U, Marahiel MA (2005) Utility of epimerization domains for the redesign of nonribosomal peptide synthetases. FEBS J 272: 4506–4520.</p>
| |
| - | <p>13. Hur GH, Meier JL, Baskin J, Codelli JA, Bertozzi CR, et al. (2009) Crosslinking studies of protein-protein interactions in nonribosomal peptide biosynthesis. Chem Biol 16: 372–381.</p>
| |
| - | <p>14. Hahn M, Stachelhaus T (2004) Selective interaction between nonribosomal peptide synthetases is facilitated by short communication-mediating domains. Proc Natl Acad Sci USA 101: 15585–15590.</p>
| |
| - | <p>15. Brachmann AO, Kirchner F, Kegler C, Kinski SC, Schmitt I, et al. (2012) Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol 157: 96–99.</p>
| |
| - | <p>16. Perzborn M, Syldatk C, Rudat J (2013) Separation of Cyclic Dipeptides (Diketopiperazines) from Their Corresponding Linear Dipeptides by RP-HPLC and Method Validation. Chromatography Research International 2013.</p>
| |
| - | <p>17. Symmank H, Franke P, Saenger W, Bernhard F (2002) Modification of biologically active peptides: production of a novel lipohexapeptide after engineering of Bacillus subtilis surfactin synthetase. Protein engineering 15: 913–921.</p>
| |
| - | <p>18. Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, et al. (1991) Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. Journal of Biological Chemistry 266: 20922–20927.</p>
| |
| - | <p>19. Francke C, Siezen RJ, Teusink B (2005) Reconstructing the metabolic network of a bacterium from its genome. TRENDS in Microbiology 13: 550–558.</p>
| |
| - | <p>20. Nguyen KT, Ritz D, Gu J-Q, Alexander D, Chu M, et al. (2006) Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc Natl Acad Sci USA 103: 17462–17467.</p>
| |
| - | <p>21. Reverchon S, Rouanet C, Expert D, Nasser W (2002) Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J Bacteriol 184: 654–665.</p>
| |
| - | <p>22. Doekel S, Marahiel MA (2000) Dipeptide formation on engineered hybrid peptide synthetases. Chem Biol 7: 373–384.</p>
| |
| - | <p>23. Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, et al. (2005) Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proceedings of the National Academy of Sciences of the United States of America 102: 15815–15820.</p>
| |
| - | <p>24. Zhou Z, Cironi P, Lin AJ, Xu Y, Hrvatin S, et al. (2007) Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS chemical biology 2: 337–346.</p>
| |
| - | <p>25. Butz D, Schmiederer T, Hadatsch B, Wohlleben W, Weber T, et al. (2008) Module extension of a non-ribosomal peptide synthetase of the glycopeptide antibiotic balhimycin produced by Amycolatopsis balhimycina. Chembiochem 9: 1195–1200.</p>
| |
| - | <p>26. Mootz HD, Schwarzer D, Marahiel MA (2000) Construction of hybrid peptide synthetases by module and domain fusions. Proc Natl Acad Sci USA 97: 5848–5853.</p>
| |
| - | <p>27. Franke R, Doll C, Eichler J (2005) Peptide ligation through click chemistry for the generation of assembled and scaffolded peptides. Tetrahedron letters 46: 4479–4482.</p>
| |
| - | <p>28. Kolb HC, Finn MG, Sharpless KB (2001) Click chemistry: diverse chemical function from a few good reactions. Angewandte Chemie International Edition 40: 2004–2021.</p>
| |
| - | </div>
| |
| - |
| |
| - | </div>
| |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | </div> |
| | | | |
| | <script type="text/javascript"> | | <script type="text/javascript"> |
| Line 274: |
Line 236: |
| | </script> | | </script> |
| | </html> | | </html> |
| - | [[File:Heidelberg_IndPD_Fig1.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]]
| + | |
| - | [[File:Heidelberg_IndPD_Fig2.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]]
| + | |
| - | [[File:Heidelberg_IndPD_Fig3.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]]
| + | |
| | [[File:Heidelberg_IndPD_Fig4.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]] | | [[File:Heidelberg_IndPD_Fig4.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]] |
| | [[File:Heidelberg_IndPD_Fig5.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]] | | [[File:Heidelberg_IndPD_Fig5.png|640px|Figure 1: NRPS module and domain structure and activation of T-domains]] |













 "
"