Team:Heidelberg/Templates/Project
From 2013.igem.org
Nils.kurzawa (Talk | contribs) |
|||
| Line 115: | Line 115: | ||
</div> | </div> | ||
<div class="container"> | <div class="container"> | ||
| - | <div class="row | + | <div class="row"> |
<div id="tyrocidineText" class="col-sm-12"> | <div id="tyrocidineText" class="col-sm-12"> | ||
<div id="delftibactinText" class="col-sm-12"> | <div id="delftibactinText" class="col-sm-12"> | ||
| Line 156: | Line 156: | ||
<p>We aimed to introduce the large Del cluster into the commonly used, easy-to-culture model organism <em>E. coli</em> to produce Delftibactin. This target bacterium already possesses many components needed for the functionality of non-ribosomal-peptide synthetases. Nevertheless, we introduce nearly the entire Del cluster into <em>E. coli</em> except for DelC (native PPTase). This function is covered by the sfp phosphopanteinyl transferaseintroduced from <em>Bacillus subtilis</em>. As DelF is a PKS, it requires methylmalonyl-CoA as substrate, which is not produced by <em>E. coli</em>. Therefore, the MMCoA synthesis pathway from <em>B. subtilus</em> which is able to activate a wide variety of PKSs including those from <em>Saccharomyces cerevisiae</em> <span class="citation">[9]</span> was transferred, too.</p> | <p>We aimed to introduce the large Del cluster into the commonly used, easy-to-culture model organism <em>E. coli</em> to produce Delftibactin. This target bacterium already possesses many components needed for the functionality of non-ribosomal-peptide synthetases. Nevertheless, we introduce nearly the entire Del cluster into <em>E. coli</em> except for DelC (native PPTase). This function is covered by the sfp phosphopanteinyl transferaseintroduced from <em>Bacillus subtilis</em>. As DelF is a PKS, it requires methylmalonyl-CoA as substrate, which is not produced by <em>E. coli</em>. Therefore, the MMCoA synthesis pathway from <em>B. subtilus</em> which is able to activate a wide variety of PKSs including those from <em>Saccharomyces cerevisiae</em> <span class="citation">[9]</span> was transferred, too.</p> | ||
<p>The resulting engineered <em>E. coli</em> could be used as host for the delftibactin synthesis pathway, possibly also eliminating the need to introduce DelC. As promoters of the Del cluster were only predicted <span class="citation">[10]</span> for Daci_4750 (DelK) and Daci_4760 (DelA) and the cluster is transcribed starting with Daci_4760, we assumed that the entire sequence stretch of approximately 40 kbp is transcribed as single polycistronic mRNA.</p> | <p>The resulting engineered <em>E. coli</em> could be used as host for the delftibactin synthesis pathway, possibly also eliminating the need to introduce DelC. As promoters of the Del cluster were only predicted <span class="citation">[10]</span> for Daci_4750 (DelK) and Daci_4760 (DelA) and the cluster is transcribed starting with Daci_4760, we assumed that the entire sequence stretch of approximately 40 kbp is transcribed as single polycistronic mRNA.</p> | ||
| - | <p>Facing these challenges, we decided to approach the project straight forward by cultivation of <em>D. acidovorans</em | + | <p>Facing these challenges, we decided to approach the project straight forward by cultivation of <em>D. acidovorans</em |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 16:48, 23 October 2013


Delftibactin.Recycling Gold from Electronic Waste.
Highlights
- Production and purification of delftibactin, a gold-precipitating NRP, from its native, cultured host Delftia acidovorans.
- Recovery of pure gold from electronic waste by Delftia acidovorans and purified delftibactin.
- Optimization of the Gibson assembly method for the creation of large plasmids (> 30 kbp) with high GC content.
- Amplification and cloning of all components required for recombinant delftibactin production.
- Transfer of the entire pathway from D. acidovorans for the synthesis of delftibactin to E. coli.
Abstract
Efficient recycling of gold from electronic waste using recombinant delftibactin
Undoubtedly, gold is one of the most precious materials on earth. Besides its common use in art and jewelry, gold is also an essential component of our modern computers and cell-phones. Due to the fast turn-over of today’s high-tech equipment, millions of tons of electronic waste accumulate each year containing tons of this valuable metal. The main approach nowadays to recycle gold from electronic waste is by electrolysis. Unfortunately, this is a highly inefficient and expensive procedure, preventing most of the gold from being recovered.
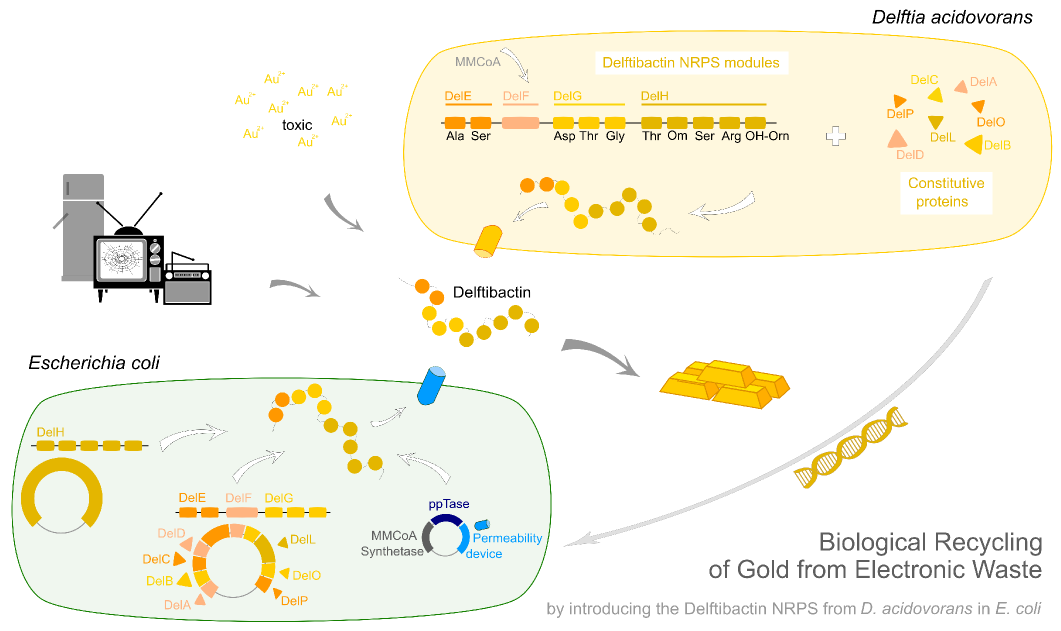
Earlier this year, a publication in Nature Chemical Biology reported the existence of a non-ribosomal peptide – delftibactin - which has the astonishing property to specifically precipitate elemental gold from gold-ion containing solutions. Naturally, delftibactin is produced by Delftia acidovorans, an extremophile bacterium, which secretes delftibactin to complexate and dispose of toxic gold ions present in its environment. Although the exact delftibactin production pathway is not known, bioinformatic predictions claim a non-ribosomal peptide synthesis pathway encoded on a large, 59 kb gene cluster (the del-cluster) to be responsible for delftibactin production.
In this subproject we want to demonstrate that the natural secondary metabolite delftibactin can be efficiently produced in E. coli and used for the recycling of gold from electronic waste. To this end, we developed a cloning strategy based on an optimized Gibson Assembly protocol, enabling the cloning of large, GC-rich genomic regions onto regular low-copy plasmids. We thereby engineered three different plasmids (about 70 kb in total size) enabling the expression of the predicted del-cluster from regular E. coli promoters along with the methylmalonyl-CoA pathway providing the basic delftibactin building blocks and a NRPS activating PPTase, Sfp from Bacillus subtilis.
We want to show that these large constructs can be potentially inserted and expressed by E. coli with the promising perspective that delftibactin could readily be used as an efficient way of gold recycling from electronic waste.
Introduction
The quest for a magical substance to generate gold from inferior metals stirred the imagination of generations. However, this substance, the Philosopher’s Stone, stands for more than just the desire to produce gold. In the old days, the fabled Philosopher’s Stone also represented wisdom, rejuvenation and health. Nowadays, gold is still of great importance for us as it is needed for most of our electronic devices.
In 2007, more than two tons of gold were discarded hidden in electronic waste in Germany. Most of the precious element end up on waste disposal sites as only a minor fraction of 28 % of the gold is recycled also due to the small amounts per devide. Since our planet’s gold supplies are limited, the metal is more and more depleted and the value of gold continously reaches all-time highs. In order to satisfy our society’s need for gold, we have to develop heavy mining techniques involving strong acids, causing devastating impact on human and environment [1] [2] [3].
Besides economical usage of the resource gold, one way to reduce global demands for gold is elevation of gold recovery [4]. Intriguingly, nature itself offers a structure that has been reported to efficiently extract pure gold from solutions containing gold ions. This fascinating molecule is called Delftibactin and is in fact a small peptide secreted by a metal-tolerant bacterium called Delftia acidovorans.
This extremophile has the incredible ability to withstand toxic amounts of gold ions in contaminated soil . What is the special feature of Delftibactin enabling precipitation of gold that efficiently? If one could culture these bacteria and produce Delftibactin in large scales, could one potentially recover gold from electronic waste in a cost- and energy-efficient way? But what is the special feature of Delfibactin to precipitate gold that efficiently?

Delftibactin is no ordinary peptide but a non-ribosomal peptide (NRP) [5] [6]. The efficient and non-polutative large-scale production of this NRP in E. coli could revolutionize the recovery of gold from electronic waste and additionally highlight the plethora of versatile applications for non-ribosomal peptide synthetases (NRPSs). The most sriking feature of these non-ribosomal synthetases is their ablity to incorporate far more than the 21 common amino acids into peptides. They make use of numerous modified and even non-proteinogenic amino acids [7] to assembly peptides of diverse functions.
Delftibactin is a NRP produced by a hybrid NRPS/ polyketide synthase (PKS) system. In their recent publication, Johnston et al. [5] predicted that the enzymes responsible for producing delftibactin are encoded on a single gene cluster, hereafter referred to as Del cluster. It comprises 59 kbp encoding for 21 genes. DelE, DelF, DelG and DelH constitute the hybrid NRPS/ PKS system producing delftibactin, with DelE, DelG and DelH being NRPS and DelF the PKS. The remaining enzymes involved in the Delftibactin synthesis pathway are required for NPRS/ PKS maturation or post-synthesis modification of Delftibactin. The predicted activities of the assumed proteins are:
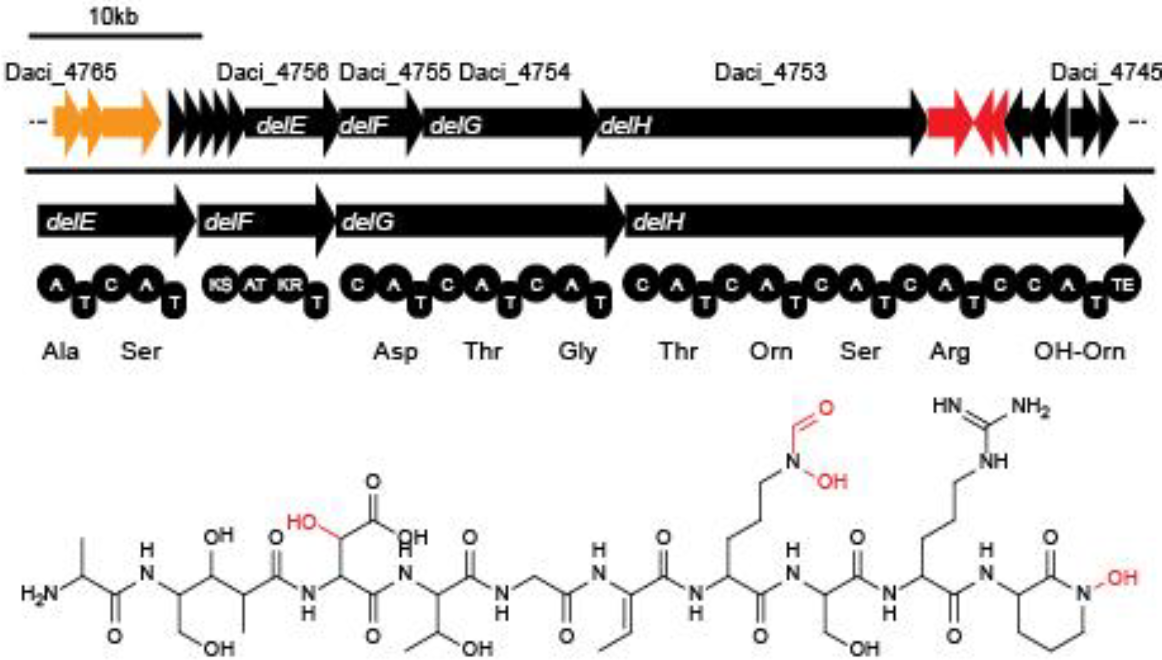
- DelA: MbtH-like protein, most likely required for efficient delftibactin synthesis [8]
- DelB: thioesterase
- DelC: 4’-phosphopanteinyl transferase: required for maturation of ACP/PCP subunits
- DelD: taurine dioxygenase
- DelL: Ornithine N-monooxygenase
- DelP: N5-hydroxyornithine formyltransferase
We aimed to introduce the large Del cluster into the commonly used, easy-to-culture model organism E. coli to produce Delftibactin. This target bacterium already possesses many components needed for the functionality of non-ribosomal-peptide synthetases. Nevertheless, we introduce nearly the entire Del cluster into E. coli except for DelC (native PPTase). This function is covered by the sfp phosphopanteinyl transferaseintroduced from Bacillus subtilis. As DelF is a PKS, it requires methylmalonyl-CoA as substrate, which is not produced by E. coli. Therefore, the MMCoA synthesis pathway from B. subtilus which is able to activate a wide variety of PKSs including those from Saccharomyces cerevisiae [9] was transferred, too.
The resulting engineered E. coli could be used as host for the delftibactin synthesis pathway, possibly also eliminating the need to introduce DelC. As promoters of the Del cluster were only predicted [10] for Daci_4750 (DelK) and Daci_4760 (DelA) and the cluster is transcribed starting with Daci_4760, we assumed that the entire sequence stretch of approximately 40 kbp is transcribed as single polycistronic mRNA.
Facing these challenges, we decided to approach the project straight forward by cultivation of D. acidovorans
 "
"














